Abstract
Feature selection (FS) was regarded as a global combinatorial optimization problem. FS is used to simplify and enhance the quality of high-dimensional datasets by selecting prominent features and removing irrelevant and redundant data to provide good classification results. FS aims to reduce the dimensionality and improve the classification accuracy that is generally utilized with great importance in different fields such as pattern classification, data analysis, and data mining applications. The main problem is to find the best subset that contains the representative information of all the data. In order to overcome this problem, two binary variants of the whale optimization algorithm (WOA) are proposed, called bWOA-S and bWOA-V. They are used to decrease the complexity and increase the performance of a system by selecting significant features for classification purposes. The first bWOA-S version uses the Sigmoid transfer function to convert WOA values to binary ones, whereas the second bWOA-V version uses a hyperbolic tangent transfer function. Furthermore, the two binary variants introduced here were compared with three famous and well-known optimization algorithms in this domain, such as Particle Swarm Optimizer (PSO), three variants of binary ant lion (bALO1, bALO2, and bALO3), binary Dragonfly Algorithm (bDA) as well as the original WOA, over 24 benchmark datasets from the UCI repository. Eventually, a non-parametric test called Wilcoxon’s rank-sum was carried out at 5% significance to prove the powerfulness and effectiveness of the two proposed algorithms when compared with other algorithms statistically. The qualitative and quantitative results showed that the two introduced variants in the FS domain are able to minimize the selected feature number as well as maximize the accuracy of the classification within an appropriate time.
1. Introduction
The datasets from real-world applications such industry or medicine are high-dimensional and contain irrelevant or redundant features. These kind of datasets then have useless information that affects the performance of machine learning algorithms; in such cases, the learning process is affected. Feature selection (FS) is a powerful rattling technique used to select the most significant subset of features, overcoming the high-dimensionality reduction problem [1], identifying the relevant features and removing redundant ones [2]. Moreover, using the subset of features, any machine learning algorithm can be applied for classification. Therefore, several studies have taken into consideration that the FS problem is an optimization problem, hence the fitness function for the optimization algorithm has been changed to classifier’s accuracy, which may be maximized by the selected features [3]. Moreover, FS has been applied successfully to solve many classification problems in different domains, such as data mining [4,5], pattern recognition [6], information retrieval [7], information feedback [8], drug design [9,10], job-shop scheduling problem [11], maximizing lifetime of wireless sensor networks [12,13], and the others where FS can be utilized [14].
There are three main classes of FS methods: (1) The wrapper, (2) filter and (3) hybrid methods [15]. The wrapper approaches generally incorporate classification algorithms to search for and select the relevant features [16]. Filter methods calculate the relevant features without prior data classification [17]. In the hybrid techniques, the compatible strengths of the wrapper and filter methods are combined. Generally speaking, the wrapper methods outperform filter methods in terms of classification accuracy, and hence the wrapper approaches are used in this paper.
In fact, a high accuracy classification does not depend on a large selected features number for many classification problems. In this context, the classification problems can be categorized into two groups: (1) binary classification and (2) multi-class classification. In this paper, we deal with the binary classification problem. There are numerous methods that are applied for binary classification problems, such as discriminant analysis [18], decision trees (DT) [19], the K-nearest neighbor (K-NN) [20], artificial neural networks (ANN) [21], and support vector machines (SVMs) [22].
On the other hand, the traditional optimization methods suffer from some limitations in solving the FS problems [23,24], and hence nature-inspired meta-heuristic algorithms [25] such as the whale optimization algorithm (WOA) [26], moth–flame optimisation [27], Ant Lion Optimization [28], Crow Search Algorithm [29], Lightning Search Algorithm [30], Henry gas solubility optimization [31] and Lévy flight distribution [32] are widely used in the scientific community for solving complex optimization problems and several real-world applications [33,34,35]. Optimization is defined as a process of searching the optimal solutions to a specific problem. In order to address issues such as FS, several nature-inspired algorithms have been applied; some of these algorithms are hybridized with each other or used alone, others created new variants like binary methods to solve this problem. A survey on evolutionary computation [36] approaches for FS is presented in [37]. Several separate and hybrid algorithms have been proposed for FS, such as hybrid ant colony optimization algorithm [38], forest optimization algorithm [39], firefly optimization algorithm [40], hybrid whale optimization algorithm with simulated annealing [41], particle swarm optimization [42], sine cosine optimization algorithm [43], monarch butterfly optimization [44], and moth search algorithm [45].
In addition to the aforementioned studies to find solutions for the FS problem, other search strategies called the binary optimization algorithms have been implemented. Some examples are the binary flower pollination algorithm (BFPA) in [46], binary bat algorithm (BBA) in [47], binary cuckoo search algorithm (BCSA) in [48]; all of them evaluate the accuracy of the classifier as an objective function. He et al. have presented a binary differential evolution algorithm (BDEA) [49] to select the relevant subset to train a SVM with radial basis function (RBF). Moreover, Emary et al., have proposed the binary ant lion and the binary grey wolf optimization [50,51], respectively. Rashedi et al. have introduced an improved binary gravitational search algorithm version called (BGSA) [52]. In addition, a salps algorithm is used for feature selection of the chemical compound activities [53]. A binary version of particle swarm optimization (BPSO) is proposed [54]. A binary whale optimization algorithm for feature selection [55,56,57] has also been introduced. As the NO Free Lunch (NFL) theorm states, there is no algorithm that is able to solve all optimization problems. Hence, if an algorithm shows a superior performance on a class of problem, it cannot show the same performance on other classes. This is the motivation of our presented study, in which we propose two novel binary variants of the whale optimization algorithm (WOA) called bWOA-S and bWOA-V. In this regard, the WOA is a nature-inspired population-based metaheuristics optimization algorithm, which simulates the humpback whales’ social behavior [26]. The original WOA was modified in this paper for solving FS issues. The two proposed variants are (1) the binary whale optimization algorithm using S-shaped transfer function (bWOA-S) and (2) the binary whale optimization algorithm using V-shaped transfer function (bWOA-V). In both approaches, the accuracy of K-NN classifier [58] is used as an objective function that must be maximized. K-NN with leave-one-out cross-validation (LOOCV) based on Euclidean distance is also used to investigate the performance of the compared algorithms. The experiments results were evaluated on 24 datasets from UCI repository [59]. The results of the two proposed algorithms were evaluated versus different well-known algorithms famous in this domain, namely (1) particle swarm optimizer (PSO) [60], (2) three versions of binary ant lion (bALO1), bALO2, and bALO3) [51], (3) binary gray wolf Optimizer bGWO [50], (4) binary dragonfly [61] and (5) the original WOA. The reason behind choosing such algorithms is that PSO, one of the most famous and well-know algorithms, as well as bALO, bGWO, and bDA, are recent algorithms whose performance has been proved to be significant. Hence, we have implemented the compared algorithms using the original studies and then generated new results using these methods under the same circumstances. The experimental results revealed that bWOA-S and bWOA-V achieved higher classification accuracy with better feature reduction than the compared algorithms.
