Abstract
Single objective optimization algorithms are the foundation of establishing more complex methods, like constrained optimization, niching and multi-objective algorithms. Therefore, improvements to single objective optimization algorithms are important because they can impact other domains as well. This paper proposes a method using turning-based mutation that is aimed to solve the problem of premature convergence of algorithms based on SHADE (Success-History based Adaptive Differential Evolution) in high dimensional search space. The proposed method is tested on the Single Objective Bound Constrained Numerical Optimization (CEC2020) benchmark sets in 5, 10, 15, and 20 dimensions for all SHADE, L-SHADE, and jSO algorithms. The effectiveness of the method is verified by population diversity measure and population clustering analysis. In addition, the new versions (Tb-SHADE, TbL-SHADE and Tb-jSO) using the proposed turning-based mutation get apparently better optimization results than the original algorithms (SHADE, L-SHADE, and jSO) as well as the advanced DISH and the jDE100 algorithms in 10, 15, and 20 dimensional functions, but only have advantages compared with the advanced j2020 algorithm in 5 dimensional functions.
1. Introduction
The single objective global optimization problem involves finding a solution vector x = (x1, …, xD) that minimizes the objective function f(x), where D is the dimension of the problem. The task of black box optimization is to solve the global optimization problem without clear objective function form or structure, that is, f is a “black box”. This problem appears in many problems of engineering optimization, where complex simulations are used to calculate the objective function.
The differential evolution (DE) algorithm, proposed by Price and Storm in 1995, laid the foundation for a series of successful algorithms for continuous optimization. DE is a random black box search method, which was originally designed for numerical optimization problems [1], and it’s also an evolutionary algorithm that ensures that every next generation has better solutions than the previous generation: a phenomenon known as elitism. The extensive study fields of DE are summarized lately in the references [2].
Studies on DE have yielded a number of improvements [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17] to the classical DE algorithm, and the status of research on it can be easily obtained by noting the results of the Continuous Optimization Competition and the Evolutionary Computing Conference (CEC).
A popular variant of DE [18] is the algorithm proposed by Fukunaga and Tanabe called Success History-based Adaptive Differential Evolution (SHADE) [19]. In the optimization process, the scale factor F and the crossover rate CR of control parameters are adjusted to adapt to the given problem, and the “current to pbest/1” mutation strategy and the external archive of poor quality solutions in JADE [20] are combined in SHADE. The SHADE algorithm ranked third in CEC2013. In the second year, the author proposed an improved scheme, adding a linear reduction to the population size called L-SHADE to improve the convergence rate of SHADE [21]. L-SHADE won the CEC2014 competition. The winners in the subsequent years were SPS-L-SHADE-EIG [22] (CEC2015), LSHADE-EpSin [23] (joint winner of CEC2016), and jSO [24] (CEC2017). These algorithms are all based on L-SHADE, which makes it one of the most effective variants of SHADE [25]. With the exception of the jSO, the other winners benefited from general enhancements in the area [26]. Consequently, this study applies an improved method to the SHADE, L-SHADE and jSO algorithms. LSHADE-ESP [27] came in second in CEC2018 and the jDE100 [28] won CEC2019. And the j2020 [29] algorithm, which was proposed on CEC2020 recently, is also within the reference range. Enhanced versions of these DE algorithms add new mechanisms or parameters for optimization, similar to those in other optimization algorithms [30], as described in substantive surveys of these areas [25,31,32,33,34,35,36,37]. Moreover, theoretical analysis supporting DE has also been provided, such as in [38,39,40,41].
The DE consists of three main steps: mutation, crossover, and selection. Many proposals [6,10,11,14,24] have been made to improve the mutation process to improve optimization performance. For instance, four strategies of combining mutation and crossover was used in SHADE4 [6], SHADE44 [10] and L-SHADE44 [11] to create a new trial individual and realize an adaptive mechanism. A novel multi-chaotic framework was used in MC-SHADE [14] to generate random numbers for the parent selection process in mutation process. A new weighted mutation strategy with parameterization enhancement was used in jSO [24] to enhance adaptability. This paper also focuses on improving this process in the DE algorithm, especially SHADE-based algorithms.
The CEC2020 [42] single-objective boundary-constrained numerical optimization benchmark sets are designed to determine the improvement in performance obtained by increasing the number of the calculation of the fitness function of an optimization algorithm. There are thus two motivations for this study. First, we need to solve the problem of premature convergence of algorithms based on SHADE in high dimensional search spaces on CEC2020 benchmark sets, so that they can maintain a high population diversity and a longer exploration phase. Second, the improvement to the algorithm should be simple, should not excessively increase complexity, and should not render the proposed algorithm incomprehensible and less applicable, as discussed in [43]. We proposed a method using turning-based mutation, and apply it to the SHADE, L-SHADE, and jSO algorithms to yield good performance while using relatively simple algorithm structure. Through experimental analysis involving 10, 15, and 20 dimensions, the improved algorithms achieved better performance than the original algorithms as well as the advanced DISH [44] and jDE100 algorithms on CEC2020 benchmark sets, but were slightly worse than the j2020 algorithm. We also use population diversity measure and population clustering analysis to verify the effectiveness of the proposed method.
Section 2 describes the process of evolution from the DE algorithm to the SHADE, L-SHADE, and jSO algorithms, and turning-based mutation is introduced in Section 3. The experimental settings and results are described in Section 4 and Section 5, respectively. Section 6 discusses the results, and the conclusion of this paper is given in Section 7.
2. DE SHADE L-SHADE and jSO
2.1. Differential Evolution
The DE consists of three main steps: mutation, crossover, and selection. In mutation, the attribute vector of the selected individual x is combined in a simple vector operation to generate the mutated vector v. The scale factor F of the control parameter is used in this operation. In the crossover step, according to the probability given by the crossover rate CR of the control parameter, the trial vector u is created by selecting the attribute from the original vector x or mutated vector v. Finally, in the selection step, the trial vector u is evaluated by the objective function and the fitness f(u) is compared with the fitness of the selected vector f(x). The vector with the better fitness value survives to the next generation.
This paper focuses on improving the mutation process, so the paragraphs below describe the mutation process of the DE algorithm. The complete steps of DE can be referred to the literature [1]. The mutation strategy of DE/rand/1/bin can be expressed as follows:
where vi,G is the mutated vector, and xr1,G, xr2,G, and xr3,G are three different individuals randomly selected from the population. Fi is the scaling factor, and G is the index of the current generation.
If any dimension of the mutated vector vj,i,G is outside the boundary of the search range [xmin, xmax], we perform the following correction for boundary-handling to handle infeasible solutions [45]:
where j is the dimensional index and i is the individual index.
The pseudo-code of the DE/rand/1/bin algorithm is shown in Algorithm 1.
| Algorithm 1 DE/rand/1/bin |
|
It can be seen from the description of DE algorithm that users need to set three control parameters: crossover rate CR, scaling factor F and population size NP. The setting of these parameters is very important to the performance of DE.
Fine-tuning the control parameter is a time-consuming task, because of which most advanced variants of DE use parameter adaptation. This is also why Tanabe and Fukunaga proposed the SHADE [19] algorithm in 2013. Because the algorithms used in this paper are based on SHADE, it is described in more detail below.
2.2. SHADE
In the control parameters of SHADE, crossover rate CR and scaling factor F are discussed. The algorithm is based on JADE [20], proposed by Sanderson and Zhang, and so they share many mechanisms [18]. The major difference between them is in historical memories MF and MCR with their update mechanisms. The next subsections describe the historical memory update of SHADE and the difference between DE and SHADE algorithm in initialization, mutation, crossover and selection, respectively. The complete steps of SHADE can be referred to the literature [19].
2.2.1. Initialization
In SHADE, the population is initialized in the same manner as in DE, but there are two additional components—historical memory and external archive—that also need to be initialized.
Initialize the control parameters stored in the historical memory, crossover rate CR and scale factor F to 0.5:
