Transfer Learning for Stenosis Detection in X-ray Coronary Angiography
Abstract
1. Introduction
2. Background
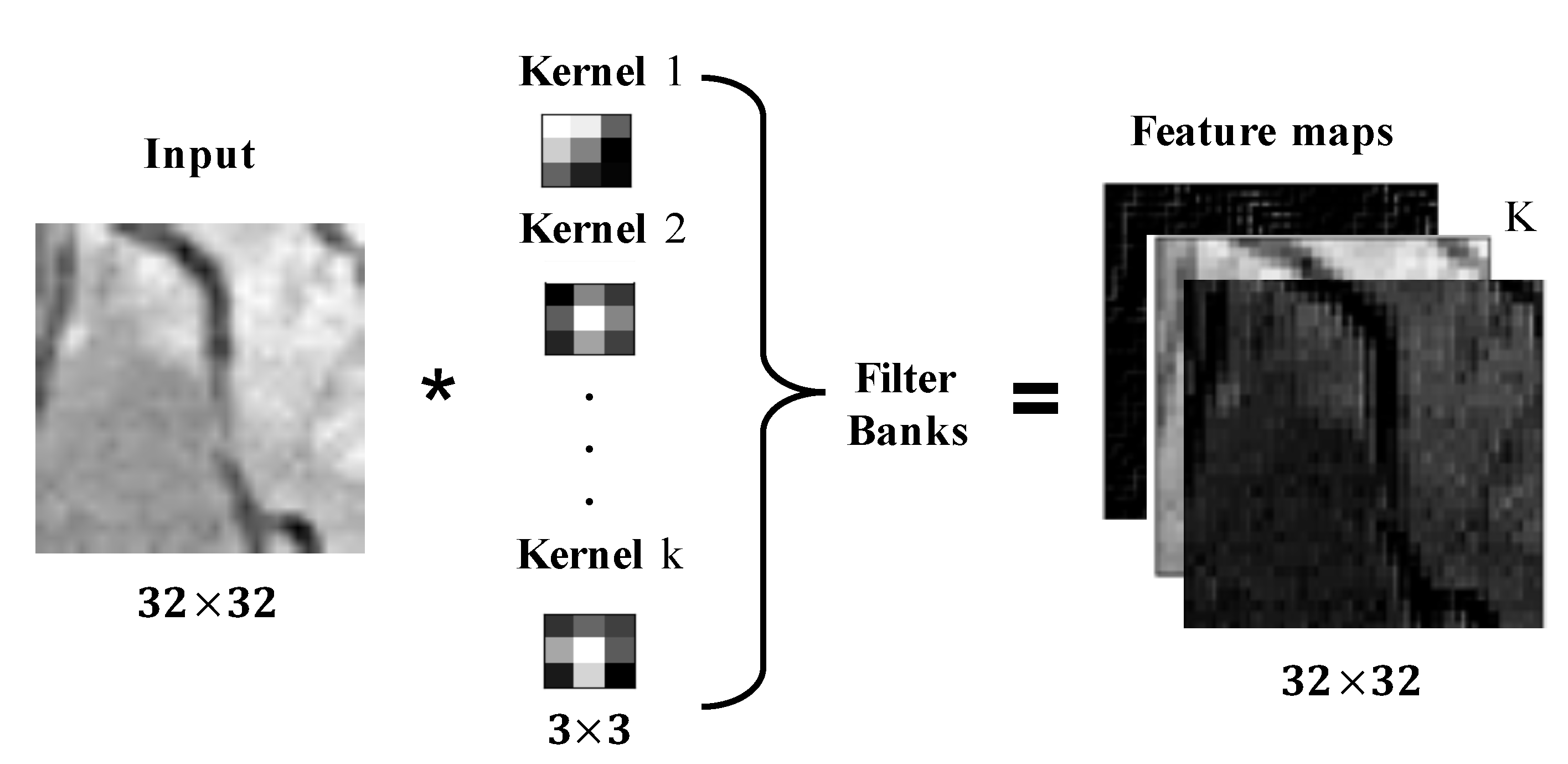
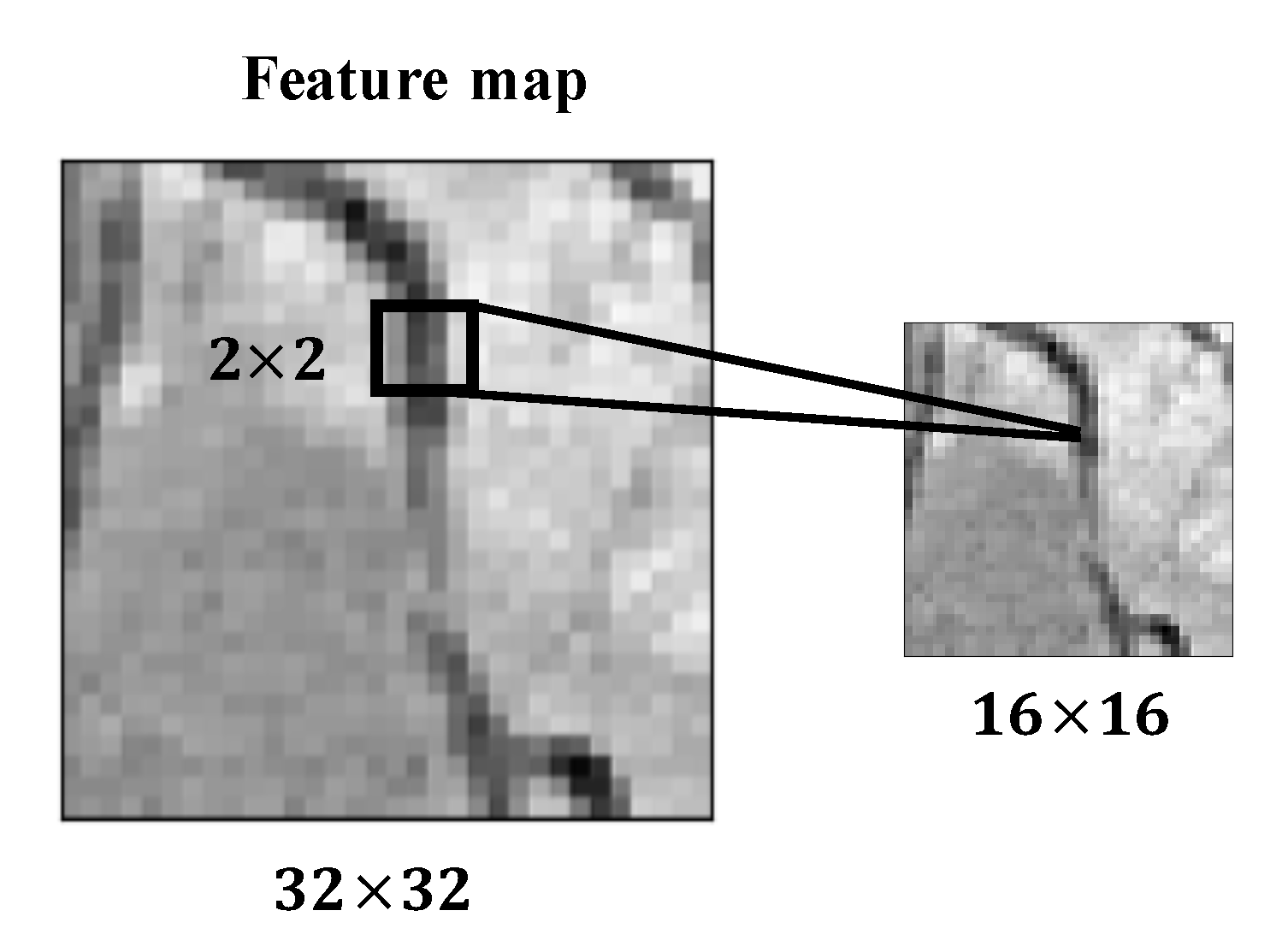
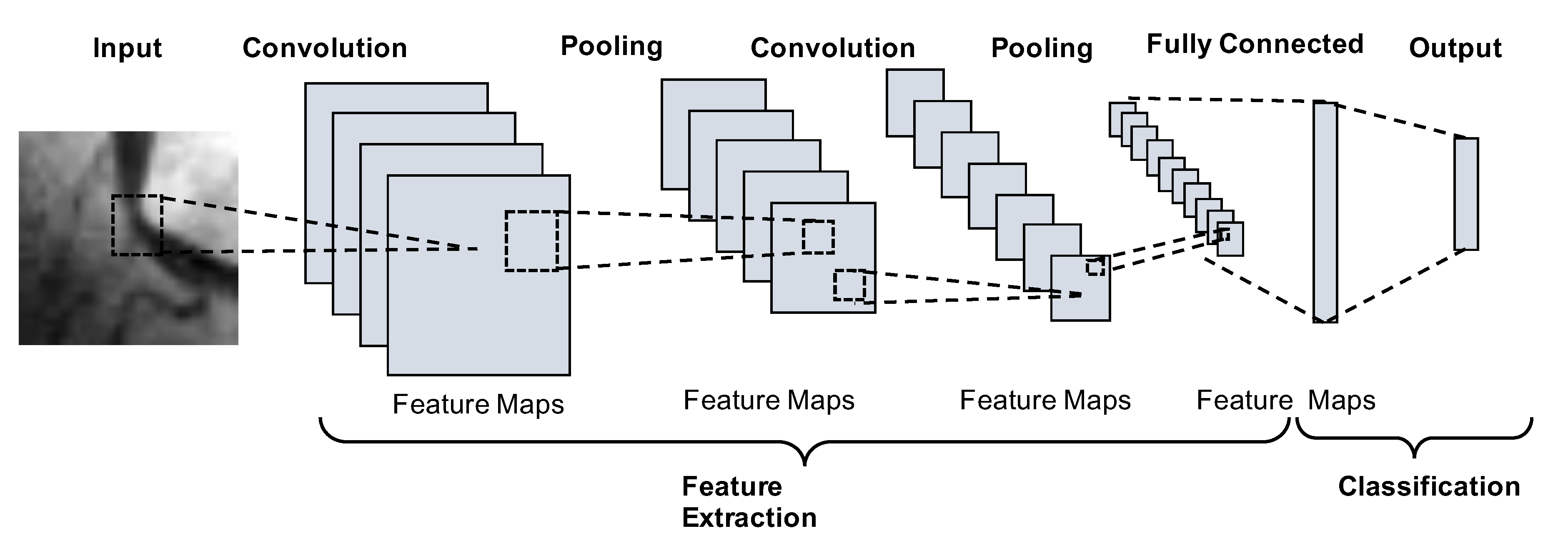
2.1. Convolutional Neural Networks
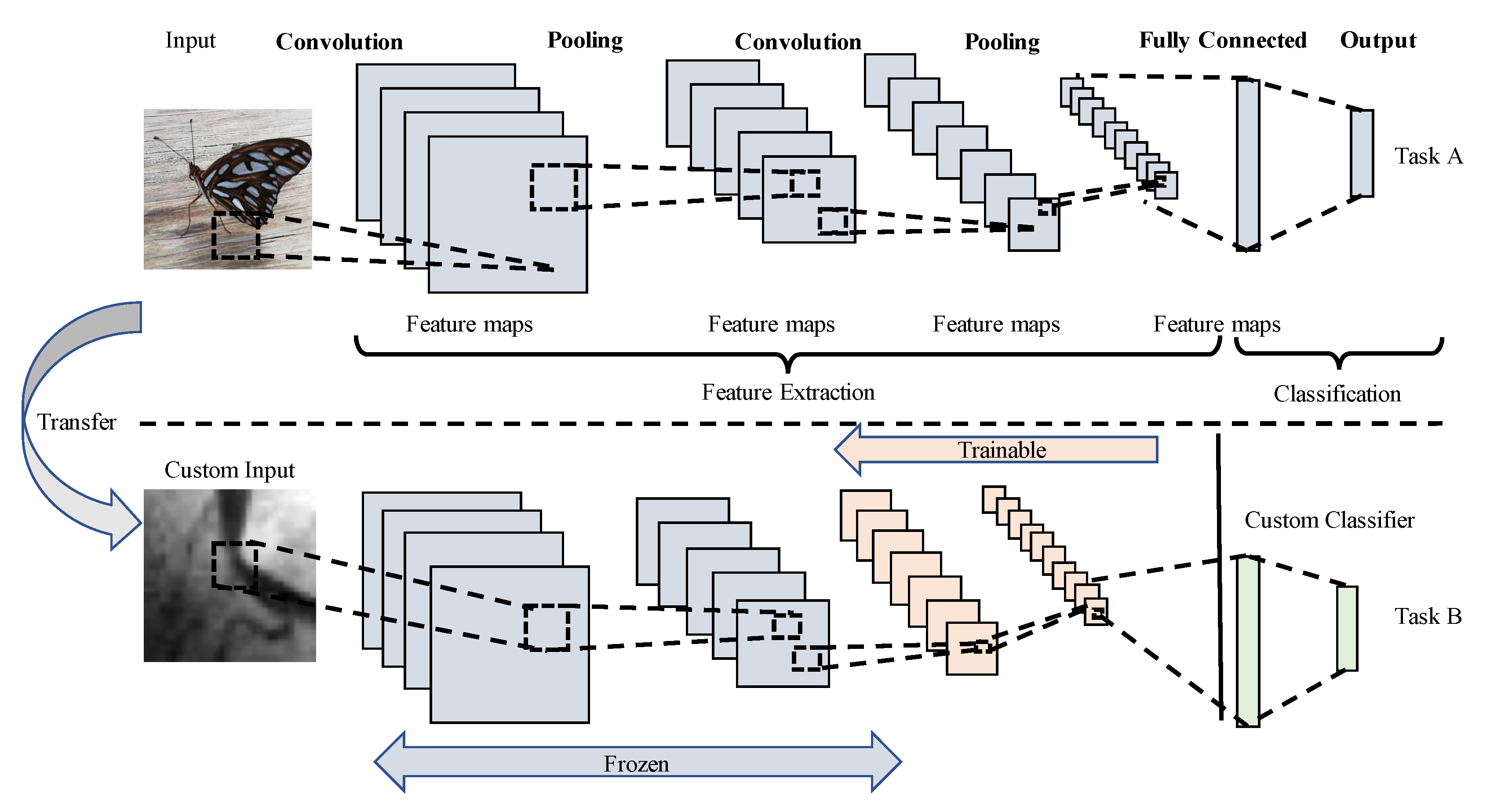
2.2. Transfer Learning
3. Proposed Method for Stenosis Detection
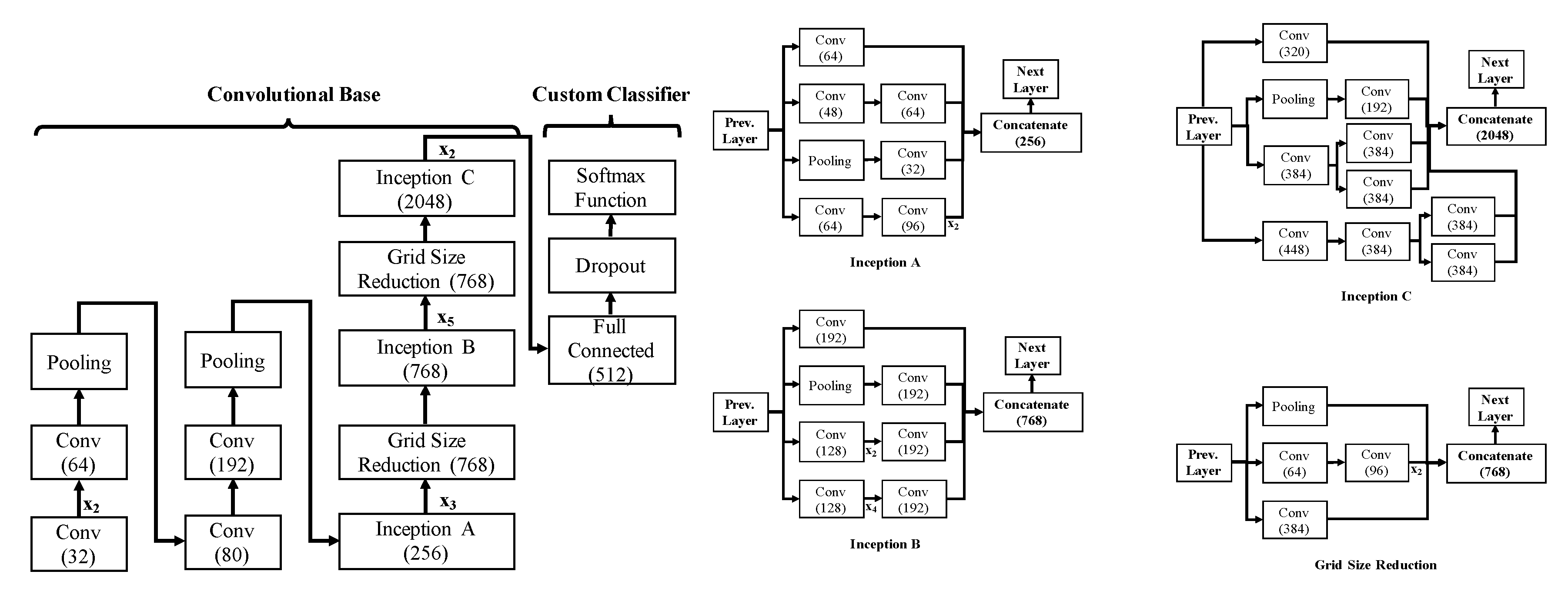
3.1. Network Architecture
3.2. Network Configuration