Therefore, the merits of the proposed algorithms versus the previous algorithms is illustrated by the following two aspects. First, bWOA-S and bWOA-V confirms not only feature reduction, but also the selection of relevant features. Second, bWOA-S and bWOA-V utilize the wrapper methods search technique for selecting prominent features, and hence the idea of these rules is based mainly on high classification accuracy regardless of a large number of selected features. The purpose of wrapper method is used to maintain an efficient balance between exploitation and exploration, so correct information of the features is provided [62]. Thus, bWOA-S and bWOA-V achieve a strong search capability that helps to select a minimum number of features as a subset from the most significant features pool.
The rest of the paper is organized as follows: Section 2 briefly introduces the WOA. Section 3, describes the two binary versions of whale optimization algorithm (bWOA), namely bWOA-S and bWOA-V, for feature selection. Section 4, discusses the empirical results for bWOA-S and bWOA-V. Eventually, conclusions and future work are drawn in Section 5.
2. Whale Optimization Algorithm
In [26], Mirjalili et al. introduced the whale optimization algorithm (WOA), based on the behaviour of whales. The special hunting method is considered the most interesting behaviour of humpack whales. This hunting technique is called bubble-net feeding. In the classical WOA, the solution of the current best candidate is set as close to either the optimum or the target prey. The other whales will update their position towards the best. Mathematically, the WOA mimics the collective movements as follows
where t refers to the current number of iterations, X refers to the position vector, is the best solution position vector. C and A are coefficient vectors and can be calculated from the following equations
where r belongs to the interval [0, 1] and a decreases linearly through the iterations from 2 to 0. WOA has two different phases: exploitation (Intensification) and exploration (diversification). In the diversification phase, the agents are moved for exploring or searching different search space regions, while in the intensification phase, the agents move in order to locally enhance the current solutions.
The intensification phase: the intensification phase is divided into two processes: the first one is the shrinking encircling technique which can be obtained by reducing a values using Equation (4). Note that a is a stochastic value in the interval . The second phase is the spiral updating position in which the distance between the whale and the prey is calculated. To model a spiral movement, the following equation is used in order to mimic the movement of the helix-shaped.
From Equation (5), l is a randomly chosen value between where b is a fixed. A 50% probability is used for choosing either the spiral model or shrinking encircling mechanism, as assumed. Consequently, the mathematical model is established as follows
where p is a random number in a uniform distribution.
The exploration phase: In the exploration phase, A used random values within to force the agent to move away from this location mathematically, formulated as in Equation (7).
3. Binary Whale Optimization Algorithm
In the classical WOA, whales move inside the continous search space in order to modify their positions, and this is called the continuous space. However, to solve FS issues, the solutions are limited to only values. In order to be able to solve feature selection problems, the continuous (free position) must be converted to their corresponding binary solutions. Therefore, two binary versions from WOA are introduced to investigate problems like FS and achieve superior results. The conversion is performed by applying specific transfer functions, either the S-shaped function or V-shaped function in each dimension [63]. Transfer functions show the probability of converting the position vectors’ from 0 to 1 and vice versa, i.e., force the search agents to move in a binary space. Figure 1 demonstrates the flow chart of the binary WOA version. Algorithm 1 shows the pseudo code of the proposed bWOA-S and bWOA-V versions.
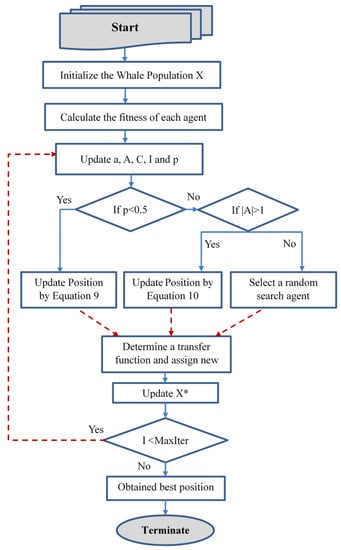
Figure 1.
Binary whale optimization algorithm flowchart.
3.1. Approach 1: Proposed bWOA-S
The common S-shaped (Sigmoid) function is used in this version. The S-shaped function is updating, as shown in Equation (11). Figure 2 illustrates the mathematical curve of the Sigmoid function.
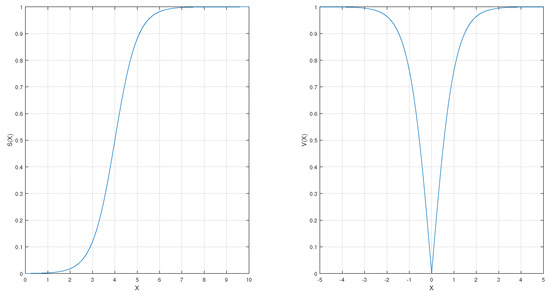
Figure 2.
S-shaped and V-shaped transfer functions.
3.2. Approach 2: Proposed bWOA-V
In this version, the hyperbolic tan function is applied. It is a common example of V-shaped functions and is given in Equations (9) and (10).
| Algorithm 1 Pseudo code of bWOA-S & bWOA-V |
|
3.3. bWOA-S and bWOA-V for Feature Selection
Two binary variants of whale optimization algorithm, called bWOA-S and bWOA-V, are employed for solving the FS problem. For a feature vector size, if N is the number of different features, then the combination number would be , which is a huge feature number to search exhaustively. Under such a situation, the proposed bWOA-S and bWOA-V algorithms are used in an adaptive feature space search and provide the best combination of features. This combination is obtained by achieving the maximum classification accuracy and the minimum selected features number. The following Equation (12) shows the fitness function accompanied by the two proposed versions to evaluate individual whale positions.
where F refers to Fitness function, R refers to the length of the selected feature subset, C refers to the total features number, refers to the classification accuracy of the condition attribute set R, and are two arguments that are symmetric to the subset length and the accuracy of the classification, and can be calculated as and . This will lead to the fitness function that achieves the maximum classification accuracy. Equation (12) can be converted to a minimization problem based on the error rate of classification and selected features. Thus, the obtained minimization problem can be calculated as in Equation (13)
where F refers to Fitness function, is the classification error rate. According to the wrapper methods characteristic in FS, the classifier was employed as an FS guide. In this study, K-NN classifier is used. Therefore, K-NN is applied to ensure that the selected features are the most relevant ones. However, bWOA is the search method that tries to explore the feature space in order to maximize the feature evaluation criteria, as shown in Equation (13).
4. Experimental Results and Discussion
The two proposed bWOA-S and bWOA-V methods are compared with a group of existing algorithms, including the PSO, three variants of binary ant lion (bALO1, bALO2, and bALO3), and the original WOA. Table 1 reports the parameter settings for the cometitior algorithms. In order to provide a fair comparison, three initialization scenarios are used and the experimental results are performed using 24 different datasets from the UCI repository.