where H is the size of the user-defined historical memory, and the index k to update the historical memory is initialized to one.
In addition, the initialization of the external archive of poor quality solutions is empty, i.e., A = .
2.2.2. Mutation
In contrast to DE/rand/1/bin, the “current to pbest/1” mutation strategy is used in SHADE:
where, xi,G is the given individual, and xpbest,G is an individual selected from the best NP × pi (pi ∈ [0, 1]) individuals randomly in the current population. Vector xr1,G is an individual selected from the current population randomly, and xr2,G is an individual selected from a combination of the external archive A and the current population randomly. Index r1 ≠ r2 ≠ i. Fi is a scaling factor, rand [] is a uniform random distribution and NP is the size of population. The vi,G is the mutated vector and G is the index of the current generation. The greed of the “current-to-pbest/1” mutation strategy depends on the control parameter pi, which is calculated as shown in Equations (5) and (6). It balances exploration and exploitation capabilities (a small value of p is more greedy). The scaling factor Fi is generated using the following formula:
where randci () is the Cauchy distribution, and MF,ri is randomly selected from historical memory MF (index ri is a uniformly distributed random value from [1, H]). If Fi > 1, let Fi = 1. If Fi ≤ 0, Equation (7) is repeated to attempt to generate a valid value.
The boundary handling of SHADE is identical to that of DE, as shown in Equation (2).
2.2.3. Crossover
DE has 2 classic crossover strategies, i.e., binomial and exponential. The crossover strategies of SHADE is the same as that of DE/rand/1/bin, i.e., binomial crossover. However, the crossover rate of DE/rand/1/bin is set in advance while the CRi of SHADE is generated by the following formula:
where randni () is Gaussian distribution, and MCR,ri are randomly selected from historical memory MCR (index ri is a uniformly distributed random value from [1, H]). If CRi > 1, let CRi = 1; if CRi < 0, let CRi = 0.
2.2.4. Selection
The process of selection of SHADE is the same as that of DE. However, the external archive needs to be updated during selection. If a better trail individual is generated, the original individual xi,G is stored in the external archive. If the external archive exceeds capacity, one of them is randomly deleted.
2.2.5. Historical Memory Update
Historical memory update is also an important operation in SHADE. The historical memories MCR and MF are initialized by Formula (3) but their contents change with the iteration of the algorithm. These memories store the “successful” crossover rate CR and scaling factor F. “Successful” here means that the trail vector u is selected instead of the original vector x to survive to the next generation. In each generation, the values of these “successful” CR and F are first stored in arrays SCR and SF, respectively. After each generation, a unit of each of the historical memories MF and MCR is updated. The updated unit is specified by index k, which is initialized to one and increases by one after each generation. If k exceeds the memory capacity H, it is reset to one. The following formula is used to update the k-th unit of historical memory:
If all individuals in the G-th generation fail to generate a better trail vector, i.e., SF = SCR = ∅, the historical memory will not be updated. The weighted Lehmer mean WL and weighted mean WA are calculated using the following formulas, respectively:
To improve the adaptability of the parameters, the weight vector w is calculated based on the absolute value of the difference that is obtained by subtracting the objective function value of the given vector from that of the trail vector in current generation G, as follows:
where in (13).
The pseudo-code of the SHADE algorithm is shown in Algorithm 2.
| Algorithm 2 SHADE |
|
2.3. Linear Decrease in Population Size: L-SHADE
In [21], a linear reduction of population size was introduced to SHADE to improve its performance. The basic thought is to gradually reduce the population size during evolution to improve exploitation capabilities. In L-SHADE, the population size is calculated after each generation using Formula (14). If the new population size NPnew is smaller than the previous population size NP, the all individuals are sorted on the basis of the value of the objective function, and the worst NP-NPnew individuals are cut. Also, the size of external archives/A/decreases synchronously with population size:
where NPf and NPinit are the final and initial population size, respectively. MaxFES and FES are the maximum and current number of the calculation of the fitness function, respectively. And round () is a rounding function.
2.4. Weighted Mutation Strategy with Parameterization Enhancement: jSO
The jSO [24] algorithm won the CEC2017 single-objective real parameter optimization competition [46]. It is a type of iL-SHADE algorithm that uses a weighted mutation strategy [47]. The iL-SHADE algorithm extends L-SHADE by initializing all parameters in the historical memories MF and MCR to 0.8, statically initializing the last unit of historical memories MF and MCR to 0.9, updating MF and MCR with the weighted Lehmer average value, limiting the crossover rate CR and scaling factor F in the early stage, and p is calculated for the “current-to-pbest/1” mutation strategy as:
where pmin and pmax are the minimum and maximum value of p, respectively. FES and MaxFES are the current and maximum number of the calculation of the fitness function, respectively.
The jSO algorithm sets pmax = 0.25 and pmin = pmax/2, initial population size to , and the size of the historical memory to H = 5. All parameters in MF and MCR are initialized to 0.3 and 0.8, respectively, and the weighted mutation strategy current-to-pbest-w/1 is used:
where Fw is calculated as:
3. Turning-Based Mutation
The opposition-based DE (ODE) algorithm was proposed by Shahryar et al. [48]. The opposition-based learning (OBL) was used for generation jumping and population initialization, and the opposite numbers was used to improve the convergence rate of DE. Shahryar et al. let all vectors of the initial population take the opposite number in the initialization and allowed the trail vectors to take the opposite number in the selection operation. They then compared their fitness values and selected the vector with the better fitness to accelerate the convergence of the DE algorithm. We refer to the idea of “opposition” in the above algorithm, but the purpose of this paper is to change the direction of mutation under certain conditions to maintain population diversity and enable a longer exploration phase.
Suppose that the search space is two-dimensional (2D). There is a ring-shaped region, the center of which is the global suboptimal individual xpbest,G. The outer radius of the ring is OR and the inner radius is IR. If the Euclidean distance Distance between the given individual and the global suboptimal individual is smaller than the outer radius OR and larger than the inner radius IR, the differential vector dei from the mutation Formulas (1) and (4) takes the opposite number, and some dimensions are randomly selected to assign random values within the search range. Experiments have verified that better outer radius OR and inner radius IR can be calculated as:
where ORinit is the initial value of the outer radius and IR is the inner radius, which is also the minimum value of the outer radius. The outer radius OR decreases with an increase in the number of fitness evaluations. MaxFES and FES are the maximum and current number of the calculation of the fitness function, respectively, and xmax and xmin are the upper and lower bounds of the search range, respectively.
The Euclidean distance Distance between the given individual and the global suboptimal individual is calculated as:
The differential vector dei from the mutation Equations (1) and (4) is calculated as:
The pseudo-code of the operation on the differential vector dei in turning-based mutation is shown as Operation 1:
where R is the randomly disordered dimension index array, M is the number of randomly selected dimensions, and xmax and xmin are the upper and lower bounds of the search range, respectively.
| Operation 1 operation on dei |
|
Finally, the mutation operation is performed as shown in Equation (23):
If the Euclidean distance Distance between the given individual and the global suboptimal individual is smaller than the outer radius OR and larger than the inner radius IR, the improved method changes the direction of mutation of the given individual to maintain the population diversity and a longer exploration phase, thus enhancing the global search ability and the ability to escape the local optimum. Then, with an increase in number of fitness evaluations, the performance of the algorithm can be improved. If the Euclidean distance Distance between the given individual and the global suboptimal individual is smaller than or equal to the inner radius IR, the former is allowed to mutate in the original direction. This enables the given individual to quickly converge to the global optimal or suboptimal position to avoid the problem of non-convergence caused by turning-based mutation.
Since Equation (21) and Operation 1 need to be executed in the mutation process of each individual, the overall time complexity [42] of the improved algorithms is slightly higher than that of the original algorithms, as shown in Table 1, Table 2 and Table 3.

