Federated Learning for Heterogeneous Multi-Site Crop Disease Diagnosis
Abstract
1. Introduction
- We propose a federated learning framework to build a centralized model that ensures the data privacy of individual farms and improves rice disease diagnosis accuracy.
- We demonstrate how farms with limited local data can collaborate in training a global disease diagnosis model, overcoming the challenges of data scarcity.
- We also illustrate how various data-sharing approaches (for instance, large-farm, pre-training, q-fair) can be utilized to improve disease detection performance.
2. Related Works
3. Materials and Methods
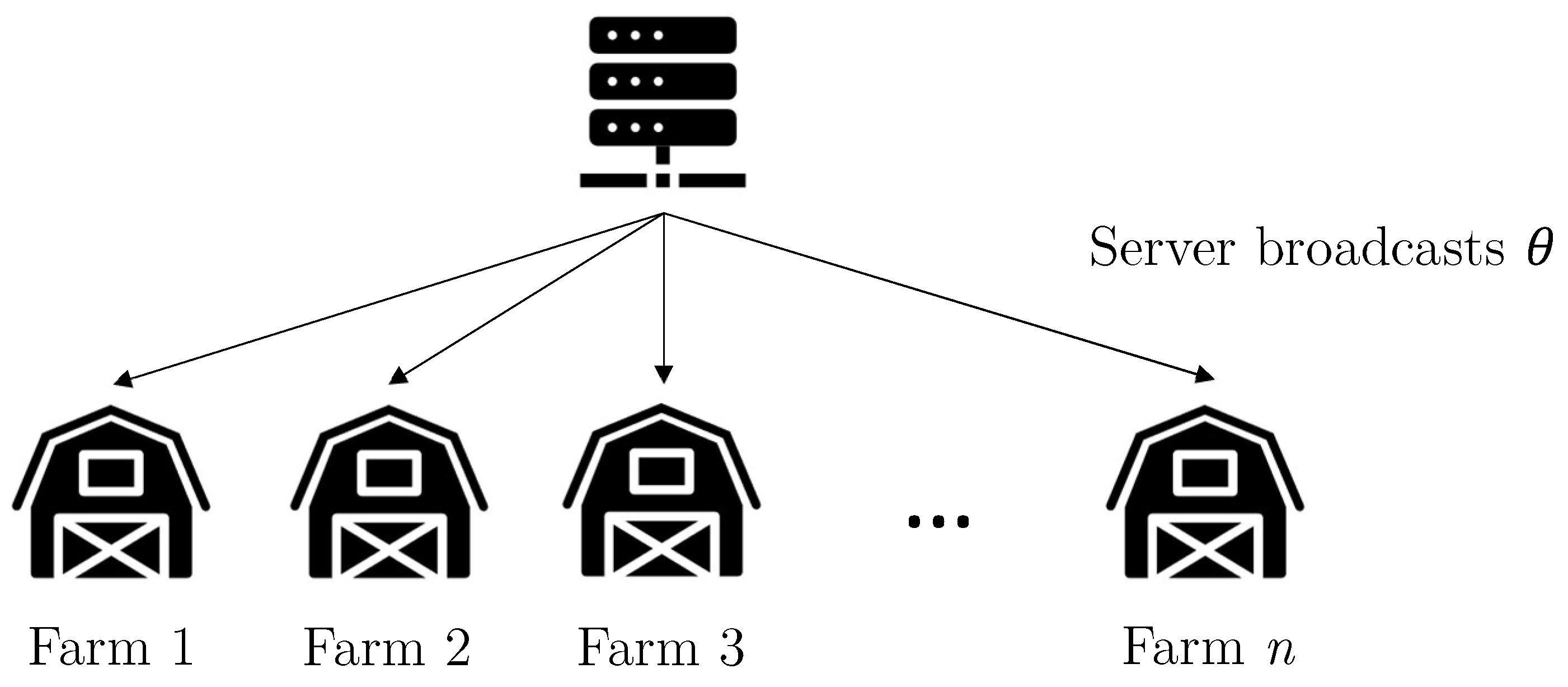
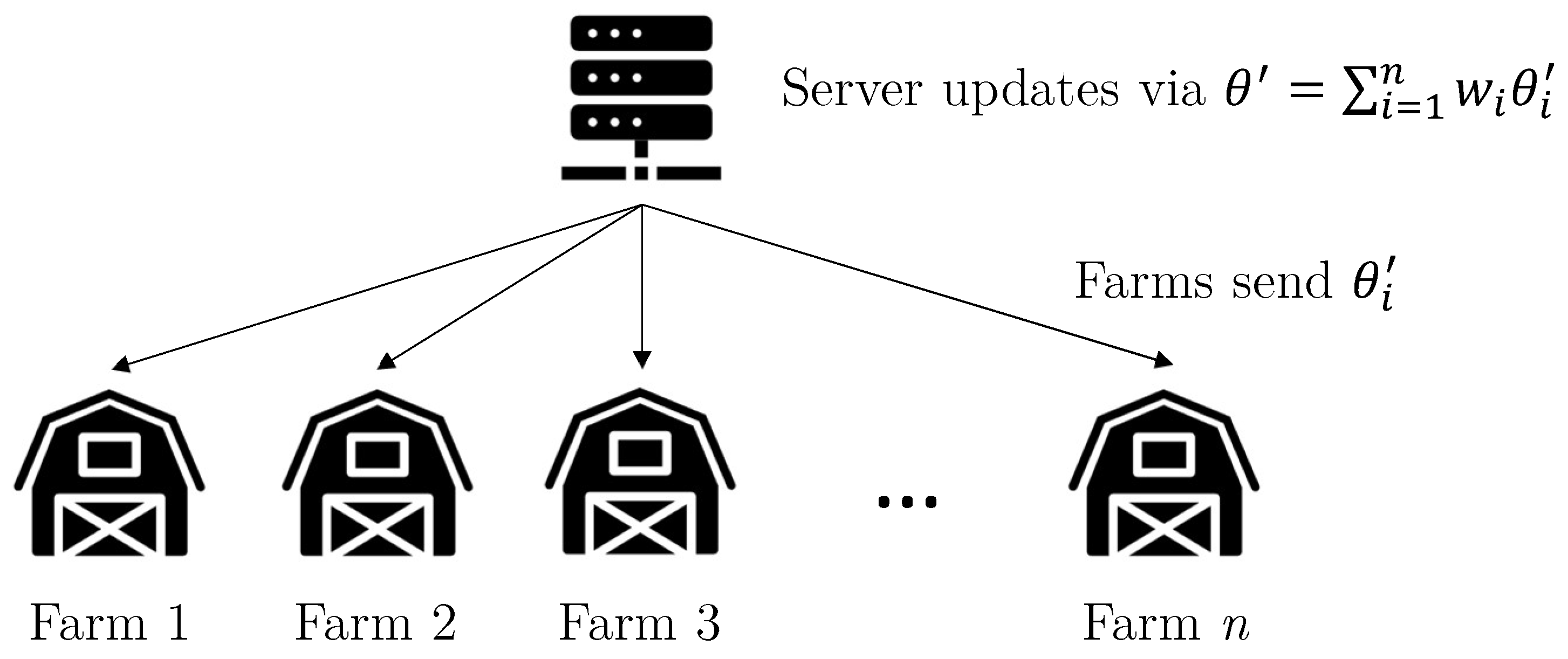
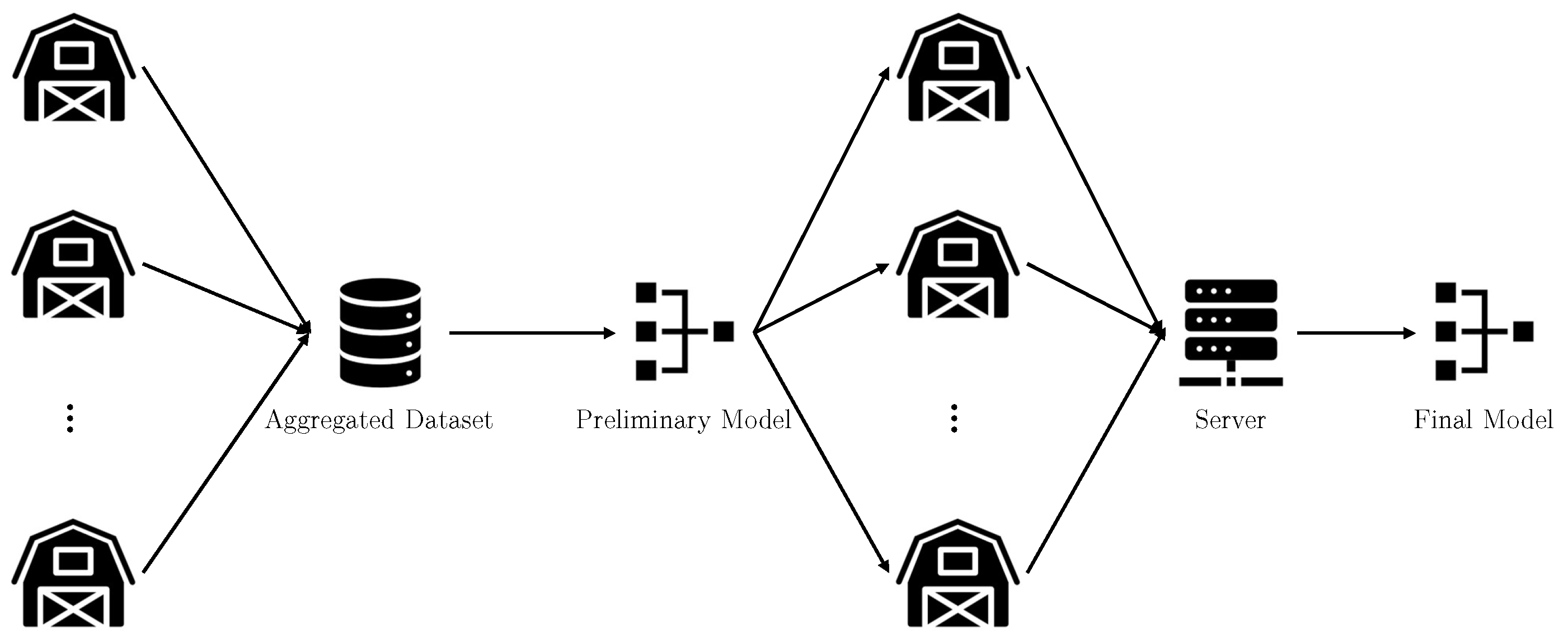
3.1. Proposed Method
3.2. Proposed Model
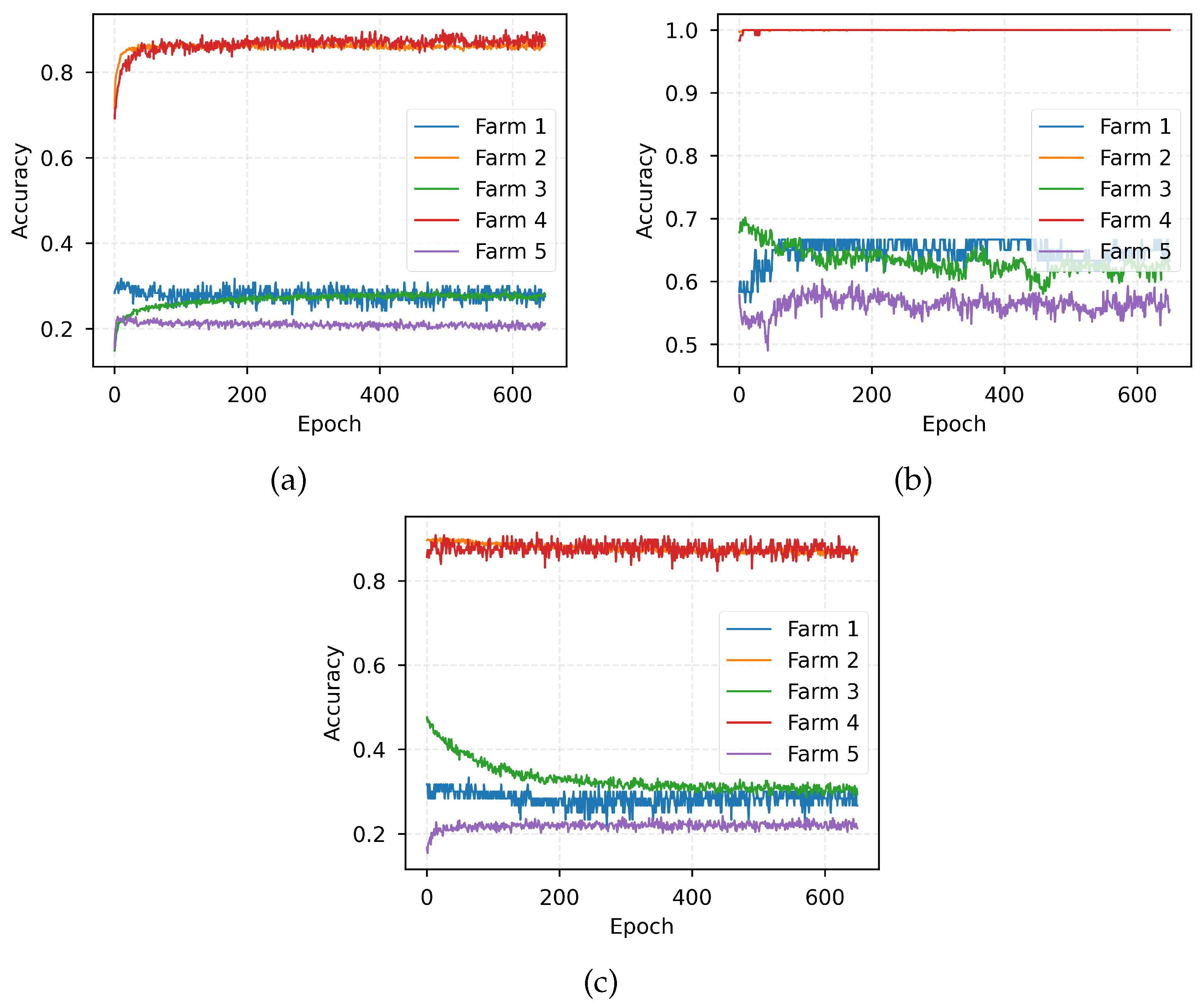
4. Experimental Results
4.1. Data Description
4.2. Implementation Details
4.3. Performance Evaluation
4.4. Performance Comparison
4.4.1. Baseline Test Without FL
4.4.2. Comparison of FL Performance
4.5. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Agrios, G. Plant Pathology, 5th ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Burlington, MA, USA, 2005; pp. 79–103. [Google Scholar]
- He, Z.; Zhang, Z.; Valè, G.; San Segundo, B.; Chen, X.; Pasupuleti, J. Disease and pest resistance in rice. Front. Plant Sci. 2023, 14, 1333904. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Yap, P.T.; Bozoki, A.; Liu, M. Federated learning for medical image analysis: A survey. Pattern Recognit. 2024, 151, 110424. [Google Scholar] [CrossRef] [PubMed]
- Shahjahan, A.; Duve, T.; Bonman, J. Climate and rice diseases. In Weather and Rice; International Rice Research Institute: Los Baños, Philippines, 1987; pp. 125–137. [Google Scholar]
- Liu, Z.; Zhao, J.; Li, Y.; Zhang, W.; Jian, G.; Peng, Y.; Qi, F. Non-uniform distribution pattern for differentially expressed genes of transgenic rice Huahui 1 at different developmental stages and environments. PLoS ONE 2012, 7, e37078. [Google Scholar] [CrossRef]
- Kaur, J.; Hazrati Fard, S.M.; Amiri-Zarandi, M.; Dara, R. Protecting farmers’ data privacy and confidentiality: Recommendations and considerations. Front. Sustain. Food Syst. 2022, 6, 903230. [Google Scholar] [CrossRef]