Table 1.
Parameter setting.
4.1. Data Acquisition
Table 2 summarizes the 24 datasets from the UCI machine learning repository [59] that were used in the experiments. The datasets were selected with different instances and attribute numbers to represent various kinds of issue (small, medium and large). In each repository, the instances are divided randomly into three different subsets, namely training, testing, and validation subsets. The proposed algorithms were tested over three gene expression datasets of colon cancer, lymphoma and the leukemia [64,65,66]. The K-NN is used in the experimental tests using the trial and error method, and 5 is the best choice of K. Meanwhile, every position of whale produces one attribute subset through the training process. The training set is used to test and evaluate the performance of the K-NN classifier in the validation subset throughout the optimization process. The bWOA is employed to simultaneously guide the FS process.

Table 2.
List of datasets used in the experiments results.
4.2. Evaluation Criteria
Each algorithm carried out 20 independent runs with a random initial positioning of the search agents. Repeated runs were used to test the capability of the convergence. Eight well-known and common measures are recorded in order to investigate the algorithms performance in a comparative way. Such metrics are listed as follows:
- Best: The minimum (or best for a minimization problem) fitness function value obtained at different independent runs, as depicted in Equation (14).
- Worst: The maximum (or worst for a minimization) fitness function value obtained at different independent operations, as shown in Equation (15).
- Mean: Average calculation performance of the optimization algorithm applied M times, as shown in Equation (16).where is the optimal solution obtained in the i-th operation;
- Standard deviation (Std) can be calculated from the following Equation (17).
- Average classification accuracy: Investigates the accuracy of the classifier and can be calculated by Equation (18).where refers to classifier output for instance i; N refers to the instance number in the test set; and refers to the reference class corresponding to instance i;
- Average selection size (Avg-Selection) measures the average reduction in selected features from all feature sets and is calculated by Equation (19)where is the total number of features in the original dataset;
- Average execution time (Avg-Time) measures the average execution time in milliseconds for all comparison optimization algorithms to obtain the results over the different runs and calculated by Equation (20)where M refers to the run number for the optimizer a, and is the computational time for optimizer a in milliseconds at run number i;
- Wilcoxon rank sum test (Wilcoxon): a non-parametric test called Wilcoxon Rank Sum (WRS) [67]. The test gives ranks to all the scores in one group, and after that the ranks of each group are added. The rank-sum test is often described as the non-parametric version of the t test for two independent groups.
The two proposed versions of whale optimization algorithm (bWOA-S and bWOA-V) are compared with three common algorithms that are famous in this domain. Four different initialization methods/techniques are used to guarantee the two proposed algorithms’ ability to converge from different initial positions. These methods are: (1) a large initialization is expected to evaluate the capability of locally searching a given algorithm, as the search agents’ positions are commonly close to the optimal solution; (2) a small initialization method is expected to evaluate the ability of a given algorithm to use global searching as the initial search; (3) mixed initialization is the case in which some search agents are close enough to the optimal solution, whereas the other search agents are apart. It will provide diversity of the population frequently. since the search agents are expected to be apart from each other. (4) random initialization.
4.3. Performance on Small Initialization
The statistical average fitness values of the different datasets obtained from the compared algorithms using the small initialization methods are shown in Table 3. Table 4 shows average classification accuracy on the test data of the compared algorithms using small initialization methods. From these tables, we can conclude that both bWOA-S and bWOA-V achieve better results compared with other algorithms.