Table 1.
Time complexity specified by CEC2020 technical document-SHADE vs. Tb-SHADE.

Table 2.
Time complexity specified by CEC2020 technical document—L-SHADE vs. TbL-SHADE.

Table 3.
Time complexity specified by CEC2020 technical document-jSO vs. Tb-jSO.
The pseudo-code of the Tb-SHADE algorithm (SHADE algorithm using turning-based mutation) is shown as Algorithm 3, that of the TbL-SHADE algorithm (L-SHADE algorithm using turning-based mutation) is shown as Algorithm 4, and that of the Tb-jSO algorithm (jSO algorithm using turning-based mutation) is shown as Algorithm 5. The improved parts of these algorithm are underlined.
| Algorithm 3 Tb-SHADE |
|
| Algorithm 4 TbL-SHADE |
|
| Algorithm 5 Tb-jSO |
|
4. Experimental Settings
To verify the improved method by experiments, the original algorithm, the improved algorithm and the advanced DISH and the jDE100 algorithms were tested on the Single Objective Bound Constrained Numerical Optimization (CEC2020) benchmark sets in 5, 10, 15 and 20 dimensions. The termination criteria, i.e., the maximum number of the calculation of the fitness function (MaxFES) and the minimum error value (Min error value), were set as in Table 4. The search range is [xmin, xmax] = [−100, 100], and 30 independent repeated experiments were conducted. The parameter setting of most algorithm [19,21,24] is shown in Table 5 and Table 6. In addition, the parameter setting of j2020 algorithm can be found in [29].

Table 4.
Termination criteria.

Table 5.
Parameter setting of some algorithms.