- Feature extractor: The entire pre-trained CNN is idle, whereas the weights of the pre-trained convolutional layers remain fixed, and only the fully connected layers are trained with the selected dataset, as in [20].
- Fine-tuning: The whole network is fine-tuned in a layer-wise manner from top to low-level blocks, where the blocks of convolutional layers pass to trainable on a descendent way. In this behavior, the number of convolutional trainable blocks is increased from one to five convolutional blocks, while the fully connected layers are always included in the training.
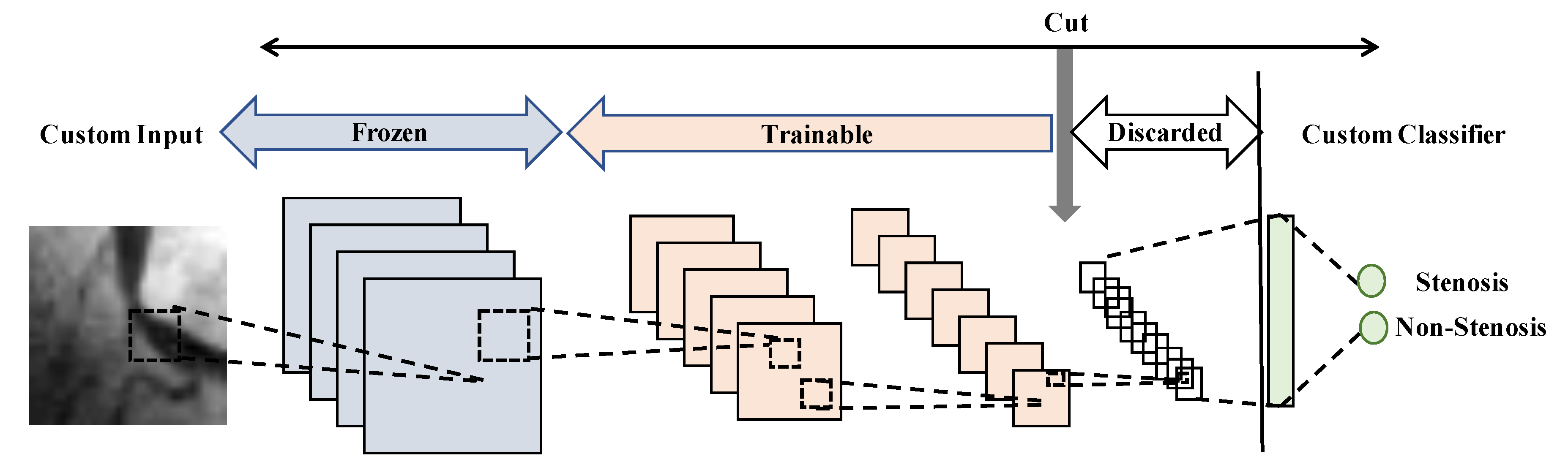
- Network-cut and feature extractor: the network is chopped up on an early convolutional block, retaining their weights. Then from this block, the last fully connected layer and softmax activation layer are added and trained. Notice that the layers after the cut block are discarded.
- Network-cut and fine-tuning: the network is chopped up on an early convolutional block, but a fine-tuning process is carried out in a layer-wise manner from new top-layers to low-level layers, as well as the fully connected layers.
