Abstract
Image denoising performs a prominent role in medical image analysis. In many cases, it can drastically accelerate the diagnostic process by enhancing the perceptual quality of noisy image samples. However, despite the extensive practicability of medical image denoising, the existing denoising methods illustrate deficiencies in addressing the diverse range of noise appears in the multidisciplinary medical images. This study alleviates such challenging denoising task by learning residual noise from a substantial extent of data samples. Additionally, the proposed method accelerates the learning process by introducing a novel deep network, where the network architecture exploits the feature correlation known as the attention mechanism and combines it with spatially refine residual features. The experimental results illustrate that the proposed method can outperform the existing works by a substantial margin in both quantitative and qualitative comparisons. Also, the proposed method can handle real-world image noise and can improve the performance of different medical image analysis tasks without producing any visually disturbing artefacts.
1. Introduction
Medical image denoising (MID) perceive as a process of improving the perceptual quality of degraded noisy images captured with specialized medical image acquisition devices. Regrettably, such imaging devices are susceptible to capture noise despite the altitude in imaging technologies [1]. However, the presence of noise in the images has a starling impact on medical image analysis as well as can convolute the decision making maneuver of an expert [1,2,3]. Hence, denoising has considered a classical yet strenuous medical image analysis task.
Typically, the MID applications examine image noise in the form of Gaussian distribution [2]. Which essentially depends on capturing conditions as well as the hardware configuration of the capturing devices [4]. Therefore, such sensor noise in MID remains blind-fold and substantially varies depending on image retrieval techniques (i.e., images capture with radiological devices employs a distinct noise factor comparing to microscopic modalities) [5]. In contrast, medical image analysis has to leverage multidisciplinary modalities in the visualization process of biological molecules for treatment purposes [6]. As a consequence, a large and diverse scale of MID is a relatively challenging task than a conventional image denoising method.
In the recent past, a substantial push has obtained in MID by introducing novel approaches such as non-local self-similarity (NSS) [7], sparse coding [8], filter-based methods [9,10,11], etc. Also, considering the massive success in various vision tasks, many recent studies [12,13,14,15] have appropriated deep learning as a viable alternative to the aforementioned MID methods. However, most of these recent studies have focused on an inadequate range of noise deviations as well as a narrow range of data diversity rather than striving to generalize their methods for multidisciplinary modalities. As a consequence, existing MID methods illustrate deficiencies in large-scale noise removal from medical images and immensely fail in numerous cases, as shown in Figure 1.
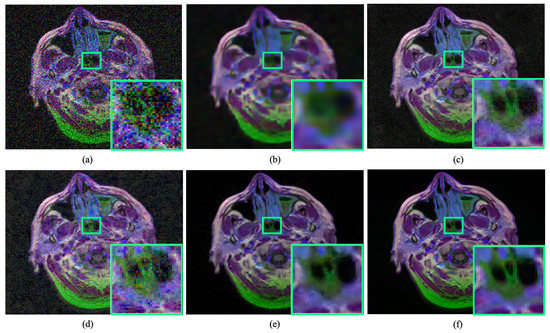
Figure 1.
Performance of existing medical image denoising methods in removing image noise. The existing denoising methods immensely failed in addressing a substantial amount of noise removal and susceptible to produce artefacts. (a) Noisy input. (b) Result obtained by BM3D [11]. (c) Result obtained by DnCNN [16]. (d) Result obtained by Residual MID [12]. (e) Result obtained by DRAN (proposed). (f) Reference sharp image. Source by: (https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation).
To alleviate the deficiencies of existing works, this study proposes a novel denoising method to learn the blind-fold residual noise from a convex set of medical images. Additionally, This study introduces a deep network for MID applications, which utilizes the feature correlation known as the attention mechanism [17,18,19,20] and combines it with refined residual learning [21,22] to illustrate supremacy over existing methods. Here, the attention mechanism leverage in such a manner that it can utilize the depth-wise feature correlation to aggregate a dynamic kernel throughout the convolution operation [23]. Also, this study proposes to refine the residual learning by using a spatial gating mechanism denoted as noise gate [19,20], which learns to control the low-level features propagation towards the top layers. To the best concern, this is the first work in the open literature, which comprehensively combines the feature correlation and refine the residual feature propagation, particularly for the MID applications. This study denotes the proposed deep model as a dynamic residual attention network (DRAN) in the rest of the sections. The feasibility of the proposed method has verified with real-world noisy medical images and fusing it with different medical image analysis tasks. The main contribution of the proposed method has summarized as follows:
- Large-scale denoising: Introduces a novel method for learning multidisciplinary medical image denoising through a single deep network. Thus, large-scale noise can be handle without illustrating any artefacts.
- Dynamic residual attention network: Proposes a novel deep network that combines feature correlation and refines the residual feature propagation for MID. The model intended to accelerate the denoising performance in a diverse dataset by recovering details. Code available: https://github.com/sharif-apu/MID-DRAN
- Dense experiments: Conducts dense experiments with a substantial amount of data samples. Therefore, the feasibility of the proposed method can identify in a diverse range of noisy images.
- Real-world applications: Illustrates the denoising performance on noisy medical images, which are collected by employing actual hardware. Also, the proposed method has combined with different medical image analysis tasks to reveal the practicability in real-world applications.
2. Related Works
This section briefly reviews the works, which are related to the proposed method.
2.1. Medical Image Denoising
A substantial amount of novel methods have been proposed in the recent past. According to their optimization strategies, the MID works can be divided into two major categories: (i) Classical approaches and (ii) Learning-based approaches.