- Sykuta, M.E. Big data in agriculture: Property rights, privacy and competition in ag data services. Int. Food Agribus. Manag. Rev. 2016, 19, 57–74. [Google Scholar]
- Liu, Z.; Wu, F.; Wang, Y.; Yang, M.; Pan, X. FedCL: Federated contrastive learning for multi-center medical image classification. Pattern Recognit. 2023, 143, 109739. [Google Scholar] [CrossRef]
- Li, T.; Sahu, A.K.; Talwalkar, A.; Smith, V. Federated learning: Challenges, methods, and future directions. IEEE Signal Process. Mag. 2020, 37, 50–60. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Peng, Z. Rice diseases detection and classification using attention based neural network and bayesian optimization. Expert Syst. Appl. 2021, 178, 114770. [Google Scholar] [CrossRef]
- Alsakar, Y.M.; Sakr, N.A.; Elmogy, M. An enhanced classification system of various rice plant diseases based on multi-level handcrafted feature extraction technique. Sci. Rep. 2024, 14, 30601. [Google Scholar] [CrossRef]
- Chaudhary, S.; Kumar, U. An efficient approach for automated system to identify the rice crop disease using intensity level based multi-fractal dimension and twin support vector machine. Arch. Phytopathol. Plant Prot. 2023, 56, 806–834. [Google Scholar] [CrossRef]
- Azim, M.A.; Islam, M.K.; Rahman, M.M.; Jahan, F. An effective feature extraction method for rice leaf disease classification. TELKOMNIKA (Telecommun. Comput. Electron. Control) 2021, 19, 463–470. [Google Scholar] [CrossRef]
- Wu, X.; Niu, J.; Liu, X.; Shi, M.; Zhu, G.; Tang, S. Tackling Feature-Classifier Mismatch in Federated Learning via Prompt-Driven Feature Transformation. arXiv 2024, arXiv:2407.16139. [Google Scholar]
- Marí, N.E.; Agripina, R.; Shen, H.; Mafukidze, B.S. Advances, Challenges & Recent Developments in Federated Learning. Open Access Libr. J. 2024, 11, e12239. [Google Scholar]
- Shivam, S.P.S.; Kumar, I. Rice Plant Infection Recognition using Deep Neural Network Systems. In Proceedings of the International Semantic Intelligence Conference (ISIC 2021), New Delhi, India, 25–27 February 2021; pp. 25–27. [Google Scholar]
- Ramesh, S.; Vydeki, D. Recognition and classification of paddy leaf diseases using Optimized Deep Neural network with Jaya algorithm. Inf. Process. Agric. 2020, 7, 249–260. [Google Scholar] [CrossRef]