Table 3.
Statistical mean fitness measure on the different datasets calculated for the compared algorithms using small initialization.

Table 4.
Average classification accuracy for the compared algorithms on the different datasets using small initialization.
4.4. Performance on Large Initialization
The statistical average fitness values of the different datasets obtained from the compared algorithms using the large initialization methods are shown in Table 5. Table 6 shows average classification accuracy of the test data of the compared algorithms using small initialization methods. From these tables, we can conclude that when using large initialization methods, both bWOA-S and bWOA-V achieve better results compared with other algorithms.

Table 5.
Statistical mean fitness measure calculated on the different datasets for the compared algorithms using large initialization.

Table 6.
Average classification accuracy on the different datasets for the compared algorithms using large initialization.
4.5. Performance on Mixed Initialization
The statistical average fitness values on the different datasets obtained from the compared algorithms using the large initialization methods are shown in Table 7. Table 8 shows average classification accuracy of the test data of the compared algorithms using small initialization methods. As is notable from this table, we can conclude that both bWOA-S and bWOA-V achieve better results compared with other algorithms.

Table 7.
Statistical mean fitness measure calculated on the different datasets for the compared algorithms using mixed initialization.

Table 8.
Average classification accuracy on the different datasets for the compared algorithms using mixed initialization.
4.6. Discussion
Figure 3 shows the effect of the initialization method on the different optimizers applied over the selected datasets. The proposed bWOA-S and bWOA-V can reach the global optimal solution in almost half of the datasets, compared to the algorithms in all initialization methods. The limited search space in the case of binary algorithms explains the enhanced performance due to the balance between global and local searching. The balance between local and global searching assists the optimization algorithm to avoid early convergence and local optimal values. The small initialization keeps away the initial search agents from the optimal solution; however, in the large initialization, the search agents are closest to the optimal solution, although they have low diversity. While the mixed initialization method improves the performance of all compared algorithms, the two proposed algorithms are superior even in a high-dimensional dataset as in Table 9.
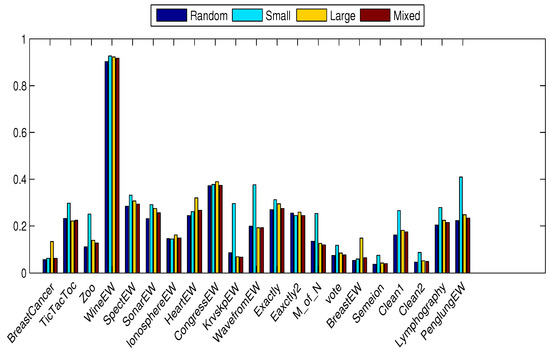
Figure 3.
Statistical mean fitness averaged on the different datasets for the different optimizers using the different initializers.

Table 9.
Results for high dimensional datasets.
The standard deviation in the obtained fitness values on the different datasets for the compared algorithms averaged over the initialization methods is given in Table 10. As shown in this table, the proposed bWOA-V can reach the optimal solution better than compared algorithms, regardless of the initialization used.

Table 10.
Standard deviation fitness function on the different datasets averaged for the compared algorithms over the three initialization methods.
With regard to the time consumption for optimization of these 11 test datasets, Table 11 presents the results of the average time obtained by the two proposed versions and other compared algorithms with 20 independent runs. As can be concluded from Table 11, bWOA-V ranks first among the algorithms. bWOA-S ranks fifth, but it is better than PSO and bALO, as it significantly outperforms the other compared algorithms with a little more time consumption.

Table 11.
Average execution time in seconds on the different datasets for the compared algorithms averaged over the three initialization methods.
On the other hand, Table 12 and Table 13 summarize the experimental results of the best and worst obtained fitness for the compared algorithms over 20 independent runs.

Table 12.
Best fitness function on the different datasets averaged for the compared algorithms over the three initialization methods.

Table 13.
Worst fitness function on the different datasets averaged for the compared algorithms over the three initialization methods.
The mean selected features obtained from the compared algorithms are shown in Table 14.

Table 14.
Average selection size on the different datasets averaged for the compared algorithms over the three initialization methods.
Table 14 reports the ratio of mean selected features obtained from the compared algorithms. In Table 14, the performance of bWOA-V is superior in keeping its good classification accuracy by selecting a lower number of features.
This reveals the outstanding performance of bWOA-V in searching for both features’ reduction and enhancing the optimization process.
In order to compare each runs results, a non-parametric statistical called Wilcoxon’s rank sum (WRS) test was carried out over the 11 UCI datasets at 5% significance level, and the p-values are given in Table 15. From this table, p-values for the bWOA-V are mostly less than 0.05, which proves that this algorithm’s superiority is statistically significant. This means that bWOA-V exhibits a statistically superior performance compared to the other compared algorithms in the pair-wise Wilcoxon signed-ranks test.