Classical approaches: The filter-based denoising method has dominated the classical medical image denoising for a long while. Most of the recent filter-based works focused on developing a low-pass filter, where the filter aims to eliminates precipitate peaks from a noisy image based on the local estimation [1]. Amongst the numerous variants of filter-based approaches, Gaussian averaging filters [24], median filters [25], mean filters [26], diffusion filters [27] are used widely for removing noise from specific types of medical imaging modalities such as ultrasound images, magnetic resonance images (MRI), computed tomography (CT) images, etc [25,26,27,28]. Despite the widespread usage of such filter-based techniques, such methods tend to smooth the given images while removing the noise.
Many recent works extended the MID by leveraging adaptive filters to address the deficiencies of previously mentioned static filter-based approaches. In these works, the authors emphasized to estimate weighted coefficients of an image by employing the statistical properties. The non-local denoising method, like Block Matching 3D (BM3D) [11], is one of the perfect examples of adaptive filter based techniques. Similarly, non-local means filters-based methods [29,30] for MR image denoising, Optimised Bayesian Non Local Mean (OBNLM) [31] and modified non local-based (MNL) [32] for ultrasound image denoising, bio-inspired bilateral filter [10] for CT image denoising are also representative of the adaptive filter-based techniques. However, such adaptive filter-based denoising methods are computationally expensive and unable to accommodate real-time results.
Another well-known classical MID genre is known to be multi-scale analysis based methods [33,34,35], where the representative techniques intended to process the noisy images in different image resolutions. In recent times, a notable amount of work exploited such denoising techniques and utilized the time-frequency analysis [1]. Nevertheless, in numerous cases, multi-scale medical image denoising illustrates deficiencies in specifying the distribution of noisy inputs in different scales. The drawbacks of the multi-scale methods have been addressed with nonlinear estimators [36] by a part. However, the performance of this classical image denoising category is still far away from the expectations.
Learning Based Methods:The learning-based image denoising method has started to draw attention in the MID domain very lately. In recent work, a feedforward autoencoder [13] has used to learn medical image denoising. However, the later study [12] on MID took the inspiration from [16] and improved the performance of their method by using residual learning. In exception, another study on MID [14] practiced a distinctive network designing strategy and used a genetic algorithm (GA) to search the hyperparameter of their deep network. Despite the satisfactory performance in specific noise levels as well as on specific datasets, none of the existing works generalize their method for multidisciplinary modalities.
2.2. Attention Guided Learning
The concept of attention mechanism has been taken from the human visual system. In deep learning, the attention mechanism has first introduced in the natural language processing domain for adaptively focuses on salient areas of a given input. By considering that approach of adaptive feature correlation is a success, many vision works quickly adopted similar concepts in computer vision applications such as super-resolution [37], in-painting [20], deblurring [38], etc. Among the recent works, a non-local spatial attention [39] formulated for video classification. Also, utilization of channel-wise interdependencies to obtain a significant performance gain over existing image classification methods [17].
In recent time, a few novel works leverage the residual learning along with the attention mechanisms. In [40], authors stacked attention modules in the feed-forward structure and combined with residual connections to train very-deep network to outperform their counterparts. Later, Ref. [41] also exploited similar residual-attention strategy in multi-scale network structure with static convolution operation to improve the accuracy of a classification task. Apart from being used in classification tasks, residual attention strategies has also illustrated a substantial push in image super-resolution. In recent work, Refs. [42,43] utilized feature attention after convolutional layers in a sequential manner to perform image super-resolution. In a later study, Ref. [44] combined spatial and temporal feature attention with residual connection after the convolutional operation to accelerate muli-image super-resolution.
Despite the success of residual-attention strategies in different computer vision tasks, the contemporary residual-attention approaches are suffering two-fold limitations. Firstly, existing residual-attention methods utilize the attention mechanism with static convolutional operation in a sequential manner. However, such stacked architecture tends to make the network designing relatively deeper and can introduce a tendency of suffering from vanishing gradient along with a larger number of trainable parameters [23]. Secondly, most of the existing DRAN utilized simple skip connections to accelerate residual learning. Notably, such straightforward skip connections can backfire the denoising by propagating unpruned low-level features. This study addresses the limitation of existing residual-attention strategies by incorporating attention-guided dynamic kernel convolutional operation known as dynamic convolution. Also, this study proposes to accelerate the residual learning by utilizing a noise gate. Which essentially aims to prune the low-level features spatial getting mechanism.
Table 1 shows a concise comparison between different methods. Here, each category has been reviewed by distinguishing strengths and weaknesses.

Table 1.
A brief comparison between existing works and the proposed work.
3. Proposed Method
This study presents a novel denoising method for addressing medical images denoising by learning from large-scale data samples. This section details the methodology of the proposed work.
3.1. Network Design
The proposed method intended to recover the clean image from a given noisy medical image by learning residual noisy image through the mapping function . Hence, learning denoising for medical images can be derived for this study as follows:
As Figure 2 shows, the proposed network is presented as an end-to-end convolutional neural network (CNN) [45]. Where the network utilized traditional convolutional operation for input and output as well as a novel dynamic residual attention block (DRAB) as the backbone of the main network. The input layer takes an normalized noisy image and generate a normalized residual noise [16] extracted from the input as . Here, and represents the height and width of the input as well as the output of the proposed DRAN.
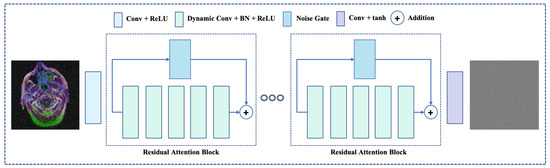
Figure 2.
The overview of proposed network architecture. The proposed network incorporates novel dynamic residual attention blocks, which utilizes dynamic convolution and a noise gate. Also, the network leverage the residual learning along with the learning feature correlation.