- Anandhan, K.; Singh, A.S. Detection of paddy crops diseases and early diagnosis using faster regional convolutional neural networks. In Proceedings of the 2021 International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE), Greater Noida, India, 4–5 March 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 898–902. [Google Scholar]
- Bari, B.S.; Islam, M.N.; Rashid, M.; Hasan, M.J.; Razman, M.A.M.; Musa, R.M.; Ab Nasir, A.F.; Majeed, A.P.A. A real-time approach of diagnosing rice leaf disease using deep learning-based faster R-CNN framework. PeerJ Comput. Sci. 2021, 7, e432. [Google Scholar] [CrossRef]
- Deng, R.; Tao, M.; Xing, H.; Yang, X.; Liu, C.; Liao, K.; Qi, L. Automatic diagnosis of rice diseases using deep learning. Front. Plant Sci. 2021, 12, 701038. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, V.K.; Pradhan, M.K.; Minz, S.; Thakur, M.P. Rice plant disease classification using transfer learning of deep convolution neural network. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 631–635. [Google Scholar] [CrossRef]
- Krishnamoorthy, N.; Prasad, L.N.; Kumar, C.P.; Subedi, B.; Abraha, H.B.; Sathishkumar, V. Rice leaf diseases prediction using deep neural networks with transfer learning. Environ. Res. 2021, 198, 111275. [Google Scholar]
- Li, D.; Wang, R.; Xie, C.; Liu, L.; Zhang, J.; Li, R.; Wang, F.; Zhou, M.; Liu, W. A recognition method for rice plant diseases and pests video detection based on deep convolutional neural network. Sensors 2020, 20, 578. [Google Scholar] [CrossRef]
- Hu, G.; Wu, H.; Zhang, Y.; Wan, M. A low shot learning method for tea leaf’s disease identification. Comput. Electron. Agric. 2019, 163, 104852. [Google Scholar] [CrossRef]
- Ahmad, I.; Hamid, M.; Yousaf, S.; Shah, S.T.; Ahmad, M.O. Optimizing pretrained convolutional neural networks for tomato leaf disease detection. Complexity 2020, 2020, 1–6. [Google Scholar] [CrossRef]
- Latif, M.R.; Khan, M.A.; Javed, M.Y.; Masood, H.; Tariq, U.; Nam, Y.; Kadry, S. Cotton leaf diseases recognition using deep learning and genetic algorithm. Comput. Mater. Contin. 2021, 69, 2917–2932. [Google Scholar]