Table 15.
The Wilcoxon test for the average fitness obtained by the compared algorithms.
Moreover, Figure 4 outlines the best and worst acquired fitness function value averaged over all the datasets, using small, mixed and large initialization. Figure 5 shows the classification accuracy average. From these figures, it can be proven that the bWOA-V performs better than other compared algorithms, such as PSO and bALO, which confirms bWOA-V’s searching capability, especially in the large initialization.
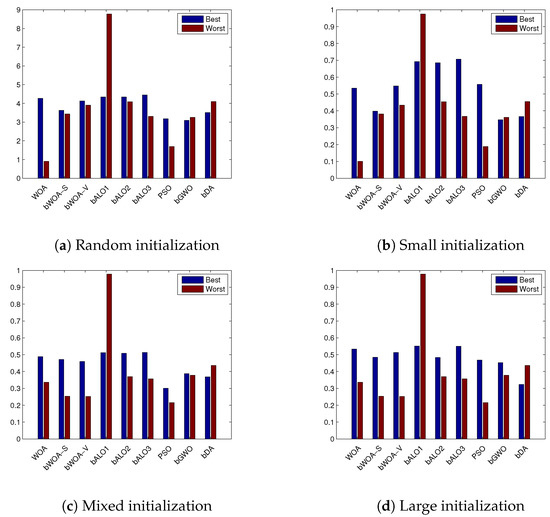
Figure 4.
Best and worst fitness obtained for the compared algorithms on the different datasets averaged over the four initialization methods.
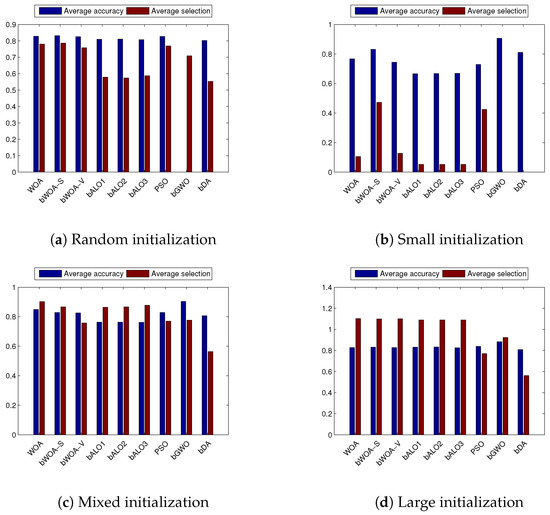
Figure 5.
Average classification accuracy and average selection size obtained on the different datasets averaged for the compared algorithms over the three initialization methods.
In order to show the merits of bWOA-S and bWOA-V qualitatively, Figure 6, Figure 7 and Figure 8, show the boxplots results for the three initialization methods obtained by all compared algorithms. According to these figures, bWOA-S and bWOA-V have superiority since the boxplot of bWOA-S and bWOA-V are extremely narrow and located under the minima of PSO, bALO, and the original WOA. In summary, the qualitative results prove that the two proposed algorithms are able to provide remarkable convergence and coverage ability in solving FS problems. Another fact worth mentioning here is that the boxplots show that bALO and PSO algorithms provide poor performance.
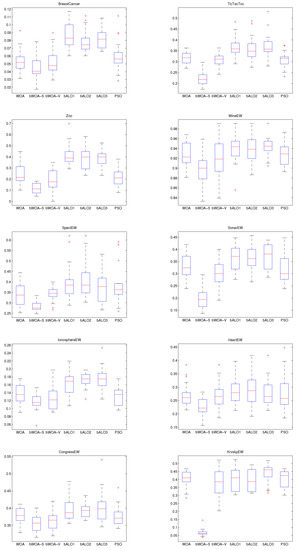
Figure 6.
Small initialization boxplot for the compared algorithms on the different datasets.
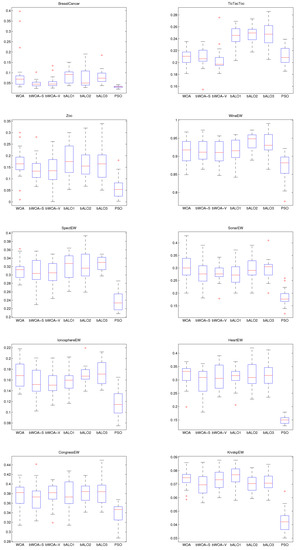
Figure 7.
Mixed initialization boxplot for the compared algorithms on the different datasets.
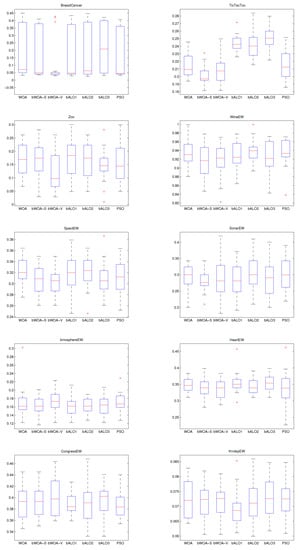
Figure 8.
Large initialization boxplot for the compared algorithms on the different datasets.
5. Conclusions and Future Work
In this paper, two binary version of the original whale optimization algorithm (WOA), called bWOA-S and bWOA-V, have been proposed to solve the FS problem. To convert the original version of WOA to a binary version, S-shaped and V-shaped transfer functions are employed. In order to investigate the performance of the two proposed algorithms, the experiments employ 24 benchmark datasets from the UCI repository and eight evaluation criteria to assess different aspects of the compared algorithms.The experimental results revealed that the two proposed algorithms achieved superior results compared to the three well-known algorithms, namely PSO, bALO (three variants), and the original WOA. Furthermore, the results proved that bWOA-S and bWOA-V both achieved smallest number of selected features with best classification accuracy in a minimum time. In addition, the Wilcoxon’s rank-sum nonparametric statistical test was carried out at 5% significance level to judge whether the results of the two proposed algorithms differ from the best results of the other compared algorithms in a statistically significant way. More specifically, the results proved that the bWOA-s and bWOA-V have merit among binary optimization algorithms. For future work, the two binary algorithms introduced here will be applied to high-dimensional real-world applications and will be used with more common classifiers such as SVM and ANN to verify the performance. The effects of different transfer functions on the performance of the two proposed algorithms are also worth investigating. This algorithm can be applied for many problems other than FS. We can also investigate a multi-objective version.
Author Contributions
A.G.H.: Software, Resources, Writing—original draft, editing. D.O.: Conceptualization, Data curation, Resources, Writing—review and editing. E.H.H.: Supervision, Methodology, Conceptualization, Formal analysis, Writing—review and editing. A.A.J.: Formal analysis, Writing—review and editing. X.Y.: Formal analysis, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external fundin.