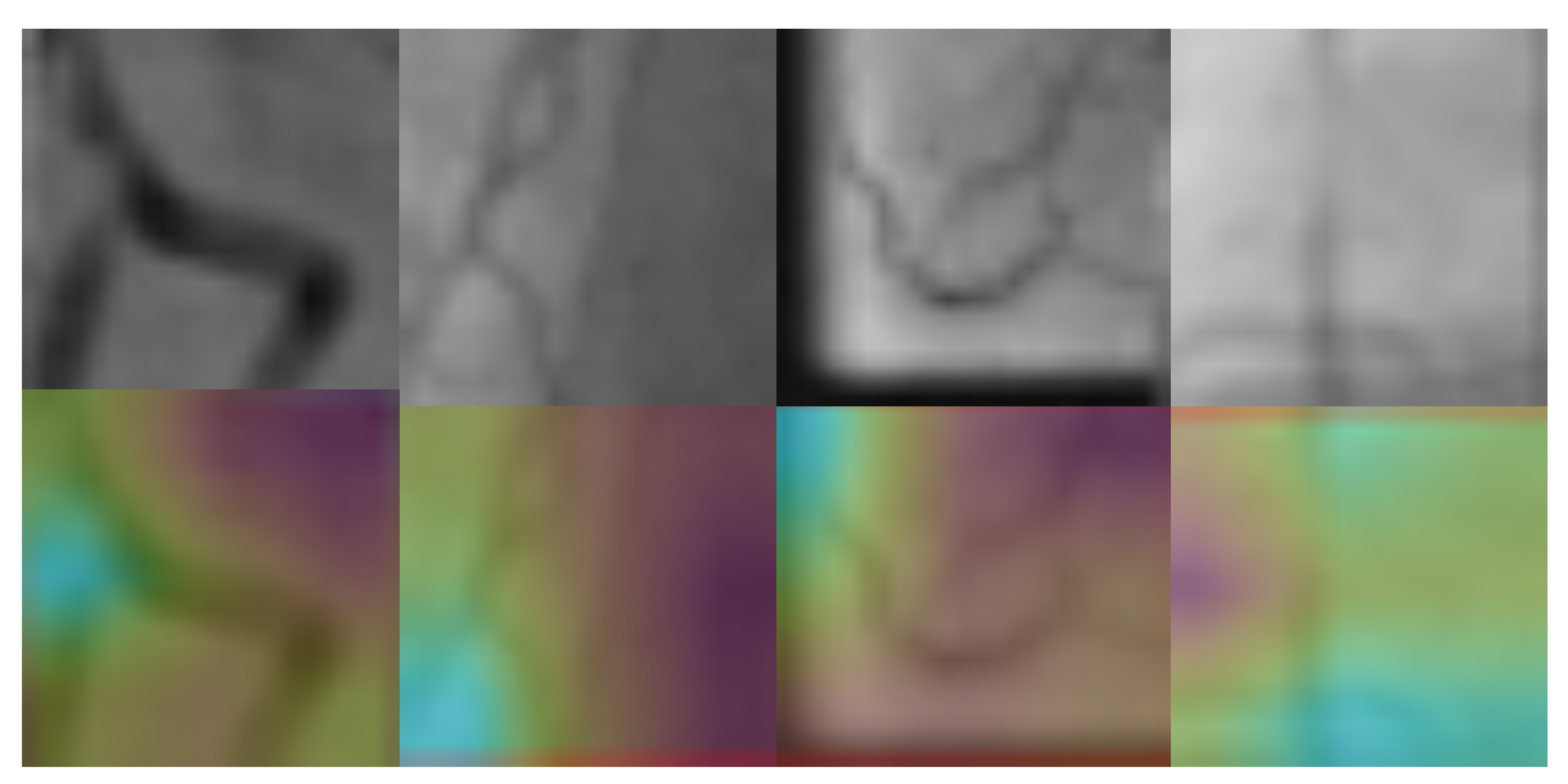
3.3. Stenosis Activation Maps
3.4. Evaluation Metrics
4. Results and Discussion
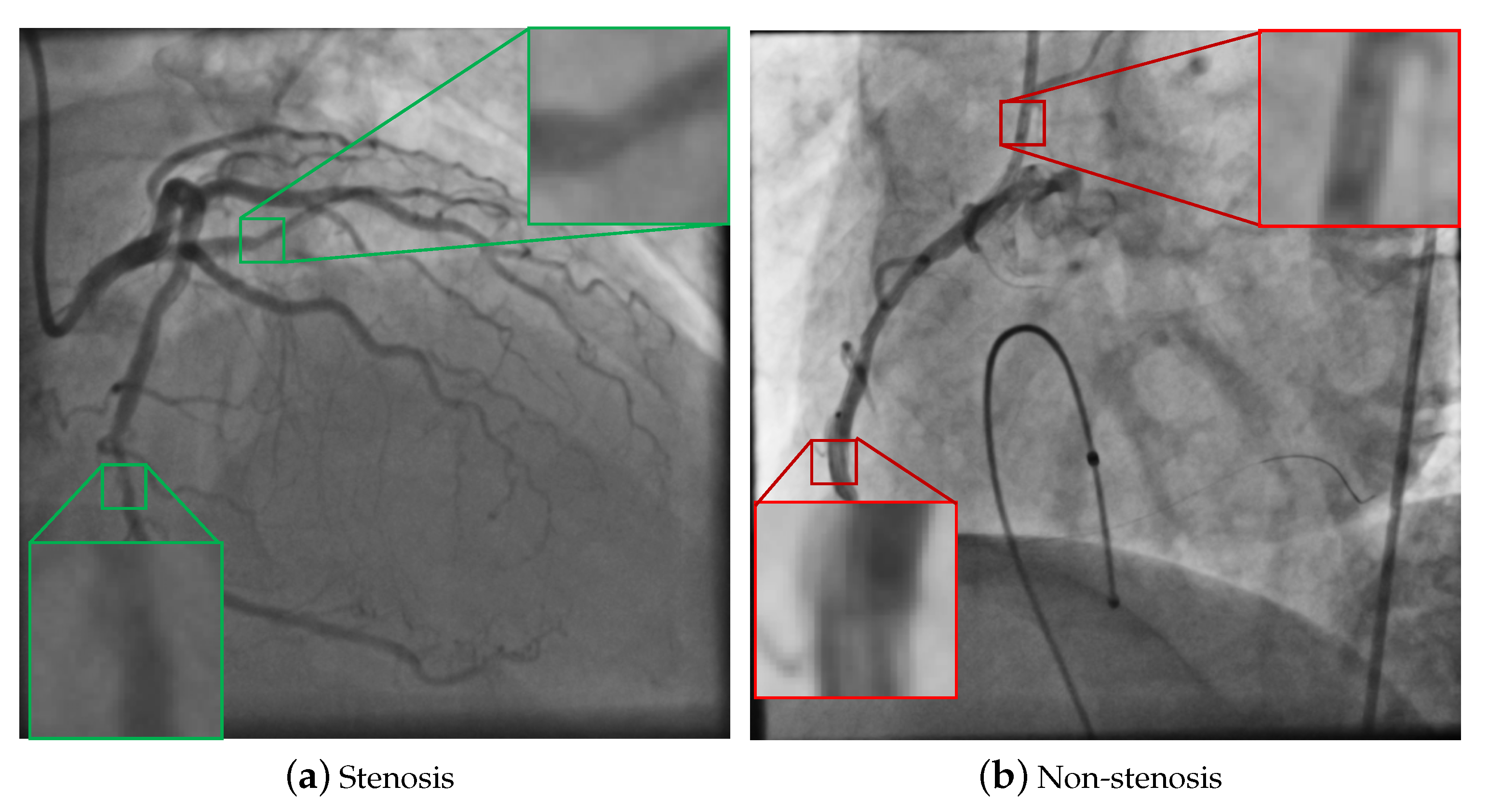
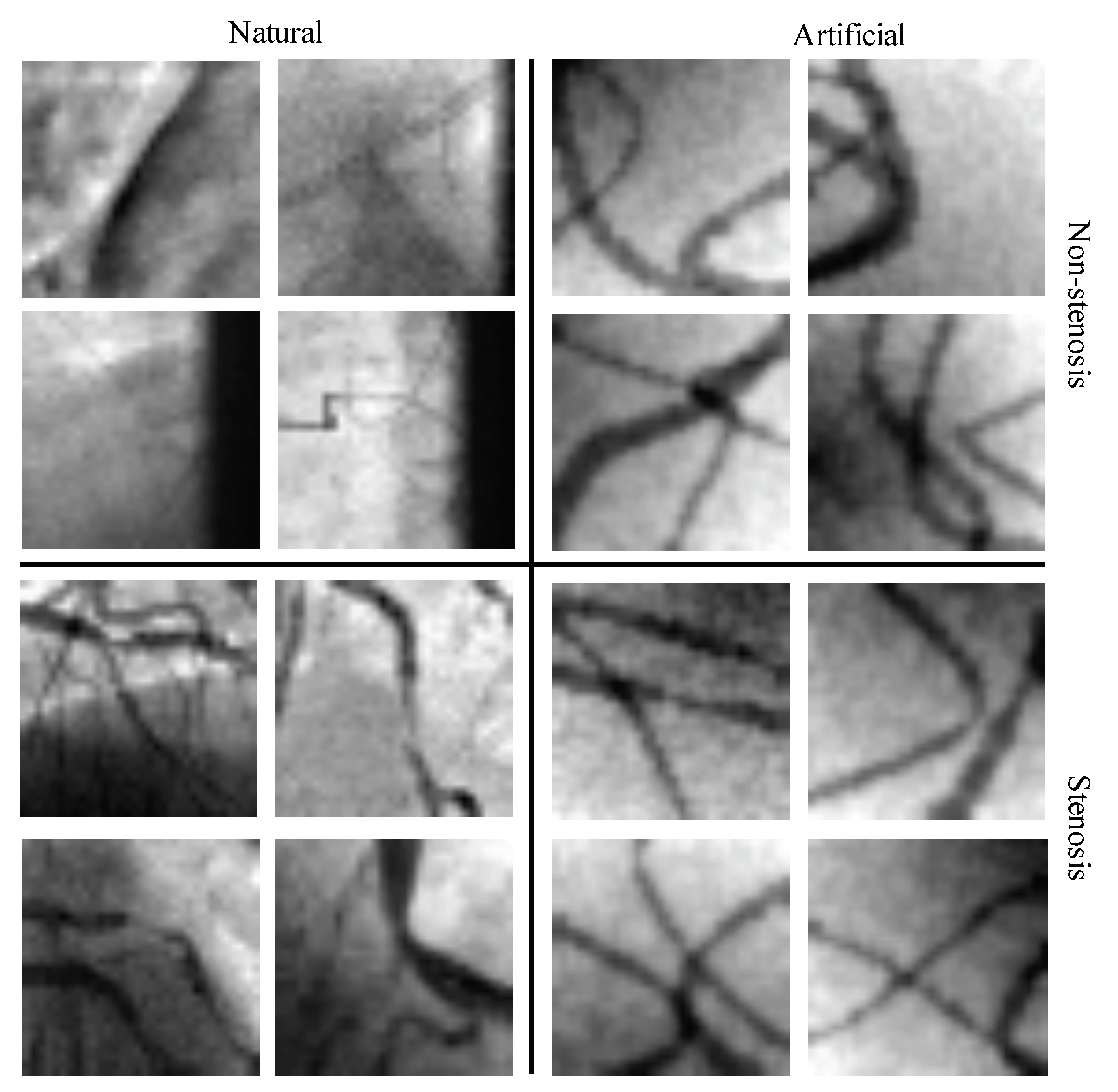
4.1. Databases of Coronary Stenosis
4.2. Network-Cut and Fine-Tuning
4.3. Detection Results
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CVDs | Cardio Vascular Diseases |
| XCA | X-ray Coronary Angiography |
| CAD | Computer-Aided Diagnosis |
| CNN(s) | Convolutional Neural Network(s) |
| ILSVRC | ImageNet Large Scale Visual Recognition Challenge |
| ACC | Accuracy |
| TP | True Positive |
| TN | True Negative |
| FP | False Positive |
| FN | False Negative |
| SN | Sensitivity |
| SP | Specificity |
| PC | Precision |
| FNFE | Full Network as Features Extractor |
| FNFT | Full Network and Fine-tuning |
| NCFE | Network-cut as Features Extractor |
| NCFT | Network-cut and Fine-tuning |
References
- Athanasiou, L.S.; Fotiadis, D.I.; Michalis, L.K. Atherosclerotic Plaque Characterization Methods Based on Coronary Imaging; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- World Health Organization. Cardiovascular Diseases (CVDs). 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 30 August 2020).