- Caldeira, R.F.; Santiago, W.E.; Teruel, B. Identification of cotton leaf lesions using deep learning techniques. Sensors 2021, 21, 3169. [Google Scholar] [CrossRef]
- Patil, B.V.; Patil, P.S. Computational method for Cotton Plant disease detection of crop management using deep learning and internet of things platforms. In Evolutionary Computing and Mobile Sustainable Networks: Proceedings of ICECMSN 2020; Springer: Berlin/Heidelberg, Germany, 2021; pp. 875–885. [Google Scholar]
- Harshitha, G.; Kumar, S.; Rani, S.; Jain, A. Cotton disease detection based on deep learning techniques. In Proceedings of the 4th Smart Cities Symposium (SCS 2021), Online, 21–23 November 2021; IET: Stevenage, UK, 2021; Volume 2021, pp. 496–501. [Google Scholar]
- Wang, C.; Du, P.; Wu, H.; Li, J.; Zhao, C.; Zhu, H. A cucumber leaf disease severity classification method based on the fusion of DeepLabV3+ and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, J.; Zhao, C.; Li, J.; Teng, G.; Wu, H. Few-shot vegetable disease recognition model based on image text collaborative representation learning. Comput. Electron. Agric. 2021, 184, 106098. [Google Scholar] [CrossRef]
- Memon, M.S.; Kumar, P.; Iqbal, R. Meta Deep Learn Leaf Disease Identification Model for Cotton Crop. Computers 2022, 11, 102. [Google Scholar] [CrossRef]
- Sudhesh, K.; Sowmya, V.; Kurian, S.; Sikha, O. AI based rice leaf disease identification enhanced by Dynamic Mode Decomposition. Eng. Appl. Artif. Intell. 2023, 120, 105836. [Google Scholar]
- Aggarwal, M.; Khullar, V.; Goyal, N.; Prola, T.A. Resource-efficient federated learning over IoAT for rice leaf disease classification. Comput. Electron. Agric. 2024, 221, 109001. [Google Scholar] [CrossRef]
- Ritharson, P.I.; Raimond, K.; Mary, X.A.; Robert, J.E.; Andrew, J. DeepRice: A deep learning and deep feature based classification of Rice leaf disease subtypes. Artif. Intell. Agric. 2024, 11, 34–49. [Google Scholar] [CrossRef]
- Padhi, J.; Mishra, K.; Ratha, A.K.; Behera, S.K.; Sethy, P.K.; Nanthaamornphong, A. Enhancing Paddy Leaf Disease Diagnosis-a Hybrid CNN Model using Simulated Thermal Imaging. Smart Agric. Technol. 2025, 10, 100814. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Kakarla, S.C.; Roberts, P. Detection of target spot and bacterial spot diseases in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precis. Agric. 2020, 21, 955–978. [Google Scholar] [CrossRef]