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- Yi, J.H.; Deb, S.; Dong, J.; Alavi, A.H.; Wang, G.G. An improved NSGA-III algorithm with adaptive mutation operator for Big Data optimization problems. Future Gener. Comput. Syst. 2018, 88, 571–585. [Google Scholar] [CrossRef]
- Neggaz, N.; Houssein, E.H.; Hussain, K. An efficient henry gas solubility optimization for feature selection. Expert Syst. Appl. 2020, 152, 113364. [Google Scholar] [CrossRef]
- Sayed, S.A.F.; Nabil, E.; Badr, A. A binary clonal flower pollination algorithm for feature selection. Pattern Recognit. Lett. 2016, 77, 21–27. [Google Scholar] [CrossRef]
- Martin-Bautista, M.J.; Vila, M.A. A survey of genetic feature selection in mining issues. In Proceedings of the 1999 Congress on Evolutionary Computation-CEC99 (Cat. No. 99TH8406), Washington, DC, USA, 6–9 July 1999; Volume 2, pp. 1314–1321. [Google Scholar]
- Piramuthu, S. Evaluating feature selection methods for learning in data mining applications. Eur. J. Oper. Res. 2004, 156, 483–494. [Google Scholar] [CrossRef]
- Gunal, S.; Edizkan, R. Subspace based feature selection for pattern recognition. Inf. Sci. 2008, 178, 3716–3726. [Google Scholar] [CrossRef]
- Lew, M.S. Principles of Visual Information Retrieval; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Wang, G.G.; Tan, Y. Improving metaheuristic algorithms with information feedback models. IEEE Trans. Cybern. 2017, 49, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Houssein, E.H.; Hosney, M.E.; Elhoseny, M.; Oliva, D.; Mohamed, W.M.; Hassaballah, M. Hybrid Harris hawks optimization with cuckoo search for drug design and discovery in chemoinformatics. Sci. Rep. 2020, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Houssein, E.H.; Hosney, M.E.; Oliva, D.; Mohamed, W.M.; Hassaballah, M. A novel hybrid Harris hawks optimization and support vector machines for drug design and discovery. Comput. Chem. Eng. 2020, 133, 106656. [Google Scholar] [CrossRef]
- Gao, D.; Wang, G.G.; Pedrycz, W. Solving fuzzy job-shop scheduling problem using de algorithm improved by a selection mechanism. IEEE Trans. Fuzzy Syst. 2020. [Google Scholar] [CrossRef]
- Houssein, E.H.; Saad, M.R.; Hussain, K.; Zhu, W.; Shaban, H.; Hassaballah, M. Optimal sink node placement in large scale wireless sensor networks based on Harris’ hawk optimization algorithm. IEEE Access 2020, 8, 19381–19397. [Google Scholar] [CrossRef]
- Ahmed, M.M.; Houssein, E.H.; Hassanien, A.E.; Taha, A.; Hassanien, E. Maximizing lifetime of large-scale wireless sensor networks using multi-objective whale optimization algorithm. Telecommun. Syst. 2019, 72, 243–259. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Liu, H.; Yu, L. Toward integrating feature selection algorithms for classification and clustering. IEEE Trans. Knowl. Data Eng. 2005, 17, 491–502. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Liu, H.; Setiono, R. A probabilistic approach to feature selection-a filter solution. In Proceedings of the 13th International Conference on Machine Learning, Bari, Italy, 3–6 July 1996; Volume 23, pp. 319–327. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2002. [Google Scholar]
- Safavian, S.R.; Landgrebe, D. hastie2002elements. IEEE Trans. Syst. Man, Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef]
- Dasarathy, B.V. Nearest Neighbor ({NN}) Norms:{NN} Pattern Classification Techniques; IEEE Computer Society Press: Washington, DC, USA, 1991. [Google Scholar]
- Verikas, A.; Bacauskiene, M. Feature selection with neural networks. Pattern Recognit. Lett. 2002, 23, 1323–1335. [Google Scholar] [CrossRef]
- Vapnik, V.N.; Vapnik, V. Statistical Learning Theory; Wiley: New York, NY, USA, 1998; Volume 1. [Google Scholar]
- Khalid, S.; Khalil, T.; Nasreen, S. A survey of feature selection and feature extraction techniques in machine learning. In Proceedings of the Science and Information Conference (SAI), London, UK, 27–29 August 2014; pp. 372–378. [Google Scholar]
- Wang, G.G.; Guo, L.; Gandomi, A.H.; Hao, G.S.; Wang, H. Chaotic krill herd algorithm. Inf. Sci. 2014, 274, 17–34. [Google Scholar] [CrossRef]