Table 6.
Parameter setting of jDE100.
The hypothesis that the turning-based mutation can maintain a longer exploration phase can be verified by analyzing the clustering and density of the population during the optimization process. These two analyses are described in more detail below.
4.1. Cluster Analysis
The clustering algorithm selected in this experiment is density based noisy application spatial clustering (DBSCAN) [49], which is based on the clustering density rather than its center, so it can find clusters of arbitrary shape. DBSCAN algorithm needs to set two control parameters and a distance measurement. The settings are as follows:
- (1)
- distance between core points, that is, Eps = 1% of the decision space; for the CEC2020 benchmark sets, Eps = 2;
- (2)
- minimum number of points forming a cluster, that is, MinPts = 4 (minimum number of mutation individuals); and
- (3)
- distance measure uses Chebyshev distance [50]—if the distance between the corresponding attributes of two individuals is greater than 1% of the decision space, they are not considered as direct density reachable.
4.2. Population Diversity
The population diversity (PD) measure is taken from [51], which is based on the square root of the deviation sum, Equation (25), of individual components and their corresponding mean values, Equation (24):
where i is the iterator of members of the population and j is that of the component (dimension).
5. Results
Table 7, Table 8, Table 9, Table 10, Table 11, Table 12, Table 13, Table 14, Table 15, Table 16, Table 17 and Table 18 compare the error values (when the error value was smaller than 10−8, the corresponding value was considered optimal) obtained by the original algorithms (SHADE, L-SHADE, and jSO) and their improved versions using turning-based mutation (Tb-SHADE, TbL-SHADE and Tb-jSO, respectively). The results of a comparison are showed in the last column of each table. If the performance of the original version was significantly better, uses the “−” sign; if the performance of the improved version was significantly better, uses the sign “+”; if their performances were similar, “=” is used. The better performance values are displayed in bold, and the last row of these tables shows the results of an overall comparison. Table 19, Table 20, Table 21 and Table 22 provide the error values obtained by the advanced algorithms DISH, jDE100 and j2020 on CEC2020. All tables provide the best, mean and std (standard deviation) values of 30 independent repetitions of the experiments.

Table 7.
SHADE vs. Tb-SHADE on CEC2020 in 5D.

Table 8.
SHADE vs. Tb-SHADE on CEC2020 in 10D.

Table 9.
SHADE vs. Tb-SHADE on CEC2020 in 15D.

Table 10.
SHADE vs. Tb-SHADE on CEC2020 in 20D.

Table 11.
L-SHADE vs. TbL-SHADE on CEC2020 in 5D.

Table 12.
L-SHADE vs. TbL-SHADE on CEC2020 in 10D.

Table 13.
L-SHADE vs. TbL-SHADE on CEC2020 in 15D.

Table 14.
L-SHADE vs. TbL-SHADE on CEC2020 in 20D.

Table 15.
jSO vs. Tb-jSO on CEC2020 in 5D.

Table 16.
jSO vs. Tb-jSO on CEC2020 in 10D.

Table 17.
jSO vs. Tb-jSO on CEC2020 in 15D.

Table 18.
jSO vs. Tb-jSO on CEC2020 in 20D.

Table 19.
DISH and jDE100 on CEC2020 in 5D.

Table 20.
DISH and jDE100 on CEC2020 in 10D.

Table 21.
DISH and jDE100 on CEC2020 in 15D.