- Konečnỳ, J.; McMahan, H.B.; Yu, F.X.; Richtárik, P.; Suresh, A.T.; Bacon, D. Federated learning: Strategies for improving communication efficiency. arXiv 2016, arXiv:1610.05492. [Google Scholar]
- McMahan, B.; Moore, E.; Ramage, D.; Hampson, S.; y Arcas, B.A. Communication-efficient learning of deep networks from decentralized data. In Proceedings of the 20th International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 20–22 April 2017; pp. 1273–1282. [Google Scholar]
- Khan, F.S.; Khan, S.; Mohd, M.N.H.; Waseem, A.; Khan, M.N.A.; Ali, S.; Ahmed, R. Federated learning-based UAVs for the diagnosis of Plant Diseases. In Proceedings of the 2022 International Conference on Engineering and Emerging Technologies (ICEET), Kuala Lumpur, Malaysia, 27–28 October 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–6. [Google Scholar]
- Deng, F.; Mao, W.; Zeng, Z.; Zeng, H.; Wei, B. Multiple diseases and pests detection based on federated learning and improved faster R-CNN. IEEE Trans. Instrum. Meas. 2022, 71, 1–11. [Google Scholar] [CrossRef]
- Li, T.; Sanjabi, M.; Beirami, A.; Smith, V. Fair resource allocation in federated learning. arXiv 2019, arXiv:1905.10497. [Google Scholar]
- Wang, J.; Chen, K.Z.; Das Gupta, S.; Huang, Z. Is small still beautiful? A comparative study of rice farm size and productivity in China and India. China Agric. Econ. Rev. 2015, 7, 484–509. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Shah, J.; Prajapati, H.; Dabhi, V. Rice Leaf Diseases. UCI Machine Learning Repository. 2019. Available online: https://doi.org/10.24432/C5R013 (accessed on 25 February 2025).
- Hossain, M.F.; Abujar, S.; Noori, S.R.H.; Hossain, S.A. Dhan-Shomadhan: A Dataset of Rice Leaf Disease Classification for Bangladeshi Local Rice. In Mendeley Data; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar] [CrossRef]
- Sethy, P.K. Rice Leaf Disease Image Samples. In Mendeley Data; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar] [CrossRef]
- Do, H.M. Rice Diseases Image Dataset. Kaggle. 2020. Available online: https://www.kaggle.com/datasets/minhhuy2810/rice-diseases-image-dataset (accessed on 25 February 2025).