- Eckert, J.; Schmidt, M.; Magedanz, A.; Voigtländer, T.; Schmermund, A. Coronary CT angiography in managing atherosclerosis. Int. J. Mol. Sci. 2015, 16, 3740–3756. [Google Scholar] [CrossRef] [PubMed]
- Kishore, A.N.; Jayanthi, V. Automatic stenosis grading system for diagnosing coronary artery disease using coronary angiogram. Int. J. Biomed. Eng. Technol. 2019, 31, 260–277. [Google Scholar] [CrossRef]
- Saad, I.A. Segmentation of Coronary Artery Images and Detection of Atherosclerosis. J. Eng. Appl. Sci. 2018, 13, 7381–7387. [Google Scholar] [CrossRef]
- Sameh, S.; Azim, M.A.; AbdelRaouf, A. Narrowed Coronary Artery Detection and Classification using Angiographic Scans. In Proceedings of the 2017 12th International Conference on Computer Engineering and Systems (ICCES), Cairo, Egypt, 19–20 December 2017; pp. 73–79. [Google Scholar]
- Wan, T.; Feng, H.; Tong, C.; Li, D.; Qin, Z. Automated Identification and Grading of Coronary Artery Stenoses with X-ray Angiography. Comput. Methods Programs Biomed. 2018, 167, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Sanchez, F.; Cruz-Aceves, I.; Hernandez-Aguirre, A. Automatic detection of coronary artery stenosis in X-ray angiograms using Gaussian filters and genetic algorithms. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2016; Volume 1747, p. 020005. [Google Scholar]
- Cruz-Aceves, I.; Cervantes-Sanchez, F.; Hernandez-Aguirre, A. Automatic Detection of Coronary Artery Stenosis Using Bayesian Classification and Gaussian Filters Based on Differential Evolution. In Hybrid Intelligence for Image Analysis and Understanding; Wiley: Hoboken, NJ, USA, 2017; pp. 369–390. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Azizpour, H.; Sharif Razavian, A.; Sullivan, J.; Maki, A.; Carlsson, S. From Generic to Specific Deep Representations for Visual Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Boston, MA, USA, 7–12 June 2015; pp. 36–45. [Google Scholar]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef]
- Xu, S.; Wu, H.; Bie, R. CXNet-m1: Anomaly Detection on Chest X-Rays With Image-Based Deep Learning. IEEE Access 2018, 7, 4466–4477. [Google Scholar] [CrossRef]
- Shen, L.; Margolies, L.R.; Rothstein, J.H.; Fluder, E.; McBride, R.; Sieh, W. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peck, D.; Hsieh, S.; Dialani, V.; Lehman, C.D.; Zhou, B.; Syrgkanis, V.; Mackey, L.; Patterson, G. Expert identification of visual primitives used by CNNs during mammogram classification. In Medical Imaging 2018: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10575, p. 105752T. [Google Scholar]
- Chakravarthy, A.D.; Abeyrathna, D.; Subramaniam, M.; Chundi, P.; Halim, M.S.; Hasanreisoglu, M.; Sepah, Y.J.; Nguyen, Q.D. An Approach Towards Automatic Detection of Toxoplasmosis using Fundus Images. In Proceedings of the 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering (BIBE), Athens, Greece, 28–30 October 2019; pp. 710–717. [Google Scholar]
- Antczak, K.; Liberadzki, Ł. Stenosis Detection with Deep Convolutional Neural Networks. In MATEC Web of Conferences; EDP Sciences: Ullis, France, 2018; Volume 210, p. 04001. [Google Scholar]
- Au, B.; Shaham, U.; Dhruva, S.; Bouras, G.; Cristea, E.; Coppi, A.; Warner, F.; Li, S.X.; Krumholz, H. Automated Characterization of Stenosis in Invasive Coronary Angiography Images with Convolutional Neural Networks. arXiv 2018, arXiv:1807.10597. [Google Scholar]
- Cong, C.; Kato, Y.; Vasconcellos, H.D.; Lima, J.; Venkatesh, B. Automated Stenosis Detection and Classification in X-ray Angiography Using Deep Neural Network. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1301–1308. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Wu, W.; Zhang, J.; Xie, H.; Zhao, Y.; Zhang, S.; Gu, L. Automatic detection of coronary artery stenosis by convolutional neural network with temporal constraint. Comput. Biol. Med. 2020, 118, 103657. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.Y.; Liu, W.; Ranga, A.; Tyagi, A.; Berg, A.C. Dssd: Deconvolutional single shot detector. arXiv 2017, arXiv:1701.06659. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 3320–3328. [Google Scholar]
- Oh, K.; Chung, Y.C.; Kim, K.W.; Kim, W.S.; Oh, I.S. Classification and Visualization of Alzheimer’s Disease using Volumetric Convolutional Neural Network and Transfer Learning. Sci. Rep. 2020, 10, 18150. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mac. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; pp. 249–256. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Antczak, K.; Liberadzki, Ł. Deep Stenosis Detection Dataset. 2020. Available online: https://github.com/KarolAntczak/DeepStenosisDetection (accessed on 30 August 2020).
