Deep Neural Network for Predicting Diabetic Retinopathy from Risk Factors
Abstract
1. Introduction
2. Literature Review
2.1. Diabetic Retinopathy Prediction
2.2. Deep Neural Network
2.3. Recursive Feature Elimination
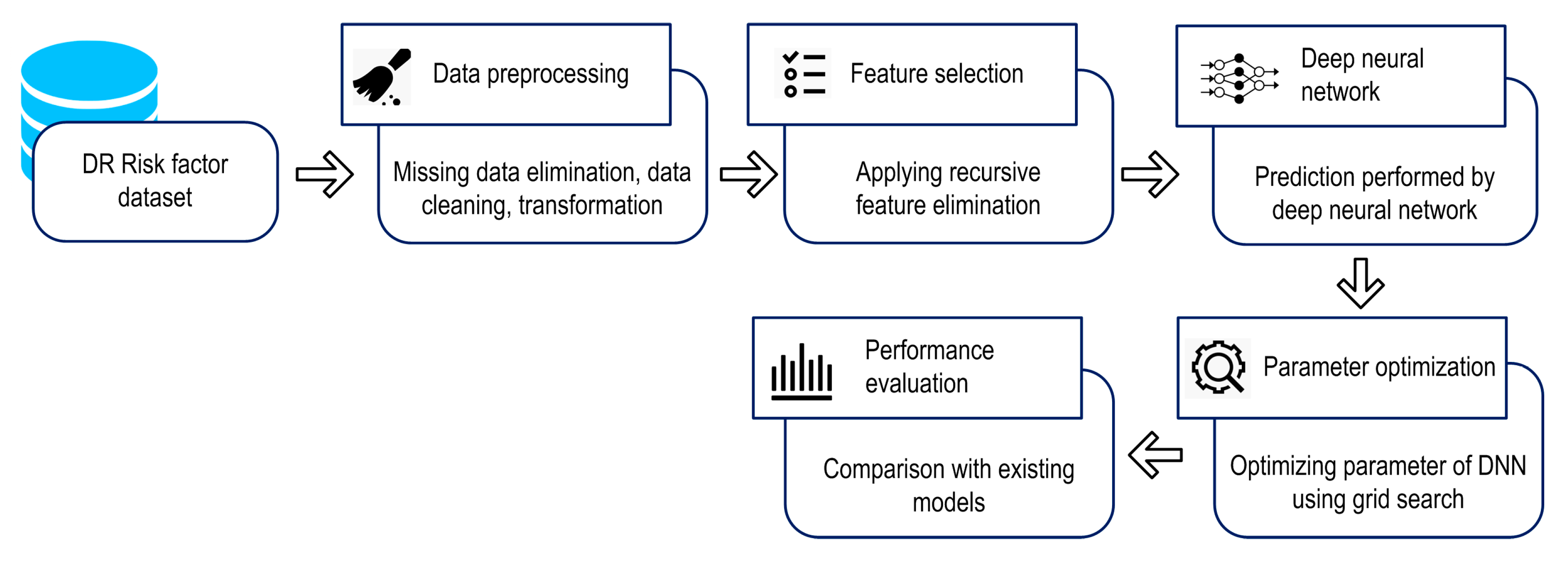
3. Methodology
3.1. Datasets
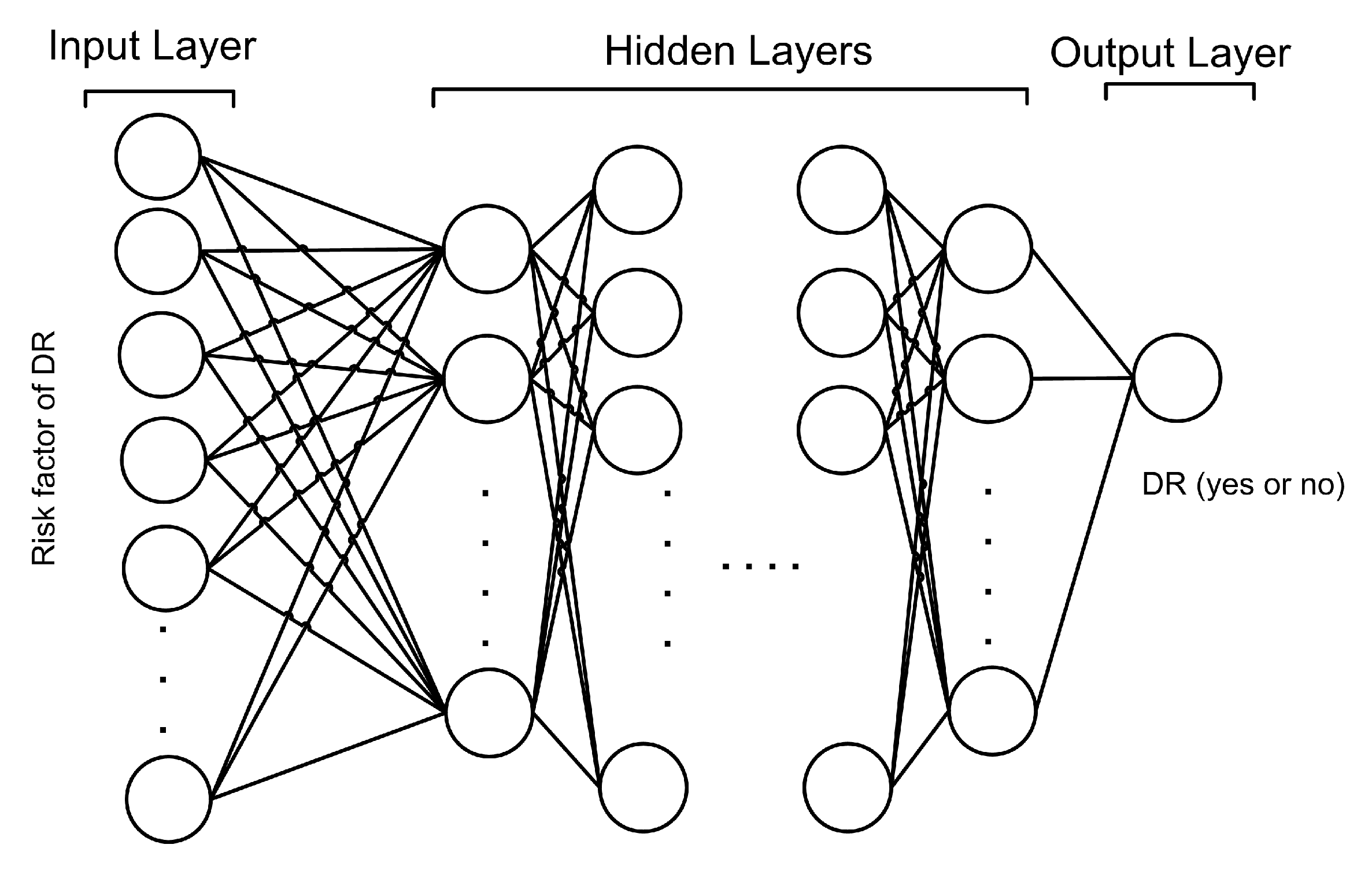
3.2. Design of Proposed Model
3.3. Recursive Feature Elimination (RFE)
| Algorithm 1. SVM-RFE pseudocode | |
| Input: | |
| Output:r | |
| whiles is not empty do | |
| end while | |
| returnr | |
3.4. Proposed Deep Neural Network
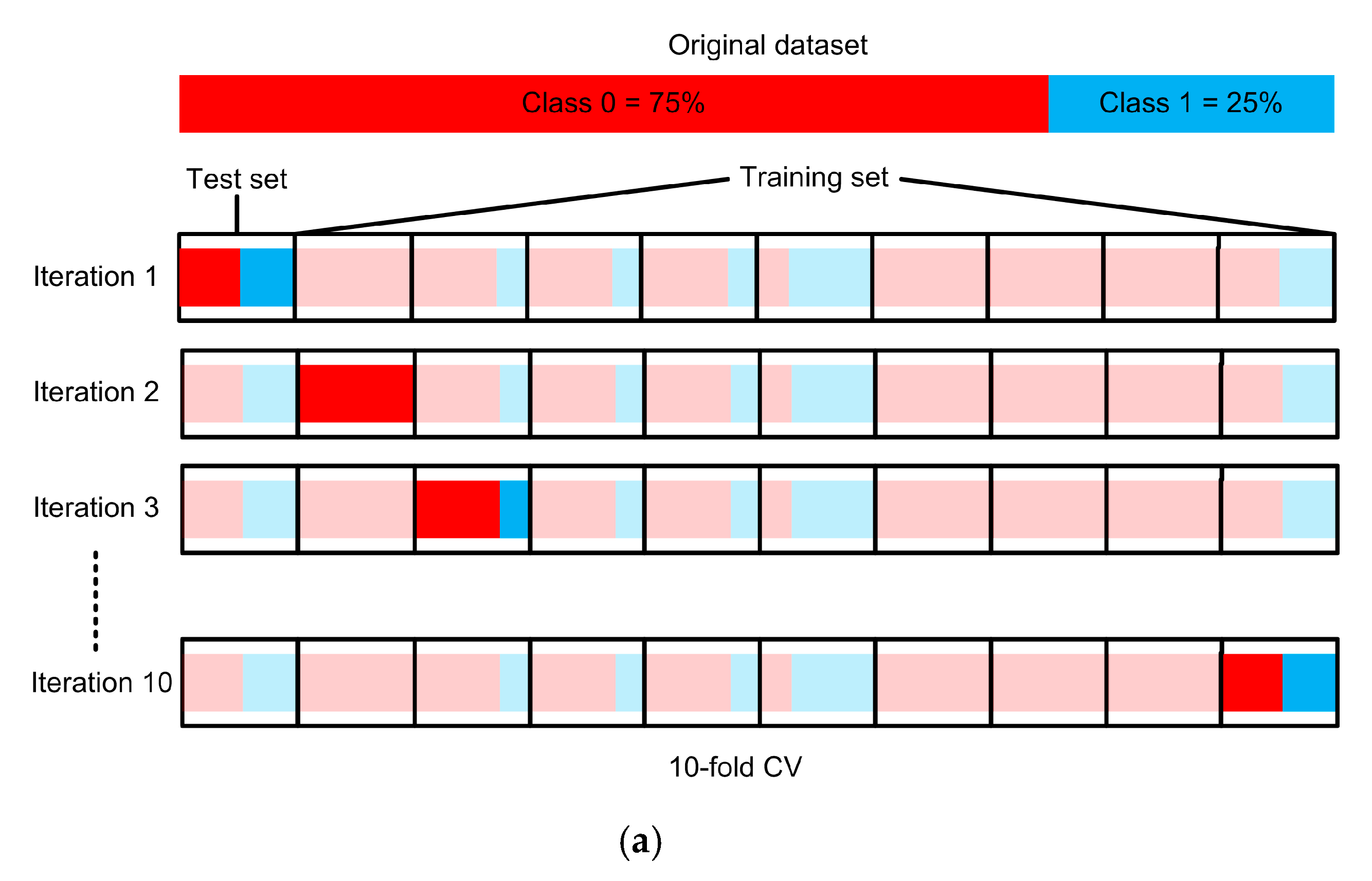
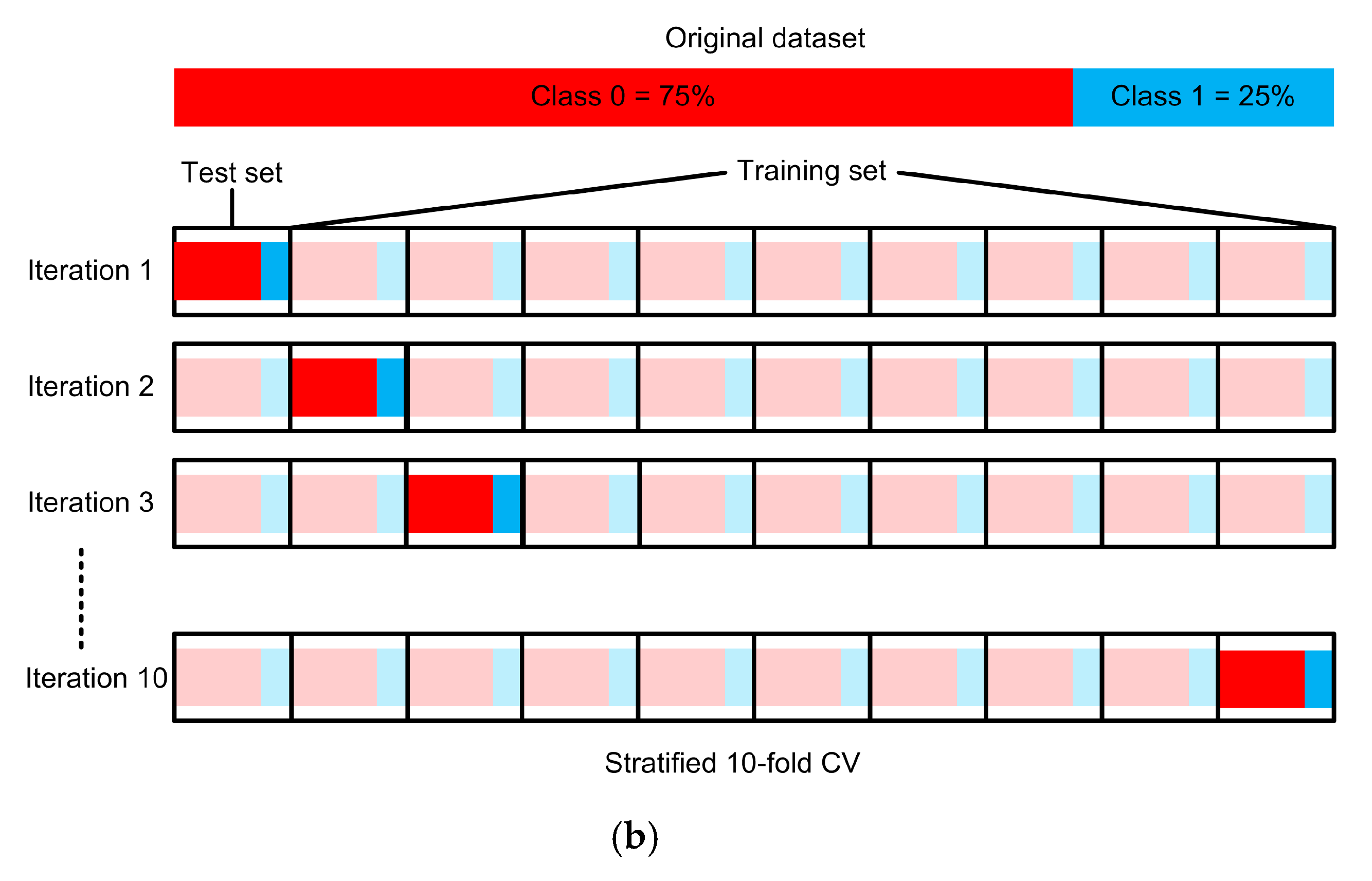
3.5. Experimental Setup
4. Results and Discussion
4.1. Prediction Model Performances
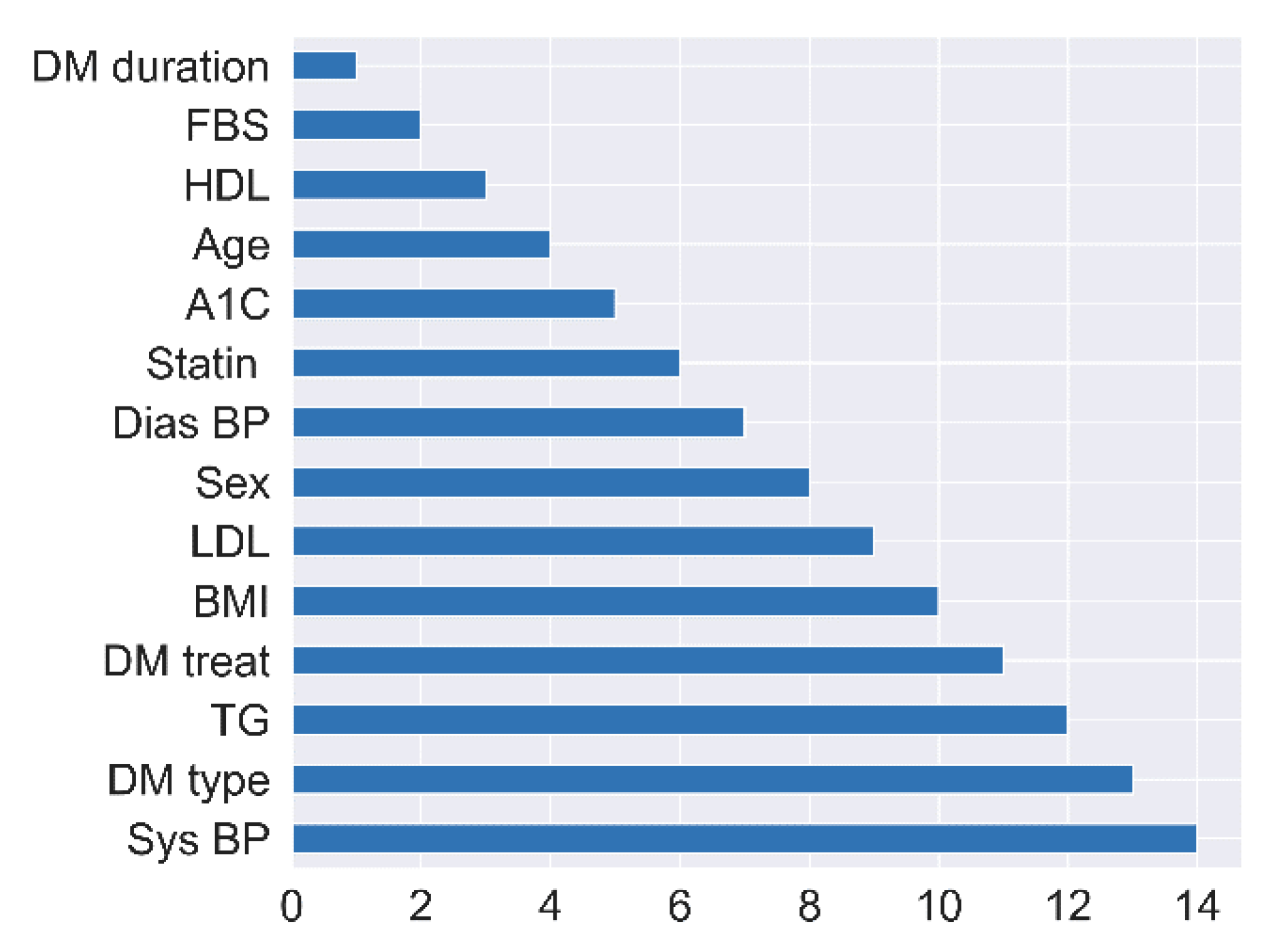
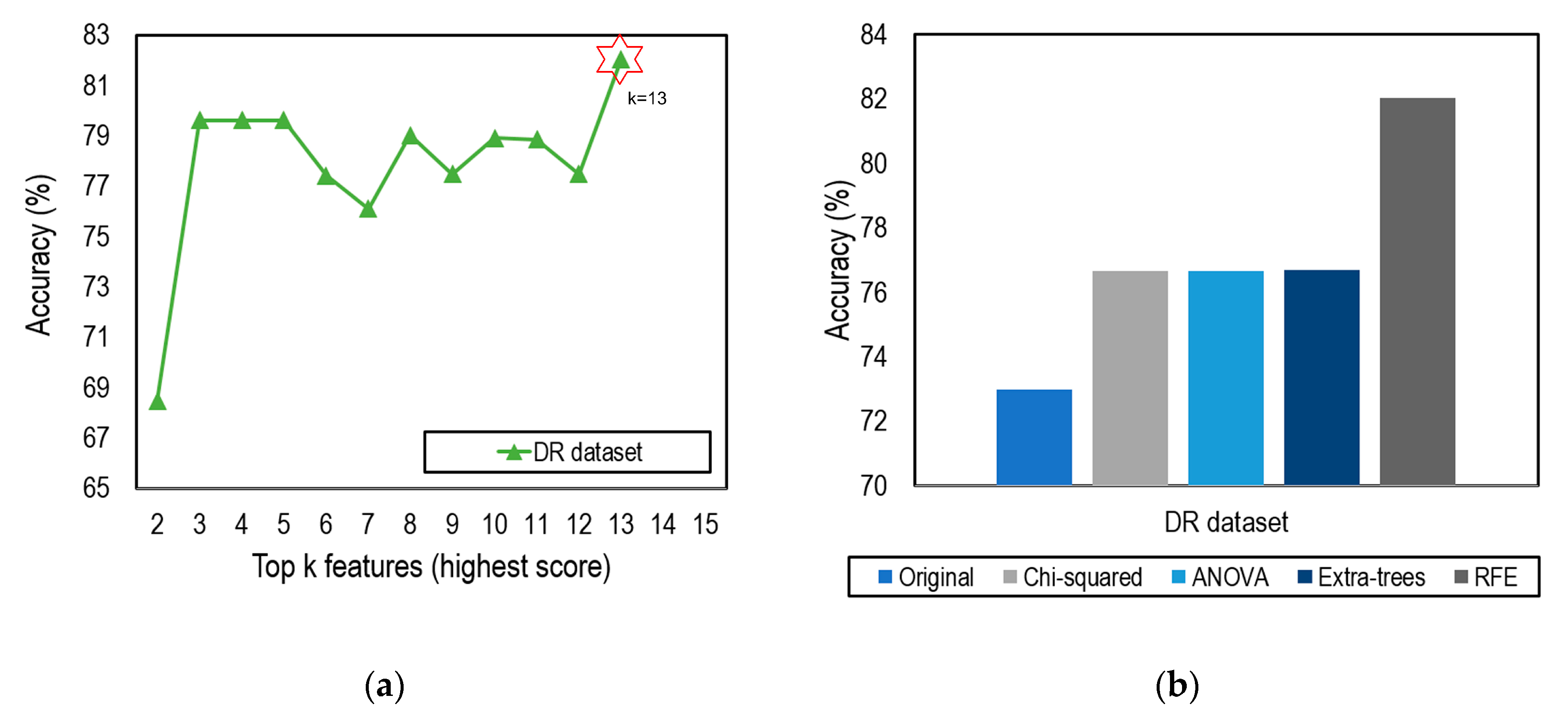
4.2. Feature Selection Impacts
4.3. Risk Factors and Previous Studies
4.4. Another Diabetes Dataset
5. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ANN | Artificial Neural Network |
| ANOVA | Analysis of Variance |
| AUC | Area under the ROC Curve |
| BG | Blood Glucose |
| BMI | Body Mass Index |
| CV | Cross Validation |
| CVD | Cardiovascular Disease |
| DBP | Diastolic Blood Pressure |
| Dias BP | Diastolic Blood Pressure |
| DM | Diabetes Mellitus |
| DN | Diabetic Nephropathy |
| DNN | Deep Neural Network |
| DR | Diabetic Retinopathy |
| DT | Decision Tree |
| FBS | Fasting Blood Sugar |
| GWO | Grey Wolf Optimization |
| HbA1c | Hemoglobin A1c |
| HDL | High-density Lipoproteins |
| KNN | K-Nearest Neighbor |
| LBFGS | Limited-memory Broyden–Fletcher–Goldfarb–Shanno |
| LDL | Low-density Lipoprotein |
| LR | Logistic Regression |
| NB | Naïve Bayes |
| NHISS | National Health Insurance Sharing Service |
| PCA | Principal Components Analysis |
| PVD | Peripheral Vessel Disease |
| ReLU | Rectified Linear Units |
| RF | Random Forest |
| RFE | Recursive Feature Elimination |
| ROC | Receiver Operating Characteristic |
| SBP | Systolic Blood Pressure |
| SGD | Stochastic Gradient Descent |
| SMOTE | Synthetic Minority Over-sampling Technique |
| SVM | Support Vector Machine |
| SVM-RFE | Support Vector Machine–Recursive Feature Elimination |
| Sys BP | Systolic Blood Pressure |
| T1D | Type 1 Diabetes |
| T2D | Type 2 Diabetes |
| TG | Triglyceride |
Appendix A
| No | Attribute | Description | Type | Range |
|---|---|---|---|---|
| 1 | BMI | Subject’s body mass index | Numeric | 18–41 |