- Kein, A. Rice Diseases Dataset. GitHub. 2019. Available online: https://github.com/aldrin233/RiceDiseases-dataset (accessed on 25 February 2025).
- Beutel, D.J.; Topal, T.; Mathur, A.; Qiu, X.; Fernandez-Marques, J.; Gao, Y.; Sani, L.; Li, K.H.; Parcollet, T.; de Gusmão, P.P.B.; et al. Flower: A Friendly Federated Learning Framework. 2022. Available online: https://hal.science/hal-03601230/ (accessed on 25 February 2025).
- Ye, H.; Liang, L.; Li, G.Y. Decentralized federated learning with unreliable communications. IEEE J. Sel. Top. Signal Process. 2022, 16, 487–500. [Google Scholar] [CrossRef]
- Rice, U. How Rice Grows. Available online: https://www.usarice.com/thinkrice/discover-us-rice/how-rice-grows (accessed on 7 April 2025).
- Chorney, W.; Wang, H. Towards federated transfer learning in electrocardiogram signal analysis. Comput. Biol. Med. 2024, 170, 107984. [Google Scholar] [CrossRef]
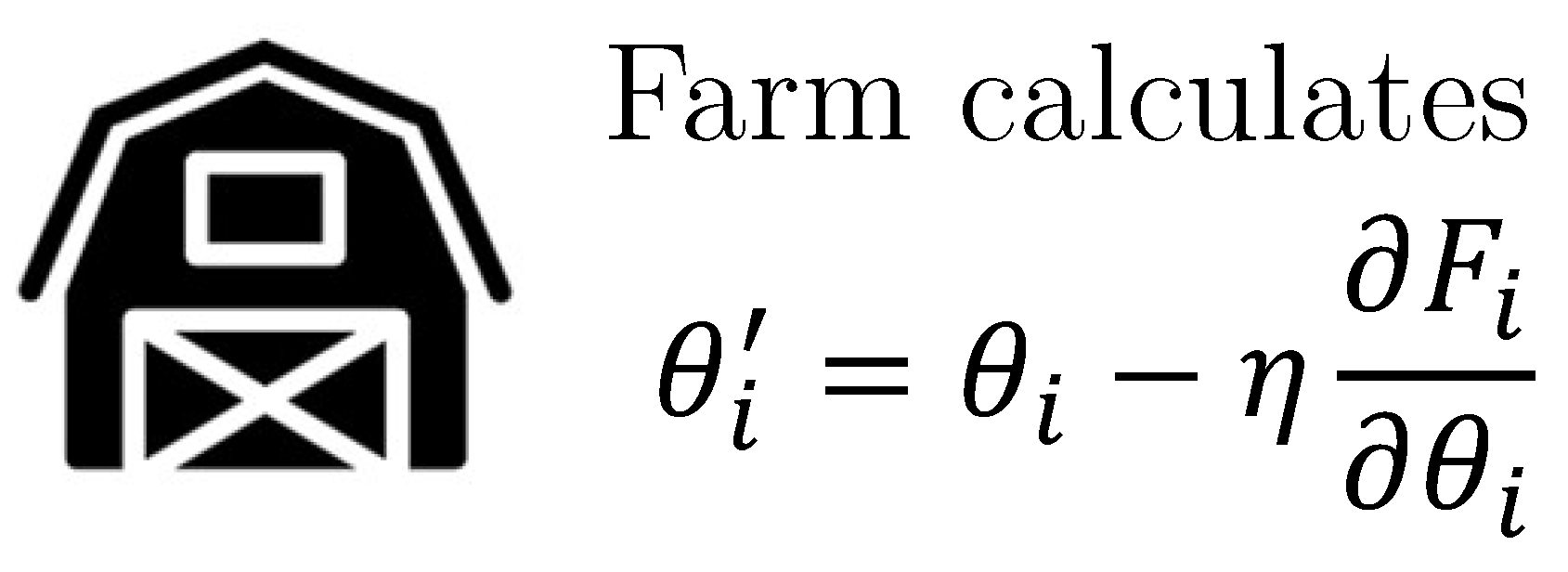

| Year | Method/Technology | Crop Species | Crop Disease | Dataset Size | Data Privacy | Reference |
|---|---|---|---|---|---|---|
| 2019 | Deep neural network with Jaya optimization algorithm (DNNJOA) | Rice | Bacterial blight, brown spot, sheath rot and blast diseases | Field self-acquired; 650 images | × | [17] |
| 2019 | CNN with SVM | Rice | Rice blast, bacterial leaf blight, sheath blight | Field self-acquired; 619 images | × | [21] |
| 2019 | Low shot learning, conditional deep convolutional generative adversarial networks (C-DCGAN) | Tea | Red leaf spot, leaf blight, red scab | Field self-acquired; 15,000 images | × | [24] |
| 2020 | Custom DCNN, YOLOv3, Faster-RCNN | Rice | Rice sheath blight, rice stem borer symptoms, rice brown spots | Field self-acquired; 5320 images; 4290 video frames from 5 videos | × | [23] |
| 2020 | VGG Net, ResNet, Inception V3 | Tomato | Late blight, septoria leaf spot, yellow-curved | Laboratory-based dataset; 2364 images | × | [25] |
| 2021 | ResNet, Genetic algorithm, Cubic SVM | Cotton | Areolate mildew, Myrothecium leaf spot, sore shin | Field self-acquired; 3000 images | × | [26] |
| 2021 | ResNet | Cotton | Leaf with lesion | Field acquired; 60,659 images | × | [27] |
| 2021 | MobileNet-based ADSNN-BO | Rice | Brown spot, rice hispa damage, Leaf blast | Public Kaggle dataset; 2370 images | × | [10] |
| 2021 | Faster R-CNN, Mask R-CNN | Rice | Brown spot, sheath blight, Blast, leaf streak | 1500 self-acquired images by camera and mobile phone | × | [18] |
| 2021 | VGG-19, LeNet-5, MobileNetV2 | Rice | Brown Spot, hispa, Leaf Blast, Bacterial Leaf, Leaf Smut | Public dataset; 2212 images | × | [16] |
| 2021 | Custom CNN | Cotton | Bacterial blight, Alternaria, Cercospora, gray mildew, fusarium wilt | Field self-acquired | × | [28] |
| 2021 | ResNet, Conv-LSTM | Cotton | Bacterial blight, Alternaria leaf spot, gray mildew, magnesium deficiency, Cercospora, fusarium wilt | Public Kaggle dataset | × | [29] |
| 2021 | DeepLabV3+, U-Net | Cucumber | Cucumber downy mildew, cucumber powdery mildew, cucumber virus disease | Field self-acquired; 1000 images | × | [30] |
| 2021 | Faster R-CNN | Rice | Brown spot, rice hispa, rice blast | Field self-acquired | × | [19] |