3.1.1. Dynamic Residual Attention Block
The proposed dynamic residual attention block (DRAB) comprises a number of dynamic convolution [23] layers stacked consecutively. Here, the dynamic convolutional were aims to improve the performance of traditional convolution by aggregating a number of dynamic kernels (each with equal dimensions) by an attention mechanism [17]. Therefore, a static convolution comprises of a weight matrix , a bias term , and activated with was replaced with . The aggregation of number of linear function was obtained as:
exploited as a ReLU activation and can be expressed as .
presents the attention mechanism, which has aggregated over the linear models for a given input x. Where the attention weights of has obtained through a global feature descriptor by applying a global average pooling [17,37]. Where depth-wise squeezed descriptors of an input feature map can be calculated as:
Here, , , and x present the global average pooling, spatial dimension, and input feature map.
The aggregated global dependencies through the gating mechanism applied as follows:
Here, present the sigmoid activation as and ReLU activation as , which were applied after and convolutional operations.
The final depth attention map achieved by rescaling the feature map as follows:
To perceive a faster convergence, each dynamic convolutional layer used in this paper normalized with a batch normalization function [46] as follows:
where, and denote the expectation of input and its variance, while and denote the learnable parameters which intended to improve the model performance.
Apart from the dynamic convolution layers, the proposed network also intended to leverage the residual learning through the skip connection [21]. However, in the denoising, the skip connection can backfire by delivering the lower-level superfluous features towards the top levels. Therefore, this study leverage a spatial attention mechanism [19,20] denoted as a noise gate, which controls the propagation of trivial features by learning spatial feature correlation. The noise gating mechanism has obtained as follows:
and presents the LeakyReLU and sigmoid activation functions as and . and represents convolutional operations.
3.1.2. Optimization
For a given the training set consisting of P pairs of images, the proposed DRAN learns to parameterized weights and intended to minimize the objective function as:
Here, denotes a pixel-wise loss, which can be calculated in the euclidean space as a form of L1 or an L2-norm [47]. However, due to the direct relation with PSNR values, the L2-norm is susceptible to produce smoother images [48]. As a consequence, this study employs an L1 norm as an objective function, which can be derived as follows:
Here, and represent the output obtain through and simulated reference noise.
3.2. Data Preparation
Data preparation plays a crucial role in learning-based denoising methods [49]. For training purposes, it is mandatory to obtain a sufficient amount of data samples. Therefore, this study has collected a substantial amount of medical images capture through different modalities. Also, the collected data samples have processed (i.e., adding noise for training purposes) carefully for further study.
3.2.1. Data Collection
It always remains a challenging task in medical image analysis to collect a diverse range of data samples [13,50]. Also, none of the existing datasets offers a collection of images accumulated by different medical imaging technologies. However, to generalize the performance of any deep method on a spacious data space, a substantial amount of training data samples is mandatory [51]. To address this contradictory condition, this study collected enormous image samples from different sources and divided them into three categories. Brief detail about each data category described below:
- Radiology: This category comprises the microscopic images collected from of four different modalities, including X-ray [52], MRI [53], CT scans [54], and ultrasound [55] images.
- Microscopy: This category includes microscopic images collected by histopathologic scan [56] and protein atlas scans [57].
- Dermatology: This category contains dermatoscopic images collected by different image acquisition methods [58].
A total of 711,223 medical images were collected by this study. Where 585,198 samples were used for model training and the rest of the 20 percent data used for performance evaluation. Figure 3 illustrates the sample images from each category.
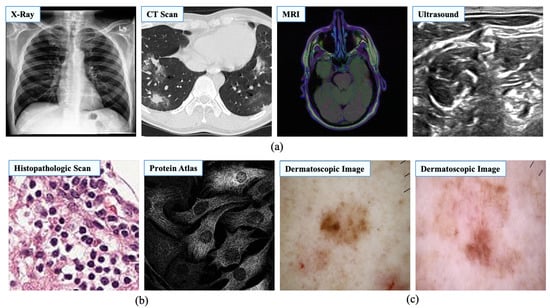
Figure 3.
Image samples from different medical image modalities. This study categorized medical images into three different categories to generalize the learning process. (a) Image samples of radiology. Source by: (https://stanfordmlgroup.github.io/competitions/chexpert/, https://www.kaggle.com/c/covid-segmentation, https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation, https://www.kaggle.com/c/ultrasound-nerve-segmentation). (b) Image samples of microscopy. Source by: (https://www.kaggle.com/c/histopathologic-cancer-detection, https://www.kaggle.com/c/human-protein-atlas-image-classification). (c) Image samples of dermatology. Source by: (https://www.kaggle.com/kmader/skin-cancer-mnist-ham10000).
3.2.2. Noise Modeling
Despite having a significant number of sample images, the collected dataset does not provide a training pair of reference and noise-contaminated input images. Therefore, reference-noisy image pairs have to be formulated by contaminating artificial noise. Here, a uniform noisy-image has generated from a given noise-free image as:
and represent the mean and the variance of a Gaussian distribution (). This Gaussian noise added to the given clean-image . The final noisy-image formed as:
Figure 4 illustrate the sample of noisy-clean image pair along with the corresponding noise (generated).
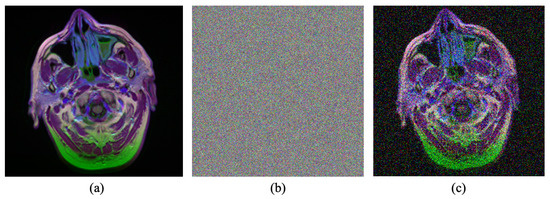
Figure 4.
Example of the noise modeling process. (a) Input clean image. (b) Generated noise. (c) Generated noisy image. Source by: (https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation).