- Hassanien, A.E.; Emary, E. Swarm Intelligence: Principles, Advances, and Applications; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Hussien, A.G.; Amin, M.; Abd El Aziz, M. A comprehensive review of moth-flame optimisation: Variants, hybrids, and applications. J. Exp. Theor. Artif. Intell. 2020, 32, 705–725. [Google Scholar] [CrossRef]
- Assiri, A.S.; Hussien, A.G.; Amin, M. Ant Lion Optimization: Variants, hybrids, and applications. IEEE Access 2020, 8, 77746–77764. [Google Scholar] [CrossRef]
- Hussien, A.G.; Amin, M.; Wang, M.; Liang, G.; Alsanad, A.; Gumaei, A.; Chen, H. Crow Search Algorithm: Theory, Recent Advances, and Applications. IEEE Access 2020, 8, 173548–173565. [Google Scholar] [CrossRef]
- Shareef, H.; Ibrahim, A.A.; Mutlag, A.H. Lightning search algorithm. Appl. Soft Comput. 2015, 36, 315–333. [Google Scholar] [CrossRef]
- Hashim, F.A.; Houssein, E.H.; Mabrouk, M.S.; Al-Atabany, W.; Mirjalili, S. Henry gas solubility optimization: A novel physics-based algorithm. Future Gener. Comput. Syst. 2019, 101, 646–667. [Google Scholar] [CrossRef]
- Houssein, E.H.; Saad, M.R.; Hashim, F.A.; Shaban, H.; Hassaballah, M. Lévy flight distribution: A new metaheuristic algorithm for solving engineering optimization problems. Eng. Appl. Artif. Intell. 2020, 94, 103731. [Google Scholar] [CrossRef]
- Hashim, F.A.; Houssein, E.H.; Hussain, K.; Mabrouk, M.S.; Al-Atabany, W. A modified Henry gas solubility optimization for solving motif discovery problem. Neural Comput. Appl. 2020, 32, 10759–10771. [Google Scholar] [CrossRef]
- Fernandes, C.; Pontes, A.; Viana, J.; Gaspar-Cunha, A. Using multiobjective evolutionary algorithms in the optimization of operating conditions of polymer injection molding. Polym. Eng. Sci. 2010, 50, 1667–1678. [Google Scholar] [CrossRef]
- Gaspar-Cunha, A.; Covas, J.A. RPSGAe—Reduced Pareto set genetic algorithm: Application to polymer extrusion. In Metaheuristics for Multiobjective Optimisation; Springer: Berlin/Heidelberg, Germany, 2004; pp. 221–249. [Google Scholar]
- Avalos, O.; Cuevas, E.; Gálvez, J.; Houssein, E.H.; Hussain, K. Comparison of Circular Symmetric Low-Pass Digital IIR Filter Design Using Evolutionary Computation Techniques. Mathematics 2020, 8, 1226. [Google Scholar] [CrossRef]
- Xue, B.; Zhang, M.; Browne, W.N.; Yao, X. A survey on evolutionary computation approaches to feature selection. IEEE Trans. Evol. Comput. 2016, 20, 606–626. [Google Scholar] [CrossRef]
- Kabir, M.M.; Shahjahan, M.; Murase, K. A new hybrid ant colony optimization algorithm for feature selection. Expert Syst. Appl. 2012, 39, 3747–3763. [Google Scholar] [CrossRef]
- Ghaemi, M.; Feizi-Derakhshi, M.R. Feature selection using forest optimization algorithm. Pattern Recognit. 2016, 60, 121–129. [Google Scholar] [CrossRef]
- Emary, E.; Zawbaa, H.M.; Ghany, K.K.A.; Hassanien, A.E.; Parv, B. Firefly optimization algorithm for feature selection. In Proceedings of the 7th Balkan Conference on Informatics Conference, Craiova, Romania, 2–4 September 2015; p. 26. [Google Scholar]
- Mafarja, M.M.; Mirjalili, S. Hybrid Whale Optimization Algorithm with simulated annealing for feature selection. Neurocomputing 2017, 260, 302–312. [Google Scholar] [CrossRef]
- Xue, B.; Zhang, M.; Browne, W.N. Particle swarm optimization for feature selection in classification: A multi-objective approach. IEEE Trans. Cybern. 2013, 43, 1656–1671. [Google Scholar] [CrossRef]
- Hafez, A.I.; Zawbaa, H.M.; Emary, E.; Hassanien, A.E. Sine cosine optimization algorithm for feature selection. In Proceedings of the 2016 International Symposium on INnovations in Intelligent Systems and Applications (INISTA), Sinaia, Romania, 2–5 August 2016; pp. 1–5. [Google Scholar]
- Wang, G.G.; Deb, S.; Cui, Z. Monarch butterfly optimization. Neural Comput. Appl. 2019, 31, 1995–2014. [Google Scholar] [CrossRef]
- Wang, G.G. Moth search algorithm: A bio-inspired metaheuristic algorithm for global optimization problems. Memetic Comput. 2018, 10, 151–164. [Google Scholar] [CrossRef]
- Rodrigues, D.; Yang, X.S.; De Souza, A.N.; Papa, J.P. Binary flower pollination algorithm and its application to feature selection. In Recent Advances in Swarm Intelligence and Evolutionary Computation; Springer: Berlin/Heidelberg, Germany, 2015; pp. 85–100. [Google Scholar]