Table 22.
DISH and jDE100 on CEC2020 in 20D.
Convergence diagrams are shown in Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12. Figure 1, Figure 2, Figure 3 and Figure 4 shows the convergence curves of SHADE and Tb-SHADE, respectively, for some test functions in 5D, 10D, 15D, and 20D, Figure 5, Figure 6, Figure 7 and Figure 8 shows those of L-SHADE and TbL-SHADE for some test functions in 5D, 10D, 15D, and 20D. and Figure 9, Figure 10, Figure 11 and Figure 12 shows those of the jSO and Tb-jSO, respectively, for some test functions in 5D, 10D, 15D and 20D. It is apparent that the red line of the turning-based mutation version of the algorithm was often slower to converge but attained better objective function values.
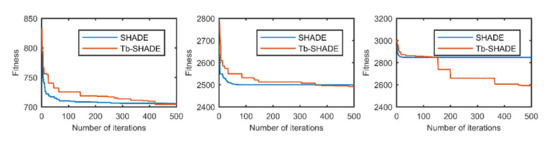
Figure 1.
The selected average convergence of SHADE and Tb-SHADE on CEC2020 in 5D is compared. From left to right f3, f9 and f10.
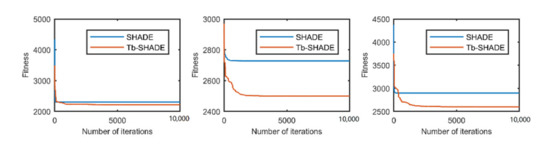
Figure 2.
The selected average convergence of SHADE and Tb-SHADE is compared on CEC2020 in 10D. From left to right f8, f9 and f10.
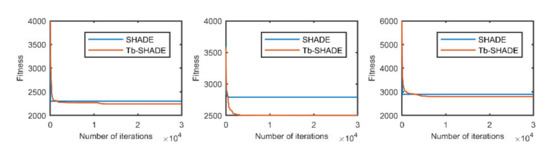
Figure 3.
The selected average convergence of SHADE and Tb-SHADE is compared on CEC2020 in 15D. From left to right f8, f9 and f10.
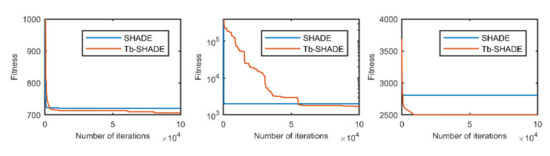
Figure 4.
The selected average convergence of SHADE and Tb-SHADE is compared on CEC2020 in 20D. From left to right f3, f5 and f9.
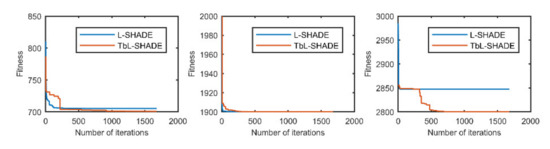
Figure 5.
The selected average convergence of L-SHADE and TbL-SHADE is compared on CEC2020 in 5D. From left to right f3, f4 and f10.
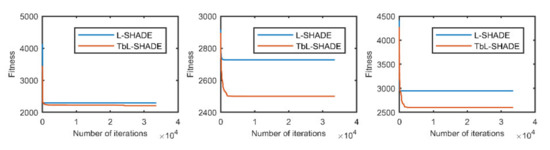
Figure 6.
The selected average convergence of L-SHADE and TbL-SHADE is compared on CEC2020 in 10D. From left to right f8, f9 and f10.
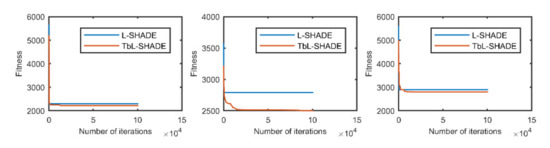
Figure 7.
The selected average convergence of L-SHADE and TbL-SHADE is compared on CEC2020 in 15D. From left to right f8, f9 and f10.
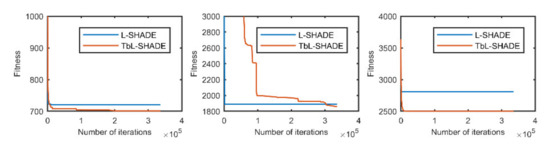
Figure 8.
The selected average convergence of L-SHADE and TbL-SHADE is compared on CEC2020 in 20D. From left to right f3, f5 and f9.
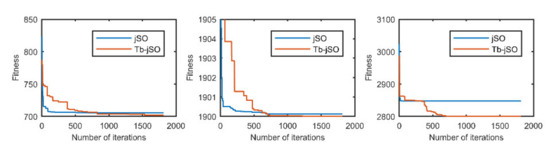
Figure 9.
The selected average convergence of jSO and Tb-jSO is compared on CEC2020 in 5D. From left to right f3, f4 and f10.
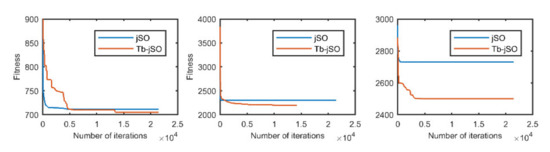
Figure 10.
The selected average convergence of jSO and Tb-jSO is compared on CEC2020 in 10D. From left to right f3, f8 and f9.
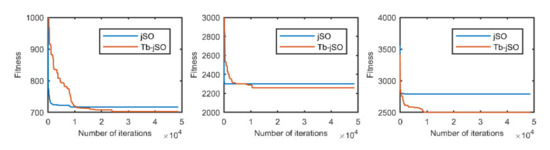
Figure 11.
The selected average convergence of jSO and Tb-jSO is compared on CEC2020 in 15D. From left to right f3, f8 and f9.
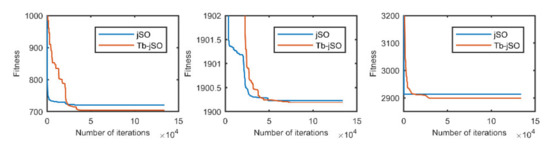
Figure 12.
The selected average convergence of jSO and Tb-jSO is compared on CEC2020 in 20D. From left to right f3, f4 and f10.
Table 23, Table 24, Table 25, Table 26, Table 27, Table 28, Table 29, Table 30, Table 31, Table 32, Table 33 and Table 34 shows the number of runs (#runs) of population aggregation, the average generation (Mean CO) of the first cluster during these runs, and the average population diversity (Mean PD) of these generations.

Table 23.
Clustering and population diversity of SHADE and Tb-SHADE on the CEC2020 in 5D.

Table 24.
Clustering and population diversity of SHADE and Tb-SHADE on the CEC2020 in 10D.

Table 25.
Clustering and population diversity of SHADE and Tb-SHADE on the CEC2020 in 15D.

Table 26.
Clustering and population diversity of SHADE and Tb-SHADE on the CEC2020 in 20D.

Table 27.
Clustering and population diversity of L-SHADE and TbL-SHADE on the CEC2020 in 5D.

Table 28.
Clustering and population diversity of L-SHADE and TbL-SHADE on the CEC2020 in 10D.

Table 29.
Clustering and population diversity of L-SHADE and TbL-SHADE on the CEC2020 in 15D.

Table 30.
Clustering and population diversity of L-SHADE and TbL-SHADE on the CEC2020 in 20D.

Table 31.
Clustering and population diversity of jSO and Tb-jSO on the CEC2020 in 5D.

Table 32.
Clustering and population diversity of jSO and Tb-jSO on the CEC2020 in 10D.

Table 33.
Clustering and population diversity of jSO and Tb-jSO on the CEC2020 in 15D.

Table 34.
Clustering and population diversity of jSO and Tb-jSO on the CEC2020 in 20D.
The rankings of the Friedman test [52] were obtained by using the average value (Mean) of each algorithm on all 10 test functions in Table 7, Table 8, Table 9, Table 10, Table 11, Table 12, Table 13, Table 14, Table 15, Table 16, Table 17, Table 18, Table 19, Table 20, Table 21 and Table 22, and are shown in Table 35, Table 36, Table 37 and Table 38. The related statistical values of the Friedman test are shown in Table 39. If the chi-square statistic was greater than the critical value, the null hypothesis was rejected. p represents the probability of the null hypothesis obtaining. The null hypothesis here was that there is no significant difference in performance among the nine algorithms considered here on CEC2020.

Table 35.
The Friedman ranks of comparative algorithms on CEC2020 in 5D.

Table 36.
The Friedman ranks of comparative algorithms on CEC2020 in 10D.

Table 37.
The Friedman ranks of comparative algorithms on CEC2020 in 15D.

Table 38.
The Friedman ranks of comparative algorithms on CEC2020 in 20D.