3.3. Implementation Details
The proposed DRAN was designed as an end-to-end convolution network and implemented using the PyTorch framework [59]. This study utilized three consecutive DRABs for making a trade-off between performance and trainable parameters. Each layer of DRAB comprises a depth size of 64, a kernel size of 3, a padding size of 1, and a stride size of 1. Thus, the network can keep the output dimension identical to the input. Also, the network was optimized with an Adam optimizer [60] with , , and learning rate = 1e-4. The network trained on resized images with dimensions of and contaminated with random noise (). The training process carried for 100,000 steps while keeping a batch size of 24. All experiments conducted on hardware incorporates an AMD Ryzen 3200G central processing unit (CPU) clocked at 3.60 GHz and a random-access memory of 16 GB. Also, an Nvidia Geforce GTX 1060 (6GB) graphical processing unit (GPU) was exploited to accelerate the training process.
4. Result and Analysis
The performance of the proposed method has been studied and compared with state-of-the-art denoising methods. Also, the feasibility of the proposed method in different medical image analysis tasks has verified with sophisticated experiments.
4.1. Comparison with State-of-the-Art Methods
In this study, three existing works have been selected for the comparison. These methods are: (i) BM3D [11], (ii) DnCNN [16], and (iii) Residual MID [12]. DnCNN [16] and Residual MID [12] both utilized residual learning for image denoising, while BM3D [11] is known to be one of the pioneers of the image denoising works. It worth noting, none of these works have been developed for addressing a diverse range of noise removal from medical images as intended in this work. Nevertheless, to make the comparison as fair as possible, both learning-based methods [12,16] have been trained and tested with the hyperparameters suggested in the actual implementations. Additionally, each model trained for 4∼5 days until they converge with the collected dataset mentioned in Section 3.2. Oppositely, the optimization-based method [11] studied for the comparison does not require any additional training similar to its counterparts. Subsequently, this study used the official implementation publicly available for the fair comparison.
4.1.1. Quantitative Comparison
This study incorporates a distinct evaluation strategy to study the feasibility of the compared denoising method in different noisy environments using two widely-used image assessment matrices: peak-signal-to-noise ratio (PSNR) [61] and structural-similarity-index metrics (SSIM) [62]. Such evaluation metrics meant to evaluate the reconstructed image quality by comparing it with the reference image, similar to the human visual system. Therefore, the higher value of such metric indicates the better performance of the target method [2,63]. Notably, this work leverage the PSNR metrics to calculate the noise ratio between reference and denoised image, while SSIM intends to evaluate the structural similarity, luminance, and contrast distortions. Overall, the performance of MID methods has summarized with the mean scores obtained from the evaluation metrics over individual categories as well as different noise deviations. Table 2 shows the quantitative comparison between different denoising methods for medical images.

Table 2.
Quantitative comparison between different medical image denoising methods. Results are obtained by calculating the mean on two evaluation metrics. In all comparing categories, the proposed method illustrates the consistency and outperforms the existing denoising methods.
As the Table 2 depicts, the proposed method outperforms the existing MID methods by a distinguished margin in all compared combinations. Most notably, depending on the noise deviations, the proposed method can exceed its counterpart on the dermatology images by up to 13.75 dB in the PSNR metric and 0.0992 in the SSIM metric. Similarly, it is illustrated the supremacy over existing denoising methods by up to 10.91 dB in PSNR metric and 0.1137 in the SSIM metric on radiology images as well as 11.17 dB in PSNR metric and 0.3065 in SSIM metric on microscopy images. It worth noting the increment of noise in the images can deteriorate the performance of MID methods. However, the proposed method illustrates it’s consistency in all categories. Overall, the Table 2 reveals a new dimension of multidisciplinary MID. Also, it demonstrates the practicability of a sophisticated denoising method for medical image analysis.
4.1.2. Qualitative Comparison
Qualitative evaluation plays an important role in medical image analysis [1,64]. Therefore, this study focused on qualitative comparison along with quantitative comparisons. Figure 5, Figure 6 and Figure 7 illustrates the visual comparison between the proposed method and existing MID methods.
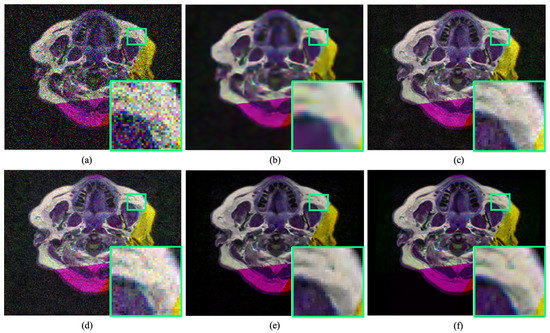
Figure 5.
Qualitative comparison of radiology image denoising at . The proposed method illustrates significant improvement over the existing denoising method by improving perceptual image quality. (a) Noisy input. (b) Result obtained by BM3D [11]. (c) Result obtained by DnCNN [16]. (d) Result obtained by Residual MID [12]. (e) Result obtained by DRAN (proposed). (f) Reference sharp image. Source by: (https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation).
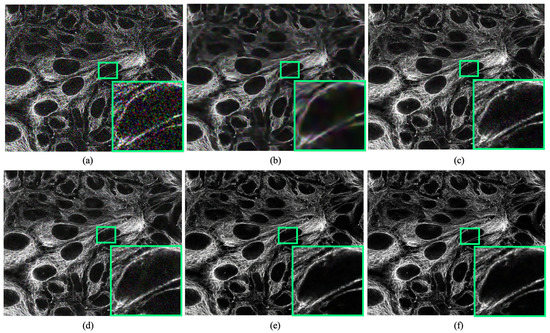
Figure 6.