| 2 | DM duration | Subject’s diabetes duration (y) | Numeric | 0–30 |
| 3 | A1c | Subject’s average blood glucose level over the past 3 months (mg/dL) | Numeric | 6.5–13.3 |
| 4 | Age | Subject’s age (y) | Numeric | 16–79 |
| 5 | FBS | Subject’s fasting blood sugar level (mg/dL) | Numeric | 80–510 |
| 6 | LDL | Subject’s low-density lipoprotein level (mg/dL) | Numeric | 36–267 |
| 7 | HDL | Subject’s high-density lipoprotein level (mg/dL) | Numeric | 20–62 |
| 8 | TG | Subject’s triglyceride level (mg/dL) | Numeric | 74–756 |
| 9 | Sys BP | Subject’s systolic blood pressure (mmHg) | Numeric | 105–180 |
| 10 | Dias BP | Subject’s diastolic blood pressure (mmHg) | Numeric | 60–120 |
| 11 | Sex | Subject’s sex | Categorical | 0 = Female 1 = Male |
| 12 | DM type | Subject’s diabetes type | Categorical | 0 = T1D 1 = T2D |
| 13 | DM treat | Subject’s diabetes treatment | Categorical | 0 = Both (Insulin and oral agent) 1 = Insulin 2 = Oral agent |
| 14 | Statin | Subject’s statin status (frequently used as part of diabetes care) | Categorical | 0 = Ator (atorvastatin) 1 = No statin 2 = ROS (rosuvastatin) |
| 15 | Nephropathy (class) | Subject’s nephropathy status | Categorical | 0 = No (60) 1 = Yes (73) |
| No | Attribute | Description | Type | Range |
|---|---|---|---|---|
| 1 | BTH_G | Age group of a subject | Categorical | 0 = 20–24 1 = 25–26 2 = 27–28 … 26 = greater than 75 |
| 2 | SBP | Subject’s systolic blood pressure (mmHg) | Numeric | 84–190 |
| 3 | DBP | Subject’s diastolic blood pressure (mmHg) | Numeric | 50–120 |
| 4 | BMI | Subject’s body mass index | Numeric | 15.6–39.9 |
| 5 | SEX | Subject’s sex | Categorical | 0 = Male 1 = Female |
| 6 | DIS (class) | Subject’s disease (hypertension, diabetes) status | Categorical | 0 = No (761) 1 = Yes (239) |
References
- American Diabetes Association. Standards of medical care in diabetes. Diabetes Care 2006, 29 (Suppl. 1), S4–S42. [Google Scholar]
- American Diabetes Association. Introduction: Standards of Medical Care in Diabetes. Diabetes Care 2018, 41, S1–S2. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, J.; Klingensmith, G.; Copeland, K.; Plotnick, L.; Kaufman, F.; Laffel, L.; Deeb, L.; Grey, M.; Anderson, B.; Holzmeister, L.A.; et al. Care of Children and Adolescents with Type 1 Diabetes: A statement of the American Diabetes Association. Diabetes Care 2005, 28, 186–212. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.E.; Little, R.R.; Lorenz, R.A.; Malone, J.I.; Nathan, D.; Peterson, C.M.; Sacks, D.B. Tests of Glycemia in Diabetes. Diabetes Care 2004, 27, 1761–1773. [Google Scholar] [CrossRef] [PubMed]