Table 39.
Related statistical values obtained of Friedman test for α = 0.05.
6. Results and Discussion
The results on the CEC2020 benchmark sets are first discussed. As shown in Table 7, Table 8, Table 9, Table 10, Table 11, Table 12, Table 13, Table 14, Table 15, Table 16, Table 17 and Table 18, the scores were two improvements against two instances of worsening (5D), four improvements and two instances of worsening (10D), five improvements and two instances of worsening (15D), and four improvements no instances of worsening (20D) in the case of SHADE; three improvements against zero instances of worsening (5D), four improvements and one worsening (10D), six improvements no worsening (15D), and five improvements and no worsening (20D) in the case of L-SHADE; and one improvement against no worsening (5D), four improvements and one worsening (10D), four improvements and one worsening (15D), and two improvements two instances of worsening (20D) in the case of jSO. In some test functions, the improved algorithm even escaped the local optimum and found the optimal value (if the error was smaller than 10−8, the relevant value was considered optimal). Examples are f3 in Table 10 and Table 13, and Table 14, f8 in Table 9, Table 13 and Table 16, and Table 17, f9 in Table 12, and f10 in Table 11. In most cases, the improved version was clearly better than the original algorithm except for Tb-SHADE (5D) and Tb-jSO (20D).
According to the convergence curves in Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12, in most cases, the improved algorithm showed similar convergence to the original in the early stage of the optimization process, but it clearly maintained a longer exploration phase and achieved better values of the objective function in the middle and late stages; in a few cases (such as f4 in Figure 5), the improved algorithm had slower convergence but did not achieve a better objective function value than the original.
As the numerical analyses in Table 23, Table 24, Table 25, Table 26, Table 27, Table 28, Table 29, Table 30, Table 31, Table 32, Table 33 and Table 34 show, in most cases, the improved algorithms exhibited fewer clusters (#runs), later clustering (mean CO), and higher population density (mean PD) than the original algorithm. But Tb-SHADE (5D) had a lower population density on f6–f9, as did TbL-SHADE (all dimensions) on f2–f7, where this might have been related to the linear decrease in the population size. Tb-jSO showed similar numbers of clusters in all dimensions and a lower population density on some test functions in 5D. Therefore, in most cases, the improved versions maintained the diversity of population and a longer exploration phase in the optimization process.
The significant improvements in Table 7, Table 8, Table 9, Table 10, Table 11, Table 12, Table 13, Table 14, Table 15, Table 16, Table 17 and Table 18 and the clustering analysis in Table 23 and Table 24 can be linked. The results in the former set of tables with the “+” symbol were always connected with the occurrence of later clustering, none at all, or fewer instances of clusters of 30 (for the last option, see, for example, column #runs in Table 24, Table 25 and Table 26, f3). Consequently, the improvement in the performance effected by the updated version was related to the maintenance of population diversity and a longer exploration phase.
According to the Friedman ranking in Table 35, Table 36, Table 37 and Table 38, Tb-SHADE, TbL-SHADE, and Tb-jSO were clearly better than the original algorithms and the advanced DISH and jDE100 in 10D, 15D, and 20D. But Tb-SHADE did not perform as well as SHADE in 5D and did not perform as well as DISH in 5D, 10D and 20D. In addition, the j2020 algorithm delivered the best performance and ranked first in 10D, 15D and 20D and one of the improved versions, TbL-SHADE, only delivered the best performance and ranked first in 5D. And jDE100 (winner of CEC2019), which ranks last in Table 35, Table 36, Table 37 and Table 38, did not seem suitable for CEC2020. Table 39 shows that the null hypothesis was rejected in all dimensions, and thus the Friedman ranking was correct. All in all, the three improved algorithms obtained good optimization results in contrast to the original algorithm as well as the advanced DISH and jDE100 algorithms but were slightly worse than the advanced j2020 algorithm.
7. Conclusions
In this paper, a relatively simple and direct method using turning-based mutation was proposed and tested on Single Objective Bound Constrained Numerical Optimization (CEC2020) benchmark sets in 5, 10, 15, and 20 dimensions against the SHADE, L-SHADE, and jSO algorithms. The basic thought of the proposed method is to change the direction of mutation under certain conditions to maintain the population diversity and a longer exploration phase. It can thus avoid premature convergence and escape the local optimum to get better optimization results. The results of experiments showed that this method is effective on CEC2020 benchmark sets in 10, 15, and 20 dimensions. The strong point of the proposed method is that it can be applied to variants of SHADE easily. A disadvantage is that it increases the time complexity and its effectiveness lacks theoretical proof. Our future research in the area will focus on further experiments, and on applying the proposed method to more algorithms. For example, the improved method may be useful for some practical problems featuring constraints.
Author Contributions