Qualitative comparison of microscopy image denoising at . The proposed method illustrates significant improvement over the existing denoising method by improving perceptual image quality. (a) Noisy input. (b) Result obtained by BM3D [11]. (c) Result obtained by DnCNN [16]. (d) Result obtained by Residual MID [12]. (e) Result obtained by DRAN (proposed). (f) Reference sharp image. Source by: (https://www.kaggle.com/c/human-protein-atlas-image-classification).
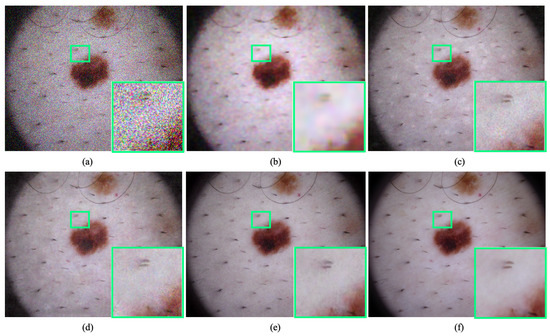
Figure 7.
Qualitative comparison of dermatology image denoising at . The proposed method illustrates significant improvement over the existing denoising method by improving perceptual image quality. (a) Noisy input. (b) Result obtained by BM3D [11]. (c) Result obtained by DnCNN [16]. (d) Result obtained by Residual MID [12]. (e) Result obtained by DRAN (proposed). (f) Reference sharp image. Source by: (https://www.kaggle.com/kmader/skin-cancer-mnist-ham10000).
As can be seen, the proposed method is proficient in improving the perceptual quality of degraded noisy images dramatically. It can remove a substantial amount of noise while maintaining the details of a degraded input image. Most notably, the method shows its consistency over the existing MID techniques in all image categories without producing any visually disturbing artefacts.
4.2. Real-World Noise Removal
In the real-world scenario, the noise that appeared in the medical images can differ from the synthesized data. Therefore, to push the MID a furthermore, the feasibility of the proposed method has been studied with real-world noisy medical images. As Figure 8 illustrates, the proposed method can notoriously handle the real-world noise and substantially improve the perceptual image quality of noisy medical images by removing blind-fold noise.
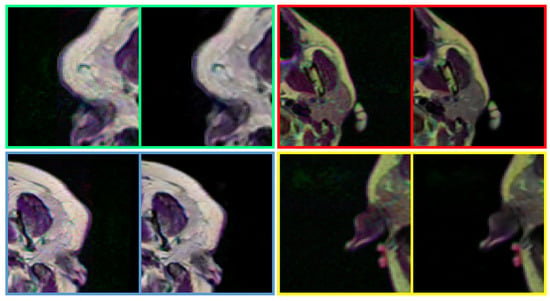
Figure 8.
Performance of the proposed method in removing noise from real-world medical images. The proposed method can handle real-world noise without producing any visually disturbing artefacts. In each pair, left: Noisy input, Right: Denoised image (obtained by DRAN). Source by: (https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation).
4.3. Applications
A sophisticated denoising method can drastically accelerate the performance of computer-aided detection (CAD) and medical image analysis tasks by improving the perceptual quality of target images. To study further, the propose DRAN has combined with existing state-of-the-art medical image analysis methods to investigate the consequences.
4.3.1. Abnormalities Detection
Computer-aided detection has obtained momentum in medical image analysis by observing the oversights of given images [65,66]. However, the presence of sensor noise in a given image can misguide the detection system, as shown in Figure 9. Here, the effect of image noise has studied over tumour detection and localization on brain MRIs using state-of-the-art Mask R-CNN [67]. It can be apparent that even the well-known learning-based method struggle in localizing the abnormalities on a noisy image. Contrarily, the addition of proposed DRAN drastically improve the localization performance of the respective detection method by performing denoising.
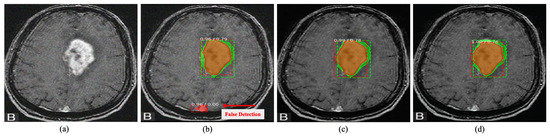
Figure 9.
Tumour detection on brain MRI image. The green box indicates the ground truth region, while the red box indicates the detected area. The proposed DRAN can improve the localization performance of the detection method to localize the abnormalities by removing the noise present in the target image. (a) Noisy image. (b) Noisy image + Mask R-CNN [67]. (c) Denoised image obtained with DRAN + Mask R-CNN [67]. (d) Reference clean image + Mask R-CNN [67]. Source by: (https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation).
4.3.2. Medical Image Segmentation
The image noise can also startlingly effect the medical image segmentation process similar to the detection methods, as shown in Figure 10. Here, segmentation has performed on brain MRIs using well-known U-Net architecture [68]. It has observed that image noise make the segmentation process substantially unsatisfactory. However, a sophisticated MID method like the proposed DRAN can assist the segmentation method by mitigating image noise.
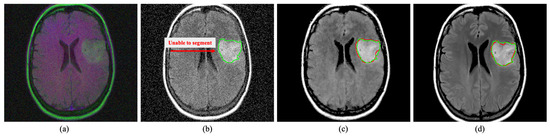
Figure 10.
Segmentation on brain MRI image. The green and red outline indicate the ground truth and the segmented area. The proposed DRAN can drastically improve the segmentation performance by excluding the noise present in the target image. (a) Noisy image. (b) Noisy image + U-Net [68]. (c) Denoised image obtained with DRAN + U-Net [68]. (d) Reference clean image + U-Net [68]. Source by: (https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation).