- Klein, R. Relationship of Hyperglycemia to the Long-term Incidence and Progression of Diabetic Retinopathy. Arch. Intern. Med. 1994, 154, 2169. [Google Scholar] [CrossRef]
- Mohamed, S.; Murray, J.C.; Dagle, J.M.; Colaizy, T. Hyperglycemia as a risk factor for the development of retinopathy of prematurity. BMC Pediatr. 2013, 13, 78. [Google Scholar] [CrossRef]
- Schrijvers, B.F.; De Vriese, A.S.; Flyvbjerg, A. From Hyperglycemia to Diabetic Kidney Disease: The Role of Metabolic, Hemodynamic, Intracellular Factors and Growth Factors/Cytokines. Endocr. Rev. 2004, 25, 971–1010. [Google Scholar] [CrossRef]
- Alicic, R.Z.; Rooney, M.T.; Tuttle, K.R. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin. J. Am. Soc. Nephrol. 2017, 12, 2032–2045. [Google Scholar] [CrossRef]
- Selvin, E.; Marinopoulos, S.; Berkenblit, G.; Rami, T.; Brancati, F.L.; Powe, N.R.; Golden, S.H. Meta-Analysis: Glycosylated Hemoglobin and Cardiovascular Disease in Diabetes Mellitus. Ann. Intern. Med. 2004, 141, 421. [Google Scholar] [CrossRef]
- Ormazabal, V.; Nair, S.; Elfeky, O.; Aguayo, C.; Salomon, C.; Zuñiga, F.A. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 2018, 17, 122. [Google Scholar] [CrossRef]
- Cheung, N.; Mitchell, P.; Wong, T.Y. Diabetic retinopathy. Lancet 2010, 376, 124–136. [Google Scholar] [CrossRef]
- Golubnitschaja, O. Advanced Diabetes Care: Three Levels of Prediction, Prevention & Personalized Treatment. Curr. Diabetes Rev. 2010, 6, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.M.; Maracy, M.R.; Amini, M.; Baradaran, H.R. A risk score development for diabetic retinopathy screening in Isfahan-Iran. J. Res. Med. Sci. 2009, 14, 105–110. [Google Scholar] [PubMed]
- Oh, E.; Yoo, T.K.; Park, E.-C. Diabetic retinopathy risk prediction for fundus examination using sparse learning: A cross-sectional study. BMC Med. Inform. Decis. Mak. 2013, 13, 106. [Google Scholar] [CrossRef]
- Ogunyemi, O.; Kermah, D. Machine Learning Approaches for Detecting Diabetic Retinopathy from Clinical and Public Health Records. AMIA Annu. Symp. Proc. 2015, 2015, 983–990. [Google Scholar]