- Nakamura, R.Y.; Pereira, L.A.; Costa, K.; Rodrigues, D.; Papa, J.P.; Yang, X.S. BBA: A binary bat algorithm for feature selection. In Proceedings of the 2012 25th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Ouro Preto, Brazil, 22–25 August 2012; pp. 291–297. [Google Scholar]
- Rodrigues, D.; Pereira, L.A.; Almeida, T.; Papa, J.P.; Souza, A.; Ramos, C.C.; Yang, X.S. BCS: A binary cuckoo search algorithm for feature selection. In Proceedings of the 2013 IEEE International Symposium on Circuits and Systems (ISCAS), Beijing, China, 19–23 May 2013; pp. 465–468. [Google Scholar]
- He, X.; Zhang, Q.; Sun, N.; Dong, Y. Feature selection with discrete binary differential evolution. In Proceedings of the 2009 International Conference on Artificial Intelligence and Computational Intelligence, Shanghai, China, 7–8 November 2009; Volume 4, pp. 327–330. [Google Scholar]
- Emary, E.; Zawbaa, H.M.; Hassanien, A.E. Binary ant lion approaches for feature selection. Neurocomputing 2016, 213, 54–65. [Google Scholar] [CrossRef]
- Emary, E.; Zawbaa, H.M.; Hassanien, A.E. Binary grey wolf optimization approaches for feature selection. Neurocomputing 2016, 172, 371–381. [Google Scholar] [CrossRef]
- Rashedi, E.; Nezamabadi-Pour, H.; Saryazdi, S. BGSA: Binary gravitational search algorithm. Nat. Comput. 2010, 9, 727–745. [Google Scholar] [CrossRef]
- Hussien, A.G.; Hassanien, A.E.; Houssein, E.H. Swarming behaviour of salps algorithm for predicting chemical compound activities. In Proceedings of the 2017 Eighth International Conference on Intelligent Computing and Information Systems (ICICIS), Cairo, Egypt, 5–7 December 2017; pp. 315–320. [Google Scholar]
- Nezamabadi-pour, H.; Rostami-Shahrbabaki, M.; Maghfoori-Farsangi, M. Binary particle swarm optimization: Challenges and new solutions. CSI J. Comput. Sci. Eng. 2008, 6, 21–32. [Google Scholar]
- Hussien, A.G.; Houssein, E.H.; Hassanien, A.E. A binary whale optimization algorithm with hyperbolic tangent fitness function for feature selection. In Proceedings of the 2017 Eighth International Conference on Intelligent Computing and Information Systems (ICICIS), Cairo, Egypt, 5–7 December 2017; pp. 166–172. [Google Scholar]
- Hussien, A.G.; Hassanien, A.E.; Houssein, E.H.; Bhattacharyya, S.; Amin, M. S-shaped Binary Whale Optimization Algorithm for Feature Selection. In Recent Trends in Signal and Image Processing; Springer: Berlin/Heidelberg, Germany, 2019; pp. 79–87. [Google Scholar]
- Hussien, A.G.; Hassanien, A.E.; Houssein, E.H.; Amin, M.; Azar, A.T. New binary whale optimization algorithm for discrete optimization problems. Eng. Optim. 2020, 52, 945–959. [Google Scholar] [CrossRef]
- Chuang, L.Y.; Chang, H.W.; Tu, C.J.; Yang, C.H. Improved binary PSO for feature selection using gene expression data. Comput. Biol. Chem. 2008, 32, 29–38. [Google Scholar] [CrossRef]
- Dua, D.; Graff, C. UCI Machine Learning Repository. 2017. Available online: http://archive.ics.uci.edu/ml (accessed on 28 September 2020).
- Eberhart, R.; Kennedy, J. A new optimizer using particle swarm theory. In Proceedings of the Sixth International Symposium on Micro Machine and Human Science, Nagoya, Japan, 4–6 October 1995; pp. 39–43. [Google Scholar]
- Mafarja, M.M.; Eleyan, D.; Jaber, I.; Hammouri, A.; Mirjalili, S. Binary dragonfly algorithm for feature selection. In Proceedings of the 2017 International Conference on New Trends in Computing Sciences (ICTCS), Amman, Jordan, 11–13 October 2017; pp. 12–17. [Google Scholar]
- D’Angelo, G.; Palmieri, F. GGA: A modified Genetic Algorithm with Gradient-based Local Search for Solving Constrained Optimization Problems. Inf. Sci. 2020, 547, 136–162. [Google Scholar] [CrossRef]
- Mirjalili, S.; Hashim, S.Z.M. BMOA: Binary magnetic optimization algorithm. Int. J. Mach. Learn. Comput. 2012, 2, 204. [Google Scholar] [CrossRef]
- Alon, U.; Barkai, N.; Notterman, D.A.; Gish, K.; Ybarra, S.; Mack, D.; Levine, A.J. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1999, 96, 6745–6750. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Wilcoxon, F. Individual comparisons by ranking methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).