Conceptualization, H.K.; methodology, H.K.; project administration, L.J. and Y.S.; software, X.S.; validation, X.S. and Q.C.; visualization, L.J. and Q.C.; formal analysis, H.K.; investigation, Q.C.; resources, Y.S.; data curation, L.J.; writing—original draft preparation, L.J.; writing—review and editing, H.K. and X.S.; supervision: X.S.; funding acquisition: Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 61663046, 61876166. This research was funded by Open Foundation of Key Laboratory of Software Engineering of Yunnan Province, grant number 2015SE204.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Storn, R.; Price, K. Differential evolution–a simple and efficient heuristic for global optimization over continuous spaces. J. Glob. Optim. 1997, 11, 341–359. [Google Scholar] [CrossRef]
- Eltaeib, T.; Mahmood, A. Differential evolution: A survey and analysis. Appl. Sci. 2018, 8, 1945. [Google Scholar] [CrossRef]
- Arafa, M.; Sallam, E.A.; Fahmy, M. An enhanced differential evolution optimization algorithm. In Proceedings of the 2014 Fourth International Conference on Digital Information and Communication Technology and its Applications (DICTAP), Bangkok, Thailand, 6–8 May 2014; pp. 216–225. [Google Scholar]
- Awad, N.H.; Ali, M.Z.; Suganthan, P.N. Ensemble sinusoidal differential covariance matrix adaptation with Euclidean neighborhood for solving CEC2017 benchmark problems. In Proceedings of the 2017 IEEE Congress on Evolutionary Computation (CEC), San Sebastian, Spain, 5–8 June 2017; pp. 372–379. [Google Scholar]
- Bujok, P.; Tvrdík, J. Adaptive differential evolution: SHADE with competing crossover strategies. In Proceedings of the International Conference on Artificial Intelligence and Soft Computing, Zakopane, Poland, 14–18 June 2015; pp. 329–339. [Google Scholar]
- Bujok, P.; Tvrdík, J.; Poláková, R. Evaluating the performance of shade with competing strategies on CEC 2014 single-parameter test suite. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 5002–5009. [Google Scholar]
- Liu, X.-F.; Zhan, Z.-H.; Zhang, J. Dichotomy guided based parameter adaptation for differential evolution. In Proceedings of the 2015 Annual Conference on Genetic and Evolutionary Computation, Madrid, Spain, 11–15 July 2015; pp. 289–296. [Google Scholar]
- Liu, Z.-G.; Ji, X.-H.; Yang, Y. Hierarchical differential evolution algorithm combined with multi-cross operation. Expert Syst. Appl. 2019, 130, 276–292. [Google Scholar] [CrossRef]
- Mohamed, A.W.; Hadi, A.A.; Fattouh, A.M.; Jambi, K.M. LSHADE with semi-parameter adaptation hybrid with CMA-ES for solving CEC 2017 benchmark problems. In Proceedings of the 2017 IEEE Congress on Evolutionary Computation (CEC), San Sebastian, Spain, 5–8 June 2017; pp. 145–152. [Google Scholar]
- Poláková, R.; Tvrdík, J.; Bujok, P. L-SHADE with competing strategies applied to CEC2015 learning-based test suite. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 4790–4796. [Google Scholar]
- Poláková, R.; Tvrdík, J.; Bujok, P. Evaluating the performance of L-SHADE with competing strategies on CEC2014 single parameter-operator test suite. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 1181–1187. [Google Scholar]
- Sallam, K.M.; Sarker, R.A.; Essam, D.L.; Elsayed, S.M. Neurodynamic differential evolution algorithm and solving CEC2015 competition problems. In Proceedings of the 2015 IEEE Congress on Evolutionary Computation (CEC), Sendai, Japan, 25–28 May 2015; pp. 1033–1040. [Google Scholar]
- Viktorin, A.; Pluhacek, M.; Senkerik, R. Network based linear population size reduction in SHADE. In Proceedings of the 2016 International Conference on Intelligent Networking and Collaborative Systems (INCoS), Ostrawva, Czech Republic, 7–9 September 2016; pp. 86–93. [Google Scholar]
- Viktorin, A.; Pluhacek, M.; Senkerik, R. Success-history based adaptive differential evolution algorithm with multi-chaotic framework for parent selection performance on CEC2014 benchmark set. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 4797–4803. [Google Scholar]
- Viktorin, A.; Senkerik, R.; Pluhacek, M.; Kadavy, T. Distance vs. Improvement Based Parameter Adaptation in SHADE. In Artificial Intelligence and Algorithms in Intelligent Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 455–464. [Google Scholar]
- Viktorin, A.; Senkerik, R.; Pluhacek, M.; Kadavy, T.; Zamuda, A. Distance based parameter adaptation for differential evolution. In Proceedings of the 2017 IEEE Symposium Series on Computational Intelligence (SSCI), Honolulu, HI, USA, 27 November–1 December 2017; pp. 1–7. [Google Scholar]
- Zhao, F.; He, X.; Yang, G.; Ma, W.; Zhang, C.; Song, H. A hybrid iterated local search algorithm with adaptive perturbation mechanism by success-history based parameter adaptation for differential evolution (SHADE). J. Eng. Optim. 2020, 52, 367–383. [Google Scholar] [CrossRef]
- Al-Dabbagh, R.D.; Neri, F.; Idris, N.; Baba, M.S. Algorithmic design issues in adaptive differential evolution schemes: Review and taxonomy. Swarm Evol. Comput. 2018, 43, 284–311. [Google Scholar] [CrossRef]
- Tanabe, R.; Fukunaga, A. Success-history based parameter adaptation for differential evolution. In Proceedings of the 2013 IEEE Congress on Evolutionary Computation, Cancun, Mexico, 20–23 June 2013; pp. 71–78. [Google Scholar]
- Zhang, J.; Sanderson, A.C. JADE: Adaptive differential evolution with optional external archive. IEEE Trans. Evol. Comput. 2009, 13, 945–958. [Google Scholar] [CrossRef]
- Tanabe, R.; Fukunaga, A.S. Improving the search performance of SHADE using linear population size reduction. In Proceedings of the 2014 IEEE Congress on Evolutionary Computation (CEC), Beijing, China, 6–11 July 2014; pp. 1658–1665. [Google Scholar]
- Guo, S.-M.; Tsai, J.S.-H.; Yang, C.-C.; Hsu, P.-H. A self-optimization approach for L-SHADE incorporated with eigenvector-based crossover and successful-parent-selecting framework on CEC 2015 benchmark set. In Proceedings of the 2015 IEEE Congress on Evolutionary Computation (CEC), Sendai, Japan, 25–28 May 2015; pp. 1003–1010. [Google Scholar]