- Ogunyemi, O.I.; Gandhi, M.; Tayek, C. Predictive Models for Diabetic Retinopathy from Non-Image Teleretinal Screening Data. AMIA Jt. Summits Transl. Sci. Proc. 2019, 2019, 472–477. [Google Scholar]
- Tsao, H.-Y.; Chan, P.-Y.; Su, E.C.-Y. Predicting diabetic retinopathy and identifying interpretable biomedical features using machine learning algorithms. BMC Bioinform. 2018, 19, 283. [Google Scholar] [CrossRef]
- Mallick, P.K.; Ryu, S.H.; Satapathy, S.K.; Mishra, S.; Nguyen, G.N.; Tiwari, P. Brain MRI Image Classification for Cancer Detection Using Deep Wavelet Autoencoder-Based Deep Neural Network. IEEE Access 2019, 7, 46278–46287. [Google Scholar] [CrossRef]
- Akyol, K. Comparing of deep neural networks and extreme learning machines based on growing and pruning approach. Expert Syst. Appl. 2020, 140, 112875. [Google Scholar] [CrossRef]
- De Falco, I.; De Pietro, G.; Della Cioppa, A.; Sannino, G.; Scafuri, U.; Tarantino, E. Evolution-based configuration optimization of a Deep Neural Network for the classification of Obstructive Sleep Apnea episodes. Future Gener. Comput. Syst. 2019, 98, 377–391. [Google Scholar] [CrossRef]
- Koshimizu, H.; Kojima, R.; Kario, K.; Okuno, Y. Prediction of blood pressure variability using deep neural networks. Int. J. Med. Inform. 2020, 136, 104067. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.; Kwak, M.J.; Bajaj, M. Genetic prediction of type 2 diabetes using deep neural network. Clin. Genet. 2018, 93, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Kannadasan, K.; Edla, D.R.; Kuppili, V. Type 2 diabetes data classification using stacked autoencoders in deep neural networks. Clin. Epidemiol. Glob. Health 2019, 7, 530–535. [Google Scholar] [CrossRef]
- Ramachandran, N.; Hong, S.C.; Sime, M.J.; Wilson, G.A. Diabetic retinopathy screening using deep neural network: Diabetic retinopathy screening. Clin. Exp. Ophthalmol. 2018, 46, 412–416. [Google Scholar] [CrossRef]
- Gao, Z.; Li, J.; Guo, J.; Chen, Y.; Yi, Z.; Zhong, J. Diagnosis of Diabetic Retinopathy Using Deep Neural Networks. IEEE Access 2019, 7, 3360–3370. [Google Scholar] [CrossRef]
- Parmar, R.; Lakshmanan, R.; Purushotham, S.; Soundrapandiyan, R. Detecting Diabetic Retinopathy from Retinal Images Using CUDA Deep Neural Network. In Intelligent Pervasive Computing Systems for Smarter Healthcare; Sangaiah, A.K., Shantharajah, S., Theagarajan, P., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 379–396. ISBN 978-1-119-43896-0. [Google Scholar]