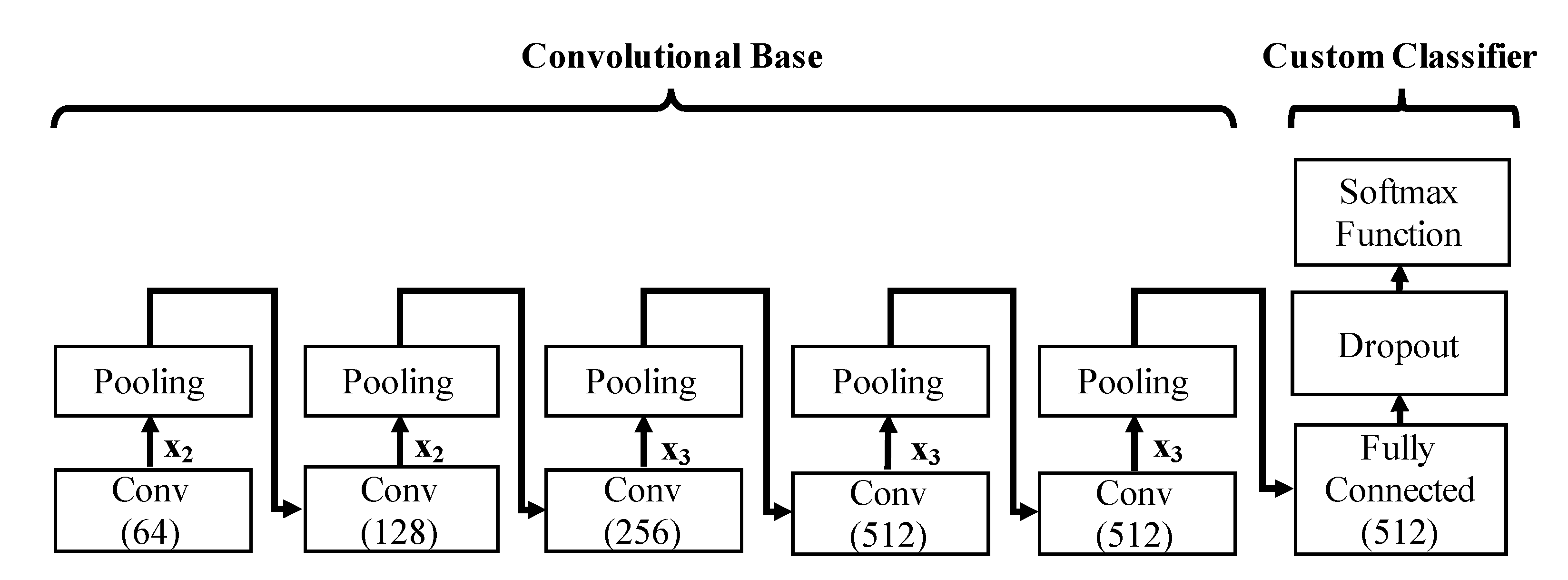
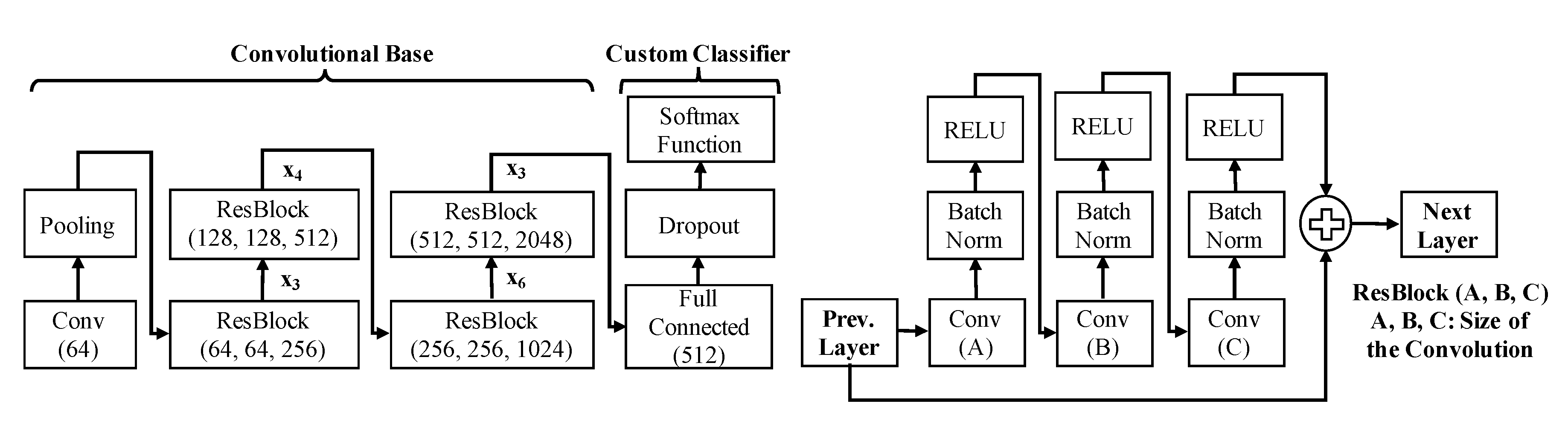
| Block Number | Convolutional Layers |
|---|---|
| 1 | {1, 2} |
| 2 | {3, 4} |
| 3 | {5, 6, 7} |
| 4 | {8, 9, 10} |
| 5 | {11, 12, 13} |
| 6 | Fully Connected Layers (Classifier) |
| Block Number | Description | Convolutional Layers |
|---|---|---|
| 1 | Convolutional Layer | {1} |
| 2 | Residual blocks | {1, 2, 3} |
| 3 | {4, 5, 6, 7} | |
| 4 | {9, 10, …, 13} | |
| 5 | {14, 15, 16} | |
| 6 | Fully Connected Layers | Classifier |
| Block Number | Description | Convolutional Layers |
|---|---|---|
| 1 | Convolutional Layer | {1, 2, 3} |
| 2 | {4, 5} | |
| 3 | Inception blocks | {1, 2, 3} |
| 4 | {4, 5, …, 8} | |
| 5 | {9, 10, 11} | |
| 6 | Fully Connected Layers | Classifier |
| Cut Block | Fine-Tuned Blocks |
|---|---|
| N/A | {6} |
| {5, 6} | |
| {4, 5, 6} | |
| {3, 4, 5, 6} | |
| {2, 3, 4, 5, 6} | |
| {1, 2, 3, 4, 5, 6} | |
| 4 | {6} |
| {4, 6} | |
| {3, 4, 6} | |
| {2, 3, 4, 6} | |
| {1, 2, 3, 4, 6} | |
| 3 | {6} |
| {3, 6} | |
| {2, 3, 6} | |
| {1, 2, 3, 6} | |
| 2 | {6} |
| {2, 6} | |
| {1, 2, 6} | |
| 1 | {6} |
| {1, 6} |
| Network | Initialization | Fine-Tuning Strategy | Best Configuration | Cut Block | Fine-Tuned Blocks |
|---|---|---|---|---|---|
| VGG16 | Random | Only real data | FNFE | N/A | N/A |
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 2 | {1, 2, 6} | |||
| Only artificial data | FNFE | N/A | N/A | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 2 | {1, 2, 6} | |||
| Artificial + real data | FNFE | N/A | N/A | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 1 | {1, 6} | |||
| VGG16 | ImageNet | Only real data | FNFE | N/A | {6} |
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | 4 | {6} | |||
| NCFT | 3 | {2, 6} | |||
| Only artificial data | FNFE | N/A | {6} | ||
| FNFT | N/A | {4, 5, 6} | |||
| NCFE | 3 | {6} | |||
| NCFT | 3 | {1, 2, 3, 6} | |||
| Artificial + real data | FNFE | N/A | {6} | ||
| FNFT | N/A | {2, 3, …, 6} | |||
| NCFE | 4 | {6} | |||
| NCFT | 3 | {3, 6} | |||
| ResNet50 | Only real data | FNFE | N/A | N/A | |
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 2 | {1, 2, 6} | |||
| Only artificial data | FNFE | N/A | N/A | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 2 | {1, 2, 6} | |||
| Artificial + real data | FNFE | N/A | N/A | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 4 | {1, 2, 3, 4, 6} | |||
| ImageNet | Only real data | FNFE | N/A | {6} | |
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | 3 | {6} | |||
| NCFT | 2 | {1, 2, 6} | |||
| Only artificial data | FNFE | N/A | {6} | ||
| FNFT | N/A | {5, 6} | |||
| NCFE | 3 | {6} | |||
| NCFT | 2 | {1, 2, 6} | |||
| Artificial + real data | FNFE | N/A | {6} | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | 2 | {6} | |||
| NCFT | 2 | {1, 2, 6} | |||
| Inception-v3 | Random | Only real data | FNFE | N/A | N/A |
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 3 | {1, 2, 3, 6} | |||
| Only artificial data | FNFE | N/A | N/A | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 2 | {1, 2, 6} | |||
| Artificial + real data | FNFE | N/A | N/A | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | N/A | N/A | |||
| NCFT | 2 | {1, 2, 6} | |||
| ImageNet | Only real data | FNFE | N/A | {6} | |
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | 3 | {6} | |||
| NCFT | 3 | {1, 2, 3, 6} | |||
| Only artificial data | FNFE | N/A | {6} | ||
| FNFT | N/A | {3, 4, 5, 6} | |||
| NCFE | 3 | {6} | |||
| NCFT | 3 | {2, 3, 6} | |||
| Artificial + real data | FNFE | N/A | {6} | ||
| FNFT | N/A | {1, 2, …, 6} | |||
| NCFE | 3 | {6} | |||
| NCFT | 3 | {1, 2, 3, 6} |
| Network | Initialization | Fine-Tuning Strategy | Best Configuration | Detection Performance | ||||
|---|---|---|---|---|---|---|---|---|
| ACC | PC | SN | SP | |||||
| Antczak and Liberadzki [18] | Random | Only real data | FNFT | 0.60 | 0.60 | 0.66 | 0.63 | 0.53 |
| Only artificial data | FNFT | 0.59 | 0.55 | 0.87 | 0.68 | 0.33 | ||
| Artificial + real data | FNFT | 0.89 | 0.87 | 0.90 | 0.89 | 0.88 | ||
| VGG16 [25] | Random | Only real data | FNFE | N/A | N/A | N/A | N/A | N/A |
| FNFT | 0.55 | 0.53 | 0.86 | 0.66 | 0.24 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.50 | 0.50 | 1.00 | 0.67 | 0.00 | |||
| Only artificial data | FNFE | N/A | N/A | N/A | N/A | N/A | ||