4.4. Network Analysis
Despite being deeper, the proposed network comprises 2,458,944 parameters. It worth noting, the number of parameters can be altered by substituting the number of DRABs. The reduction of blocks can also deteriorate the performance of the proposed network while reducing inference time. However, for any quantity of DRABs, the proposed denoising network can take any dimension of images for the inference.
The DRAB also has a significant impact on the learning processes. Expressly, the noise gate introduced in the DRAB plays a crucial role in the network stability. Figure 11 illustrates the impact of the noise gate on the training phase. Notably, the noise gate allowed the proposed DRAN to perceive a faster convergence even with a very complex set of medical images.
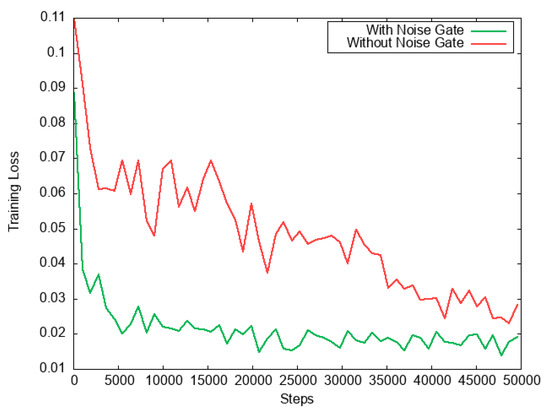
Figure 11.
Graph of training loss. It is visible that the noise gate used in DRAB illustrates far more training stability. Also, it helps the proposed method to encounter faster convergence time.
Apart from the training stability, the noise gate also has a clear impact on the performance gain. Table 3 demonstrates that the noise gate improves the performance of the proposed DRAN drastically among all medical image categories. Here, the performance metrics calculated by exploiting random noise. Both models have evaluated the same noisy images during their training phases. Also, the evaluation has repeated over every 5000 steps for consistency.

Table 3.
Impact of noise gate in network performance. It can be seen that the noise gate drastically improves the performance of the proposed method compared to its counterpart.
4.5. Discussion
Despite the extensive experiments, it is undeniable that the proposed study encounters a few limitations. Contrastly, the experimental results of this study reveals a dimension of medical image denoising an apart.
Similar to the existing works, one of the limitations of this study identified as lacking real-world training data. Although the data samples used in this study are synthesized with artificial noise, nonetheless, in numerous instances, the simulated data can differ from the real world noise. Also, due to the lacking of reference images, the quantitative performance of the proposed method remains underexplored, particularly on real-world noisy medical images.
The limitation and observation perceive through the proposed study reveal an interesting future dimension of MID methods. Despite the sophisticated preprocessing, it has found that the reference images can contain noise. Therefore, it would be interesting to extend this work in an unsupervised manner. Also, for the generalization, the proposed study conducted all testing and training on three-channel RGB images. However, depending on the application, the proposed DRAN can be optimized by incorporating a single-channel dataset. In the foreseeable future, it has planned to study the feasibility of the proposed study in one channel data as well as by exploiting unsupervised learning.
5. Conclusions
This work presented a novel end-to-end learning-based denoising method for medical image analysis. Additionally, it has illustrated that MID can be generalized by utilizing large-scale multidisciplinary images rather than learning from a small range of homogeneous data samples. The proposed method also incorporates a novel deep network, which combines the attention mechanism and spatially-refine residual learning in a feed-forward manner. Notably, such a comprehensive learning strategy allowed this study in drastically improving the denoising performance, particularly for medical images. The experimental results illustrate that the proposed method can outperform the existing works by a distinguishable margin while maintaining details. Also, the practicability of the proposed denoising method has explicitly inspected by employing sophisticated experiments. It has planned to extend the proposed method by exploiting unsupervised learning in the foreseeable future.
Author Contributions
SMAS. Conceptualization, Methodology, Software, Visualization, Writing—Original Draft. R.A.N. Resources, Validation, Investigation, Funding acquisition, Project administration, Supervision. M.B. Data Curation, Resources, Software, Editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research is an independent work and didn’t receive any additional funding.
Acknowledgments
The authors would like to thank Dildar Hussain, Amir Haider, and Omar M. Aldossary for their constructive discussion during the manuscript revision.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sagheer, S.V.M.; George, S.N. A review on medical image denoising algorithms. Biomed. Signal Process. Control 2020, 61, 102036. [Google Scholar] [CrossRef]
- Kollem, S.; Reddy, K.R.L.; Rao, D.S. A review of image Denoising and segmentation methods based on medical images. Int. J. Mach. Learn. Comput. 2019, 9, 288–295. [Google Scholar] [CrossRef]
- Rodrigues, I.; Sanches, J.; Bioucas-Dias, J. Denoising of medical images corrupted by Poisson noise. In Proceedings of the 2008 15th IEEE International Conference on Image Processing, San Diego, CA, USA, 12–15 October 2008; pp. 1756–1759. [Google Scholar]
- Hansen, M.S.; Sørensen, T.S. Gadgetron: An open source framework for medical image reconstruction. Magn. Reson. Med. 2013, 69, 1768–1776. [Google Scholar] [CrossRef] [PubMed]