- Shankar, K.; Perumal, E.; Vidhyavathi, R.M. Deep neural network with moth search optimization algorithm based detection and classification of diabetic retinopathy images. SN Appl. Sci. 2020, 2, 748. [Google Scholar] [CrossRef]
- Ayhan, M.S.; Kühlewein, L.; Aliyeva, G.; Inhoffen, W.; Ziemssen, F.; Berens, P. Expert-validated estimation of diagnostic uncertainty for deep neural networks in diabetic retinopathy detection. Med. Image Anal. 2020, 64, 101724. [Google Scholar] [CrossRef]
- Gadekallu, T.R.; Khare, N.; Bhattacharya, S.; Singh, S.; Maddikunta, P.K.R.; Srivastava, G. Deep neural networks to predict diabetic retinopathy. J. Ambient Intell. Human Comput. 2020. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Gysels, E.; Renevey, P.; Celka, P. SVM-based recursive feature elimination to compare phase synchronization computed from broadband and narrowband EEG signals in Brain–Computer Interfaces. Signal Process. 2005, 85, 2178–2189. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, J. Operator functional state classification using least-square support vector machine based recursive feature elimination technique. Comput. Methods Programs Biomed. 2014, 113, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Senturk, Z.K. Early diagnosis of Parkinson’s disease using machine learning algorithms. Med. Hypotheses 2020, 138, 109603. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Dey, D.; Munshi, S. Integration of morphological preprocessing and fractal based feature extraction with recursive feature elimination for skin lesion types classification. Comput. Methods Programs Biomed. 2019, 178, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xiao, Z.; Wang, B.; Wu, J. Identification of Autism Based on SVM-RFE and Stacked Sparse Auto-Encoder. IEEE Access 2019, 7, 118030–118036. [Google Scholar] [CrossRef]
- Richhariya, B.; Tanveer, M.; Rashid, A.H. Diagnosis of Alzheimer’s disease using universum support vector machine based recursive feature elimination (USVM-RFE). Biomed. Signal Process. Control 2020, 59, 101903. [Google Scholar] [CrossRef]
- Kumar, A.; Sharmila, D.J.S.; Singh, S. SVMRFE based approach for prediction of most discriminatory gene target for type II diabetes. Genom. Data 2017, 12, 28–37. [Google Scholar] [CrossRef]