| FNFT | 0.52 | 0.53 | 0.40 | 0.45 | 0.65 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.51 | 0.52 | 0.41 | 0.46 | 0.61 | |||
| Artificial + real data | FNFE | N/A | N/A | N/A | N/A | N/A | ||
| FNFT | 0.54 | 0.55 | 0.46 | 0.50 | 0.61 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.50 | 0.52 | 0.25 | 0.34 | 0.76 | |||
| ImageNet | Only real data | FNFE | 0.87 | 0.85 | 0.90 | 0.88 | 0.84 | |
| FNFT | 0.93 | 0.91 | 0.95 | 0.93 | 0.90 | |||
| NCFE | 0.92 | 0.91 | 0.94 | 0.92 | 0.90 | |||
| NCFT | 0.92 | 0.90 | 0.95 | 0.92 | 0.89 | |||
| Only artificial data | FNFE | 0.70 | 0.69 | 0.71 | 0.70 | 0.68 | ||
| FNFT | 0.58 | 0.56 | 0.84 | 0.67 | 0.32 | |||
| NCFE | 0.53 | 0.52 | 0.73 | 0.61 | 0.32 | |||
| NCFT | 0.63 | 0.58 | 0.94 | 0.72 | 0.32 | |||
| Artificial + real data | FNFE | 0.87 | 0.85 | 0.90 | 0.88 | 0.84 | ||
| FNFT | 0.91 | 0.87 | 0.97 | 0.92 | 0.85 | |||
| NCFE | 0.89 | 0.86 | 0.94 | 0.89 | 0.84 | |||
| NCFT | 0.94 | 0.91 | 0.97 | 0.94 | 0.90 | |||
| ResNet50 [26] | Random | Only real data | FNFE | N/A | N/A | N/A | N/A | N/A |
| FNFT | 0.46 | 0.45 | 0.32 | 0.37 | 0.61 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.82 | 0.77 | 0.94 | 0.84 | 0.71 | |||
| Only artificial data | FNFE | N/A | N/A | N/A | N/A | N/A | ||
| FNFT | 0.52 | 0.52 | 0.63 | 0.57 | 0.40 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.48 | 0.49 | 0.70 | 0.58 | 0.26 | |||
| Artificial + real data | FNFE | N/A | N/A | N/A | N/A | N/A | ||
| FNFT | 0.86 | 0.83 | 0.90 | 0.86 | 0.81 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.83 | 0.84 | 0.83 | 0.83 | 0.84 | |||
| ImageNet | Only real data | FNFE | 0.50 | 0.00 | 0.00 | 0.00 | 1.00 | |
| FNFT | 0.79 | 0.77 | 0.84 | 0.80 | 0.74 | |||
| NCFE | 0.91 | 0.88 | 0.95 | 0.92 | 0.87 | |||
| NCFT | 0.91 | 0.88 | 0.95 | 0.92 | 0.87 | |||
| Only artificial data | FNFE | 0.52 | 0.56 | 0.22 | 0.32 | 0.82 | ||
| FNFT | 0.55 | 0.55 | 0.62 | 0.58 | 0.48 | |||
| NCFE | 0.66 | 0.69 | 0.57 | 0.63 | 0.74 | |||
| NCFT | 0.71 | 0.68 | 0.83 | 0.74 | 0.60 | |||
| Artificial + real data | FNFE | 0.50 | 0.00 | 0.00 | 0.00 | 1.00 | ||
| FNFT | 0.90 | 0.93 | 0.87 | 0.90 | 0.94 | |||
| NCFE | 0.75 | 0.92 | 0.56 | 0.69 | 0.95 | |||
| NCFT | 0.91 | 0.89 | 0.94 | 0.91 | 0.89 | |||
| Inception-v3 [21] | Random | Only real data | FNFE | N/A | N/A | N/A | N/A | N/A |
| FNFT | 0.51 | 1.00 | 0.03 | 0.06 | 1.00 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.71 | 0.70 | 0.75 | 0.72 | 0.68 | |||
| Only artificial data | FNFE | N/A | N/A | N/A | N/A | N/A | ||
| FNFT | 0.47 | 0.48 | 0.62 | 0.54 | 0.32 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.58 | 0.58 | 0.67 | 0.62 | 0.50 | |||
| Artificial + real data | FNFE | N/A | N/A | N/A | N/A | N/A | ||
| FNFT | 0.82 | 0.78 | 0.90 | 0.84 | 0.74 | |||
| NCFE | N/A | N/A | N/A | N/A | N/A | |||
| NCFT | 0.94 | 0.91 | 0.97 | 0.94 | 0.90 | |||
| ImageNet | Only real data | FNFE | 0.88 | 0.82 | 0.98 | 0.89 | 0.77 | |
| FNFT | 0.57 | 0.60 | 0.43 | 0.50 | 0.71 | |||
| NCFE | 0.95 | 0.93 | 0.98 | 0.95 | 0.92 | |||
| NCFT | 0.92 | 0.90 | 0.95 | 0.92 | 0.89 | |||
| Only artificial data | FNFE | 0.49 | 0.49 | 0.51 | 0.50 | 0.47 | ||
| FNFT | 0.38 | 0.41 | 0.49 | 0.45 | 0.27 | |||
| NCFE | 0.69 | 0.64 | 0.87 | 0.74 | 0.50 | |||
| NCFT | 0.63 | 0.58 | 0.94 | 0.72 | 0.32 | |||
| Artificial + real data | FNFE | 0.73 | 0.77 | 0.65 | 0.71 | 0.81 | ||
| FNFT | 0.88 | 0.90 | 0.86 | 0.88 | 0.90 | |||
| NCFE | 0.87 | 0.94 | 0.79 | 0.86 | 0.95 | |||
| NCFT | 0.94 | 0.95 | 0.92 | 0.94 | 0.95 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ovalle-Magallanes, E.; Avina-Cervantes, J.G.; Cruz-Aceves, I.; Ruiz-Pinales, J. Transfer Learning for Stenosis Detection in X-ray Coronary Angiography. Mathematics 2020, 8, 1510. https://doi.org/10.3390/math8091510
Ovalle-Magallanes E, Avina-Cervantes JG, Cruz-Aceves I, Ruiz-Pinales J. Transfer Learning for Stenosis Detection in X-ray Coronary Angiography. Mathematics. 2020; 8(9):1510. https://doi.org/10.3390/math8091510
Chicago/Turabian StyleOvalle-Magallanes, Emmanuel, Juan Gabriel Avina-Cervantes, Ivan Cruz-Aceves, and Jose Ruiz-Pinales. 2020. "Transfer Learning for Stenosis Detection in X-ray Coronary Angiography" Mathematics 8, no. 9: 1510. https://doi.org/10.3390/math8091510
APA StyleOvalle-Magallanes, E., Avina-Cervantes, J. G., Cruz-Aceves, I., & Ruiz-Pinales, J. (2020). Transfer Learning for Stenosis Detection in X-ray Coronary Angiography. Mathematics, 8(9), 1510. https://doi.org/10.3390/math8091510