- Arsalan, M.; Naqvi, R.A.; Kim, D.S.; Nguyen, P.H.; Owais, M.; Park, K.R. IrisDenseNet: Robust iris segmentation using densely connected fully convolutional networks in the images by visible light and near-infrared light camera sensors. Sensors 2018, 18, 1501. [Google Scholar] [CrossRef] [PubMed]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep learning applications in medical image analysis. IEEE Access 2017, 6, 9375–9389. [Google Scholar] [CrossRef]
- Wang, J.; Guo, Y.; Ying, Y.; Liu, Y.; Peng, Q. Fast non-local algorithm for image denoising. In Proceedings of the IEEE 2006 International Conference on Image Processing, Atlanta, GA, USA, 8–11 October 2006; pp. 1429–1432. [Google Scholar]
- Elad, M.; Aharon, M. Image denoising via sparse and redundant representations over learned dictionaries. IEEE Trans. Image Process. 2006, 15, 3736–3745. [Google Scholar] [CrossRef] [PubMed]
- Arif, A.S.; Mansor, S.; Logeswaran, R. Combined bilateral and anisotropic-diffusion filters for medical image de-noising. In Proceedings of the 2011 IEEE Student Conference on Research and Development, Cyberjaya, Malaysia, 19–20 December 2011; pp. 420–424. [Google Scholar]
- Bhonsle, D.; Chandra, V.; Sinha, G. Medical image denoising using bilateral filter. Int. J. Image Graph. Signal Process. 2012, 4, 36. [Google Scholar] [CrossRef]
- Dabov, K.; Foi, A.; Katkovnik, V.; Egiazarian, K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans. Image Process. 2007, 16, 2080–2095. [Google Scholar] [CrossRef]
- Jifara, W.; Jiang, F.; Rho, S.; Cheng, M.; Liu, S. Medical image denoising using convolutional neural network: A residual learning approach. J. Supercomput. 2019, 75, 704–718. [Google Scholar] [CrossRef]
- Gondara, L. Medical image denoising using convolutional denoising autoencoders. In Proceedings of the 2016 IEEE 16th International Conference on Data Mining Workshops (ICDMW), Barcelona, Spain, 12–15 December 2016; pp. 241–246. [Google Scholar]
- Liu, P.; El Basha, M.D.; Li, Y.; Xiao, Y.; Sanelli, P.C.; Fang, R. Deep evolutionary networks with expedited genetic algorithms for medical image denoising. Med. Image Anal. 2019, 54, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yan, P.; Zhang, Y.; Yu, H.; Shi, Y.; Mou, X.; Kalra, M.K.; Zhang, Y.; Sun, L.; Wang, G. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE Trans. Med. Imaging 2018, 37, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zuo, W.; Chen, Y.; Meng, D.; Zhang, L. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising. IEEE Trans. Image Process. 2017, 26, 3142–3155. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Siam, M.; Valipour, S.; Jagersand, M.; Ray, N. Convolutional gated recurrent networks for video segmentation. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 3090–3094. [Google Scholar]
- Yu, J.; Lin, Z.; Yang, J.; Shen, X.; Lu, X.; Huang, T.S. Free-form image inpainting with gated convolution. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019; pp. 4471–4480. [Google Scholar]
- Chang, Y.L.; Liu, Z.Y.; Lee, K.Y.; Hsu, W. Free-form video inpainting with 3d gated convolution and temporal patchgan. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019; pp. 9066–9075. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Naqvi, R.A.; Loh, W.K. Sclera-net: Accurate sclera segmentation in various sensor images based on residual encoder and decoder network. IEEE Access 2019, 7, 98208–98227. [Google Scholar] [CrossRef]
- Chen, Y.; Dai, X.; Liu, M.; Chen, D.; Yuan, L.; Liu, Z. Dynamic convolution: Attention over convolution kernels. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11030–11039. [Google Scholar]
- Andria, G.; Attivissimo, F.; Cavone, G.; Giaquinto, N.; Lanzolla, A. Linear filtering of 2-D wavelet coefficients for denoising ultrasound medical images. Measurement 2012, 45, 1792–1800. [Google Scholar] [CrossRef]
- Loupas, T.; McDicken, W.; Allan, P.L. An adaptive weighted median filter for speckle suppression in medical ultrasonic images. IEEE Trans. Circuits Syst. 1989, 36, 129–135. [Google Scholar] [CrossRef]
- Yang, J.; Fan, J.; Ai, D.; Wang, X.; Zheng, Y.; Tang, S.; Wang, Y. Local statistics and non-local mean filter for speckle noise reduction in medical ultrasound image. Neurocomputing 2016, 195, 88–95. [Google Scholar] [CrossRef]
- Guan, F.; Ton, P.; Ge, S.; Zhao, L. Anisotropic diffusion filtering for ultrasound speckle reduction. Sci. China Technol. Sci. 2014, 57, 607–614. [Google Scholar] [CrossRef]
- Anand, C.S.; Sahambi, J. MRI denoising using bilateral filter in redundant wavelet domain. In Proceedings of the TENCON 2008-2008 IEEE Region 10 Conference, Hyderabad, India, 19–21 November 2008; pp. 1–6. [Google Scholar]
- Manjón, J.V.; Carbonell-Caballero, J.; Lull, J.J.; García-Martí, G.; Martí-Bonmatí, L.; Robles, M. MRI denoising using non-local means. Med. Image Anal. 2008, 12, 514–523. [Google Scholar] [CrossRef]
- Dolui, S.; Kuurstra, A.; Patarroyo, I.C.S.; Michailovich, O.V. A new similarity measure for non-local means filtering of MRI images. J. Vis. Commun. Image Represent. 2013, 24, 1040–1054. [Google Scholar] [CrossRef][Green Version]
- Coupé, P.; Hellier, P.; Kervrann, C.; Barillot, C. Nonlocal means-based speckle filtering for ultrasound images. IEEE Trans. Image Process. 2009, 18, 2221–2229. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Hou, T. Speckle filtering of ultrasonic images using a modified non local-based algorithm. Biomed. Signal Process. Control 2011, 6, 129–138. [Google Scholar] [CrossRef]
- Diwakar, M.; Kumar, M. Edge preservation based CT image denoising using Wiener filtering and thresholding in wavelet domain. In Proceedings of the IEEE 2016 Fourth International Conference on Parallel, Distributed and Grid Computing (PDGC), Waknaghat, India, 22–24 December 2016; pp. 332–336. [Google Scholar]