- Chen, Z.; Pang, M.; Zhao, Z.; Li, S.; Miao, R.; Zhang, Y.; Feng, X.; Feng, X.; Zhang, Y.; Duan, M.; et al. Feature selection may improve deep neural networks for the bioinformatics problems. Bioinformatics 2020, 36, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Karthik, S.; Srinivasa Perumal, R.; Chandra Mouli, P.V.S.S.R. Breast Cancer Classification Using Deep Neural Networks. In Knowledge Computing and Its Applications; Margret Anouncia, S., Wiil, U.K., Eds.; Springer: Singapore, 2018; pp. 227–241. ISBN 978-981-10-6679-5. [Google Scholar]
- Khodadadi, B.; Mousavi, N.; Mousavi, M.; Baharvand, P.; Ahmadi, S.A.Y. Diagnosis and predictive clinical and para-clinical cutoffs for diabetes complications in Lur and Lak populations of Iran; a ROC curve analysis to design a regional guideline. J. Nephropharmacol. 2018, 7, 83–89. [Google Scholar] [CrossRef]
- Kohavi, R. A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence—Volume 2, Montreal, QC, Canada, 20–25 August 1995; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1995; pp. 1137–1143. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Jiménez, Á.B.; Lázaro, J.L.; Dorronsoro, J.R. Finding Optimal Model Parameters by Discrete Grid Search. In Innovations in Hybrid Intelligent Systems. Advances in Soft Computing; Corchado, E., Corchado, J.M., Abraham, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 44, pp. 120–127. ISBN 978-3-540-74971-4. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kim, H.-E.; Kim, H.H.; Han, B.-K.; Kim, K.H.; Han, K.; Nam, H.; Lee, E.H.; Kim, E.-K. Changes in cancer detection and false-positive recall in mammography using artificial intelligence: A retrospective, multireader study. Lancet Digit. Health 2020, 2, e138–e148. [Google Scholar] [CrossRef]
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef]
- Huang, J.; Ling, C.X. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans. Knowl. Data Eng. 2005, 17, 299–310. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An Introduction to Feature Extraction. In Feature Extraction. Studies in Fuzziness and Soft Computing; Guyon, I., Nikravesh, M., Gunn, S., Zadeh, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 207, pp. 1–25. ISBN 978-3-540-35487-1. [Google Scholar]
- Miao, J.; Niu, L. A Survey on Feature Selection. Procedia Comput. Sci. 2016, 91, 919–926. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- National Health Insurance Sharing Service (NHISS) Korea. Available online: https://nhiss.nhis.or.kr/bd/ab/bdabf003cv.do (accessed on 20 August 2020).