- Awad, N.H.; Ali, M.Z.; Suganthan, P.N.; Reynolds, R.G. An ensemble sinusoidal parameter adaptation incorporated with L-SHADE for solving CEC2014 benchmark problems. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 2958–2965. [Google Scholar]
- Brest, J.; Maučec, M.S.; Bošković, B. Single objective real-parameter optimization: Algorithm jSO. In Proceedings of the 2017 IEEE Congress on Evolutionary Computation (CEC), San Sebastian, Spain, 5–8 June 2017; pp. 1311–1318. [Google Scholar]
- Piotrowski, A.P.; Napiorkowski, J.J. Step-by-step improvement of JADE and SHADE-based algorithms: Success or failure? Swarm Evol. Comput. 2018, 43, 88–108. [Google Scholar] [CrossRef]
- Piotrowski, A.P. L-SHADE optimization algorithms with population-wide inertia. Inf. Sci. 2018, 468, 117–141. [Google Scholar] [CrossRef]
- Stanovov, V.; Akhmedova, S.; Semenkin, E. LSHADE algorithm with rank-based selective pressure strategy for solving CEC 2017 benchmark problems. In Proceedings of the 2018 IEEE Congress on Evolutionary Computation (CEC), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–8. [Google Scholar]
- Brest, J.; Maučec, M.S.; Bošković, B. The 100-Digit Challenge: Algorithm jDE100. In Proceedings of the 2019 IEEE Congress on Evolutionary Computation (CEC), Wellington, New Zealand, 10–13 June 2019; pp. 19–26. [Google Scholar]
- Brest, J.; Maučec, M.S.; Bošković, B. Differential Evolution Algorithm for Single Objective Bound-Constrained Optimization: Algorithm j2020. In Proceedings of the 2020 IEEE Congress on Evolutionary Computation (CEC), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar]
- Suganthan, P.N. Particle swarm optimiser with neighbourhood operator. In Proceedings of the 1999 Congress on Evolutionary Computation-CEC99 (Cat. No. 99TH8406), Washington, DC, USA, 6–9 July 1999; pp. 1958–1962. [Google Scholar]
- Das, S.; Maity, S.; Qu, B.-Y.; Suganthan, P.N. Real-parameter evolutionary multimodal optimization—A survey of the state-of-the-art. Swarm Evol. Comput. 2011, 1, 71–88. [Google Scholar] [CrossRef]
- Mezura-Montes, E.; Coello, C.A.C. Constraint-handling in nature-inspired numerical optimization: Past, present and future. Swarm Evol. Comput. 2011, 1, 173–194. [Google Scholar] [CrossRef]
- Neri, F.; Cotta, C. Memetic algorithms and memetic computing optimization: A literature review. Swarm Evol. Comput. 2012, 2, 1–14. [Google Scholar] [CrossRef]
- Neri, F.; Tirronen, V. Recent advances in differential evolution: A survey and experimental analysis. Artif. Intell. Rev. 2010, 33, 61–106. [Google Scholar] [CrossRef]
- Zamuda, A.; Brest, J. Self-adaptive control parameters’ randomization frequency and propagations in differential evolution. Swarm Evol. Comput. 2015, 25, 72–99. [Google Scholar] [CrossRef]
- Zhou, A.; Qu, B.-Y.; Li, H.; Zhao, S.-Z.; Suganthan, P.N.; Zhang, Q. Multiobjective evolutionary algorithms: A survey of the state of the art. Swarm Evol. Comput. 2011, 1, 32–49. [Google Scholar] [CrossRef]
- Piotrowski, A.P. Review of differential evolution population size. Swarm Evol. Comput. 2017, 32, 1–24. [Google Scholar] [CrossRef]
- Opara, K.R.; Arabas, J. Differential Evolution: A survey of theoretical analyses. Swarm Evol. Comput. 2019, 44, 546–558. [Google Scholar] [CrossRef]
- Poikolainen, I.; Neri, F.; Caraffini, F. Cluster-based population initialization for differential evolution frameworks. Inf. Sci. 2015, 297, 216–235. [Google Scholar] [CrossRef]
- Weber, M.; Neri, F.; Tirronen, V. A study on scale factor/crossover interaction in distributed differential evolution. Artif. Intell. Rev. 2013, 39, 195–224. [Google Scholar] [CrossRef]
- Zaharie, D. Influence of crossover on the behavior of differential evolution algorithms. Appl. Soft Comput. 2009, 9, 1126–1138. [Google Scholar] [CrossRef]
- Yue, C.; Price, K.; Suganthan, P.; Liang, J.; Ali, M.; Qu, B.; Awad, N.; Biswas, P. Problem definitions and evaluation criteria for the CEC 2020 special session and competition on single objective bound constrained numerical optimization. In Proceedings of the 2020 IEEE Congress on Evolutionary Computation (CEC), Glasgow, UK, 19–24 July 2020. [Google Scholar]
- Piotrowski, A.P.; Napiorkowski, J.J. Some metaheuristics should be simplified. Inf. Sci. 2018, 427, 32–62. [Google Scholar] [CrossRef]
- Viktorin, A.; Senkerik, R.; Pluhacek, M.; Kadavy, T.; Zamuda, A. Distance based parameter adaptation for success-history based differential evolution. Swarm Evol. Comput. 2019, 50, 100462. [Google Scholar] [CrossRef]
- Caraffini, F.; Kononova, A.V.; Corne, D. Infeasibility and structural bias in differential evolution. Inf. Sci. 2019, 496, 161–179. [Google Scholar] [CrossRef]
- Awad, N.; Ali, M.; Liang, J.; Qu, B.; Suganthan, P.; Definitions, P. Evaluation Criteria for the CEC 2017 Special Session and Competition on Single Objective Real-Parameter Numerical Optimization. In Proceedings of the 2017 IEEE Congress on Evolutionary Computation (CEC), San Sebastian, Spain, 5–8 June 2017. [Google Scholar]
- Brest, J.; Maučec, M.S.; Bošković, B. iL-SHADE: Improved L-SHADE algorithm for single objective real-parameter optimization. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 1188–1195. [Google Scholar]
- Rahnamayan, S.; Tizhoosh, H.R.; Salama, M.M. Opposition-based differential evolution. IEEE Trans. Evol. Comput. 2008, 12, 64–79. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In KDD-96 Proceedings; AAAI: Menlo Park, CA, USA, 1996; pp. 226–231. [Google Scholar]
- Deza, M.M.; Deza, E. Encyclopedia of distances. In Encyclopedia of Distances; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–583. [Google Scholar]
- Poláková, R.; Tvrdík, J.; Bujok, P.; Matoušek, R. Population-size adaptation through diversity-control mechanism for differential evolution. In Proceedings of the MENDEL, 22th International Conference on Soft Computing, Brno, Czech Republic, 8–10 June 2016; pp. 49–56. [Google Scholar]
- Demšar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).