- Bhuiyan, M.I.H.; Ahmad, M.O.; Swamy, M. Spatially adaptive thresholding in wavelet domain for despeckling of ultrasound images. IET Image Process. 2009, 3, 147–162. [Google Scholar] [CrossRef]
- Wood, J.C.; Johnson, K.M. Wavelet packet denoising of magnetic resonance images: Importance of Rician noise at low SNR. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 1999, 41, 631–635. [Google Scholar] [CrossRef]
- Tian, J.; Chen, L. Image despeckling using a non-parametric statistical model of wavelet coefficients. Biomed. Signal Process. Control 2011, 6, 432–437. [Google Scholar] [CrossRef]
- Dai, T.; Cai, J.; Zhang, Y.; Xia, S.T.; Zhang, L. Second-order attention network for single image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 11065–11074. [Google Scholar]
- Zhang, X.; Jiang, R.; Wang, T.; Huang, P.; Zhao, L. Attention-based interpolation network for video deblurring. Neurocomputing 2020. [Google Scholar] [CrossRef]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7794–7803. [Google Scholar]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual attention network for image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3156–3164. [Google Scholar]
- Liang, L.; Cao, J.; Li, X.; You, J. Improvement of Residual Attention Network for Image Classification. In International Conference on Intelligent Science and Big Data Engineering; Springer: Berlin, Germany, 2019; pp. 529–539. [Google Scholar]
- Shi, Z.; Chen, C.; Xiong, Z.; Liu, D.; Zha, Z.J.; Wu, F. Deep residual attention network for spectral image super-resolution. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Xue, S.; Qiu, W.; Liu, F.; Jin, X. Wavelet-based residual attention network for image super-resolution. Neurocomputing 2020, 382, 116–126. [Google Scholar] [CrossRef]
- Salvetti, F.; Mazzia, V.; Khaliq, A.; Chiaberge, M. Multi-Image Super Resolution of Remotely Sensed Images Using Residual Attention Deep Neural Networks. Remote Sens. 2020, 12, 2207. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. Handb. Brain Theory Neural Netw. 1995, 3361, 1995. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Zhao, H.; Gallo, O.; Frosio, I.; Kautz, J. Loss functions for image restoration with neural networks. IEEE Trans. Comput. Imaging 2016, 3, 47–57. [Google Scholar] [CrossRef]
- Schwartz, E.; Giryes, R.; Bronstein, A.M. DeepISP: Toward learning an end-to-end image processing pipeline. IEEE Trans. Image Process. 2018, 28, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Sharif, S.; Mahboob, M. Deep hog: A hybrid model to classify bangla isolated alpha-numerical symbols. Neural Netw. World 2019, 29, 111–133. [Google Scholar] [CrossRef]
- Arsalan, M.; Hong, H.G.; Naqvi, R.A.; Lee, M.B.; Kim, M.C.; Kim, D.S.; Kim, C.S.; Park, K.R. Deep learning-based iris segmentation for iris recognition in visible light environment. Symmetry 2017, 9, 263. [Google Scholar] [CrossRef]
- L’heureux, A.; Grolinger, K.; Elyamany, H.F.; Capretz, M.A. Machine learning with big data: Challenges and approaches. IEEE Access 2017, 5, 7776–7797. [Google Scholar] [CrossRef]
- CheXpert: A Large Chest X-Ray Dataset and Competition. Available online: https://stanfordmlgroup.github.io/competitions/chexpert/ (accessed on 4 December 2020).
- Buda, M.; Saha, A.; Mazurowski, M.A. Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm. Comput. Biol. Med. 2019, 109, 218–225. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Y.; He, X.; Xie, P. COVID-CT-Dataset: A CT scan dataset about COVID-19. arXiv 2020, arXiv:2003.13865. [Google Scholar]
- Baby, M.; Jereesh, A. Automatic nerve segmentation of ultrasound images. In Proceedings of the IEEE 2017 International conference of Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 20–22 April 2017; Volume 1, pp. 107–112. [Google Scholar]
- Veeling, B.S.; Linmans, J.; Winkens, J.; Cohen, T.; Welling, M. Rotation equivariant CNNs for digital pathology. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 2018; pp. 210–218. [Google Scholar]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Rezvantalab, A.; Safigholi, H.; Karimijeshni, S. Dermatologist level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms. arXiv 2018, arXiv:1810.10348. [Google Scholar]
- Pytorch. PyTorch Framework Code. 2016. Available online: https://pytorch.org/ (accessed on 24 August 2020).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Huynh-Thu, Q.; Ghanbari, M. Scope of validity of PSNR in image/video quality assessment. Electron. Lett. 2008, 44, 800–801. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Hore, A.; Ziou, D. Image quality metrics: PSNR vs. SSIM. In Proceedings of the IEEE 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 2366–2369. [Google Scholar]
- Nazir, T.; Irtaza, A.; Javed, A.; Malik, H.; Hussain, D.; Naqvi, R.A. Retinal Image Analysis for Diabetes-Based Eye Disease Detection Using Deep Learning. Appl. Sci. 2020, 10, 6185. [Google Scholar] [CrossRef]
- Narayanan, B.N.; Ali, R.; Hardie, R.C. Performance analysis of machine learning and deep learning architectures for malaria detection on cell images. In Applications of Machine Learning; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 11139, p. 111390W. [Google Scholar]
- Narayanan, B.N.; De Silva, M.S.; Hardie, R.C.; Kueterman, N.K.; Ali, R. Understanding Deep Neural Network Predictions for Medical Imaging Applications. arXiv 2019, arXiv:1912.09621. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 2015; pp. 234–241. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).