| No | Attribute | Description | Type | Range |
|---|---|---|---|---|
| 1 | BMI | Subject’s body mass index | Numeric | 18–41 |
| 2 | DM duration | Subject’s diabetes duration (y) | Numeric | 0–30 |
| 3 | A1c | Subject’s average blood glucose level over the past 3 months (mg/dL) | Numeric | 6.5–13.3 |
| 4 | Age | Subject’s age (y) | Numeric | 16–79 |
| 5 | FBS | Subject’s fasting blood sugar level (mg/dL) | Numeric | 80–510 |
| 6 | LDL | Subject’s low-density lipoprotein level (mg/dL) | Numeric | 36–267 |
| 7 | HDL | Subject’s high-density lipoprotein level (mg/dL) | Numeric | 20–62 |
| 8 | TG | Subject’s triglyceride level (mg/dL) | Numeric | 74–756 |
| 9 | Sys BP | Subject’s systolic blood pressure (mmHg) | Numeric | 105–180 |
| 10 | Dias BP | Subject’s diastolic blood pressure (mmHg) | Numeric | 60–120 |
| 11 | Sex | Subject’s sex | Categorical | 0 = Female 1 = Male |
| 12 | DM type | Subject’s diabetes type (T1D or T2D) | Categorical | 0 = T1D 1 = T2D |
| 13 | DM treat | Subject’s diabetes treatment | Categorical | 0 = Both (Insulin and oral agent) 1 = Insulin 2 = Oral agent |
| 14 | Statin | Subject’s statin status (frequently used as part of diabetes care) | Categorical | 0 = Ator (atorvastatin) 1 = No statin 2 = ROS (rosuvastatin) |
| 15 | Retinopathy (class) | Subject’s retinopathy status | Categorical | 0 = No (91) 1 = Yes (42) |
| No | Attribute | Feature Selection Model | |||
|---|---|---|---|---|---|
| RFE (Rank) | Chi-Squared (Score) | ANOVA (F-Value) | Extra Trees (Gini Importance) | ||
| 1 | BMI | 10 | 0.225 | 2.352 | 0.056 |
| 2 | DM duration | 1 | 5.474 | 49.028 | 0.161 |
| 3 | A1c | 5 | 1.054 | 4.780 | 0.081 |
| 4 | Age | 4 | 1.352 | 24.473 | 0.098 |
| 5 | FBS | 2 | 0.970 | 12.349 | 0.088 |
| 6 | LDL | 9 | 0.520 | 6.207 | 0.059 |
| 7 | HDL | 3 | 0.571 | 8.726 | 0.077 |
| 8 | TG | 12 | 0.643 | 6.419 | 0.077 |
| 9 | Sys BP | 14 | 1.992 | 18.519 | 0.083 |
| 10 | Dias BP | 7 | 1.734 | 18.738 | 0.070 |
| 11 | Sex | 8 | 2.669 | 4.641 | 0.045 |
| 12 | DM type | 13 | 0.127 | 1.870 | 0.009 |
| 13 | DM treat | 11 | 0.779 | 2.889 | 0.061 |
| 14 | Statin | 6 | 0.064 | 0.176 | 0.031 |
| Hyperparameter | Optimized Value |
|---|---|
| Hidden layer size | 100, 64, 128, 64, 32 |
| Activation function | ReLU |
| Alpha | 0.0001 |
| Initial learning rate | 0.01 |
| Maximum iteration | 500 |
| Optimization algorithm | SGD |
| Method | Accuracy | Precision | Sensitivity (Recall) | Specificity | F1 | AUC |