| 2021 | Inception-ResNet-v2, CNN | Rice | Leaf blast, brown spot, bacterial blight | Public dataset; 5200 images | × | [22] |
| 2021 | Ensemble model of DenseNet-121, SE-ResNet-50, ResNeSt-50 | Rice | Blast, false smut, neck blast, sheath blight, bacterial stripe disease, and brown spot | Field self-acquired; 33,026 images | × | [20] |
| 2021 | Image text collaborative representation learning (ITC-Net) | Tomato & Cucumber | Tomato powdery mildew, early blight; Cucumber powdery mildew, virus disease, downy mildew | Field self-acquired; 1516 text-described images | × | [31] |
| 2022 | Meta-learning | Cotton | Leaf spot, nutrient deficiency, powdery mildew, target spot, verticillium wilt, leaf curl | Field self-acquired; 2385 images | × | [32] |
| 2023 | DenseNet121, XceptionNet, Dynamic mode decomposition | Rice | Bacterial blight, blast, brown spot, and tungro | Public dataset; 3416 images | × | [33] |
| 2024 | Resource-efficient federated learning | Rice | Bacterial leaf blight, blast, brown spot, and tungro | Public dataset; 5932 images | [34] | |
| 2024 | Custom CNN model | Rice | Tungro, blast, bacterial blight, brown spot | Public dataset; 5932 images | × | [35] |
| 2025 | Hybrid CNN model | Rice | Tungro, blast, bacterial blight, brown spot | Public dataset; 5932 images | × | [36] |
| - | Our proposed approach | Rice | Blast, blight, brown spot, hispa, leaf scaled, leaf smut, tungro | 5 public datasets | - |
| Class | Blast | Blight | Brown Spot | Healthy | Hispa | Leaf Scaled | Leaf Smut | Tungro | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| Farm Number | ||||||||||
| 1 | ∼ | 40 | 40 | ∼ | ∼ | ∼ | 40 | ∼ | 120 | |
| 2 | 1440 | 1584 | 1600 | ∼ | ∼ | ∼ | ∼ | 1308 | 5932 | |
| 3 | 779 | ∼ | 523 | 1488 | 565 | ∼ | ∼ | ∼ | 3355 | |
| 4 | 57 | 82 | 96 | ∼ | ∼ | ∼ | ∼ | ∼ | 235 | |
| 5 | 272 | 283 | 139 | ∼ | ∼ | 217 | ∼ | ∼ | 911 | |
| Total | 2548 | 1989 | 2398 | 1488 | 565 | 217 | 40 | 1308 | 10,553 |
| Dataset | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|
| Overall (N = 10,789) | 94.07 ± 1.06 | 87.13 ± 1.50 | 85.78 ± 2.08 | 86.21 ± 1.47 |
| Farm #1 (n1 = 120) | 99.92 ± 0.08 | 99.92 ± 0.08 | 99.91 ± 0.08 | 99.92 ± 0.08 |
| Farm #2 (n2 = 5932) | 93.13 ± 0.06 | 86.84 ± 1.23 | 85.83 ± 1.82 | 85.76 ± 0.04 |
| Farm #3 (n3 = 3355) | 94.73 ± 1.13 | 86.47 ± 0.06 | 84.61 ± 1.34 | 85.44 ± 1.03 |
| Farm #4 (n4 = 276) | 93.04 ± 1.43 | 89.65 ± 4.85 | 89.43 ± 5.20 | 89.41 ± 0.49 |
| Farm #5 (n5 = 1106) | 93.03 ± 2.07 | 89.70 ± 6.26 | 89.48 ± 6.24 | 89.49 ± 6.25 |
| Dataset | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|
| Farm #2 (n2 = 5932) | ∼ | ∼ | ∼ | ∼ |
| Farm #3 (n3 = 3355) | ∼ | ∼ | ∼ | ∼ |
| Farm #1 (n1 = 120) | 2.09 ± 2.09 | 1.39 ± 1.39 | 2.09 ± 2.09 | 1.67 ± 1.67 |
| Farm #4 (n4 = 276) | 26.08 ± 1.59 | 34.06 ± 1.56 | 35.66 ± 0.76 | 19.49 ± 0.25 |
| Farm #5 (n5 = 1106) | 30.53 ± 0.08 | 14.48 ± 1.83 | 26.90 ± 1.90 | 14.70 ± 0.98 |
| Model | Algorithm | Portion | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|---|---|
| MobileNet | FedAvg | ∼ | 61.09 | 61.09 | 61.09 | 61.09 |
| q-FedAvg | ∼ | 21.52 | 23.18 | 21.52 | 18.04 | |
| LF-FedAvg | ∼ | 50.48 | 54.11 | 50.48 | 48.79 | |
| PT-FedAvg | 10% | 61.09 | 63.09 | 61.09 | 59.34 | |