|---|---|---|---|---|---|---|
| KNN | 77.418 | 79.714 | 49.500 | 69.806 | 56.492 | 0.698 |
| DT | 75.989 | 63.095 | 65.000 | 73.222 | 59.558 | 0.732 |
| SVM | 78.846 | 75.214 | 51.500 | 71.361 | 56.333 | 0.714 |
| NB | 73.022 | 54.970 | 66.000 | 70.889 | 56.939 | 0.709 |
| RF | 78.352 | 62.500 | 39.000 | 67.889 | 45.944 | 0.679 |
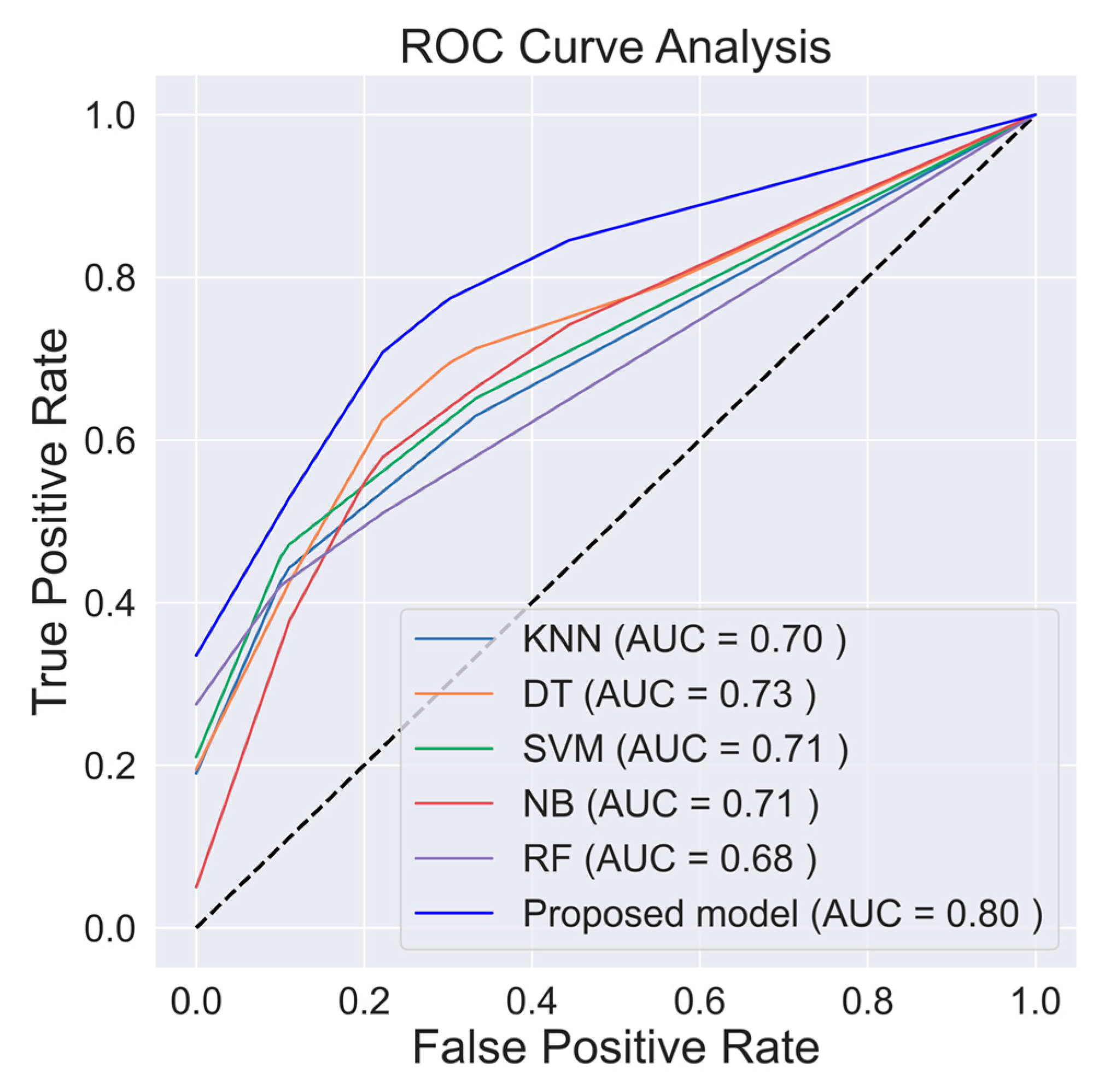
| Proposed model (DNN + RFE) | 82.033 | 72.937 | 76.000 | 80.389 | 71.820 | 0.804 |
| Dataset | Population | Study | Method | Number of Features | Model Validation | Accuracy | AUC |
|---|---|---|---|---|---|---|---|
| DR | Iran | [13] | LR | 9 | - | - | 0.704 |
| South Korea | [14] | Lasso | 19 | Holdout (67:33) | 0.736 | 0.810 | |
| United States | [15] | Ensemble RUSBoost | 11 | Holdout (80:20) | 0.735 | 0.720 | |
| Taiwan | [17] | SVM | 10 | Holdout (80:20) | 0.795 | 0.839 | |
| United States | [16] | ANN + SMOTE | 8 | Holdout (66:34) | - | 0.754 | |
| Iran | Current | DNN + RFE | 13 | Stratified 10-fold CV | 0.820 | 0.804 |
| Method | DN | NHISS | ||
|---|---|---|---|---|
| Accuracy | AUC | Accuracy | AUC | |
| KNN | 81.813 | 0.814 | 80.600 | 0.690 |
| DT | 81.978 | 0.817 | 80.900 | 0.679 |
| SVM | 83.297 | 0.834 | 80.700 | 0.665 |
| NB | 67.527 | 0.649 | 79.600 | 0.701 |
| RF | 82.747 | 0.825 | 80.775 | 0.664 |
| Proposed DNN + RFE | 84.121 | 0.839 | 81.600 | 0.702 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfian, G.; Syafrudin, M.; Fitriyani, N.L.; Anshari, M.; Stasa, P.; Svub, J.; Rhee, J. Deep Neural Network for Predicting Diabetic Retinopathy from Risk Factors. Mathematics 2020, 8, 1620. https://doi.org/10.3390/math8091620
Alfian G, Syafrudin M, Fitriyani NL, Anshari M, Stasa P, Svub J, Rhee J. Deep Neural Network for Predicting Diabetic Retinopathy from Risk Factors. Mathematics. 2020; 8(9):1620. https://doi.org/10.3390/math8091620
Chicago/Turabian StyleAlfian, Ganjar, Muhammad Syafrudin, Norma Latif Fitriyani, Muhammad Anshari, Pavel Stasa, Jiri Svub, and Jongtae Rhee. 2020. "Deep Neural Network for Predicting Diabetic Retinopathy from Risk Factors" Mathematics 8, no. 9: 1620. https://doi.org/10.3390/math8091620
APA StyleAlfian, G., Syafrudin, M., Fitriyani, N. L., Anshari, M., Stasa, P., Svub, J., & Rhee, J. (2020). Deep Neural Network for Predicting Diabetic Retinopathy from Risk Factors. Mathematics, 8(9), 1620. https://doi.org/10.3390/math8091620