| PT-FedAvg | 30% | 59.60 | 62.36 | 59.60 | 57.66 | |
| PT-FedAvg | 50% | 61.16 | 62.19 | 61.16 | 59.51 | |
| ADSNN-BO | FedAvg | ∼ | 70.66 | 72.60 | 70.66 | 71.03 |
| q-FedAvg | ∼ | 32.36 | 31.56 | 32.81 | 31.64 | |
| LF-FedAvg | ∼ | 64.79 | 68.73 | 64.79 | 64.04 | |
| PT-FedAvg | 10% | 80.19 | 78.70 | 80.19 | 78.47 | |
| PT-FedAvg | 30% | 81.77 | 81.95 | 81.77 | 79.68 | |
| PT-FedAvg | 50% | 83.14 | 84.14 | 83.14 | 82.17 |
| Farm | Metrics (%) | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | Average |
|---|---|---|---|---|---|---|---|
| Farm #1 (n1 = 120) | Accuracy | 66.66 | 66.66 | 50.00 | 66.66 | 66.66 | 63.33 |
| Precision | 55.55 | 60.00 | 60.00 | 50.00 | 50.00 | 55.11 | |
| Recall | 66.66 | 66.66 | 50.00 | 66.66 | 66.66 | 63.33 | |
| F1 score | 60.00 | 62.96 | 51.85 | 55.55 | 55.55 | 57.18 | |
| Farm #2 (n2 = 5932) | Accuracy | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Precision | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| Recall | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| F1 score | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| Farm #3 (n3 = 3355) | Accuracy | 39.40 | 65.37 | 64.47 | 65.67 | 71.68 | 61.32 |
| Precision | 43.40 | 70.87 | 71.12 | 66.57 | 73.58 | 65.11 | |
| Recall | 39.40 | 65.37 | 64.47 | 65.67 | 71.68 | 61.32 | |
| F1 score | 35.12 | 59.11 | 66.39 | 65.46 | 71.31 | 59.48 | |
| Farm #4 (n4 = 276) | Accuracy | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Precision | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| Recall | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| F1 score | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| Farm #5 (n5 = 1106) | Accuracy | 30.00 | 50.90 | 55.45 | 60.00 | 79.04 | 55.08 |
| Precision | 25.57 | 50.39 | 57.30 | 58.12 | 83.15 | 54.91 | |
| Recall | 30.00 | 50.90 | 55.45 | 60.00 | 79.04 | 55.08 | |
| F1 score | 24.11 | 46.98 | 52.37 | 55.62 | 80.13 | 51.84 |
| Farms | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|
| Farm 1 (n1 = 120) | 63.33 | 55.11 | 63.33 | 57.18 |
| Farm 2 (n2 = 5932) | 100.00 | 100.00 | 100.00 | 100.00 |
| Farm 3 (n3 = 3355) | 61.32 | 65.11 | 61.32 | 59.48 |
| Farm 4 (n4 = 276) | 100.00 | 100.00 | 100.00 | 100.00 |
| Farm 5 (n5 = 1106) | 55.08 | 54.91 | 55.08 | 51.84 |
| Weighted Average | 83.14 | 84.13 | 83.14 | 82.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chorney, W.; Rahman, A.; Wang, Y.; Wang, H.; Peng, Z. Federated Learning for Heterogeneous Multi-Site Crop Disease Diagnosis. Mathematics 2025, 13, 1401. https://doi.org/10.3390/math13091401
Chorney W, Rahman A, Wang Y, Wang H, Peng Z. Federated Learning for Heterogeneous Multi-Site Crop Disease Diagnosis. Mathematics. 2025; 13(9):1401. https://doi.org/10.3390/math13091401
Chicago/Turabian StyleChorney, Wesley, Abdur Rahman, Yibin Wang, Haifeng Wang, and Zhaohua Peng. 2025. "Federated Learning for Heterogeneous Multi-Site Crop Disease Diagnosis" Mathematics 13, no. 9: 1401. https://doi.org/10.3390/math13091401
APA StyleChorney, W., Rahman, A., Wang, Y., Wang, H., & Peng, Z. (2025). Federated Learning for Heterogeneous Multi-Site Crop Disease Diagnosis. Mathematics, 13(9), 1401. https://doi.org/10.3390/math13091401








