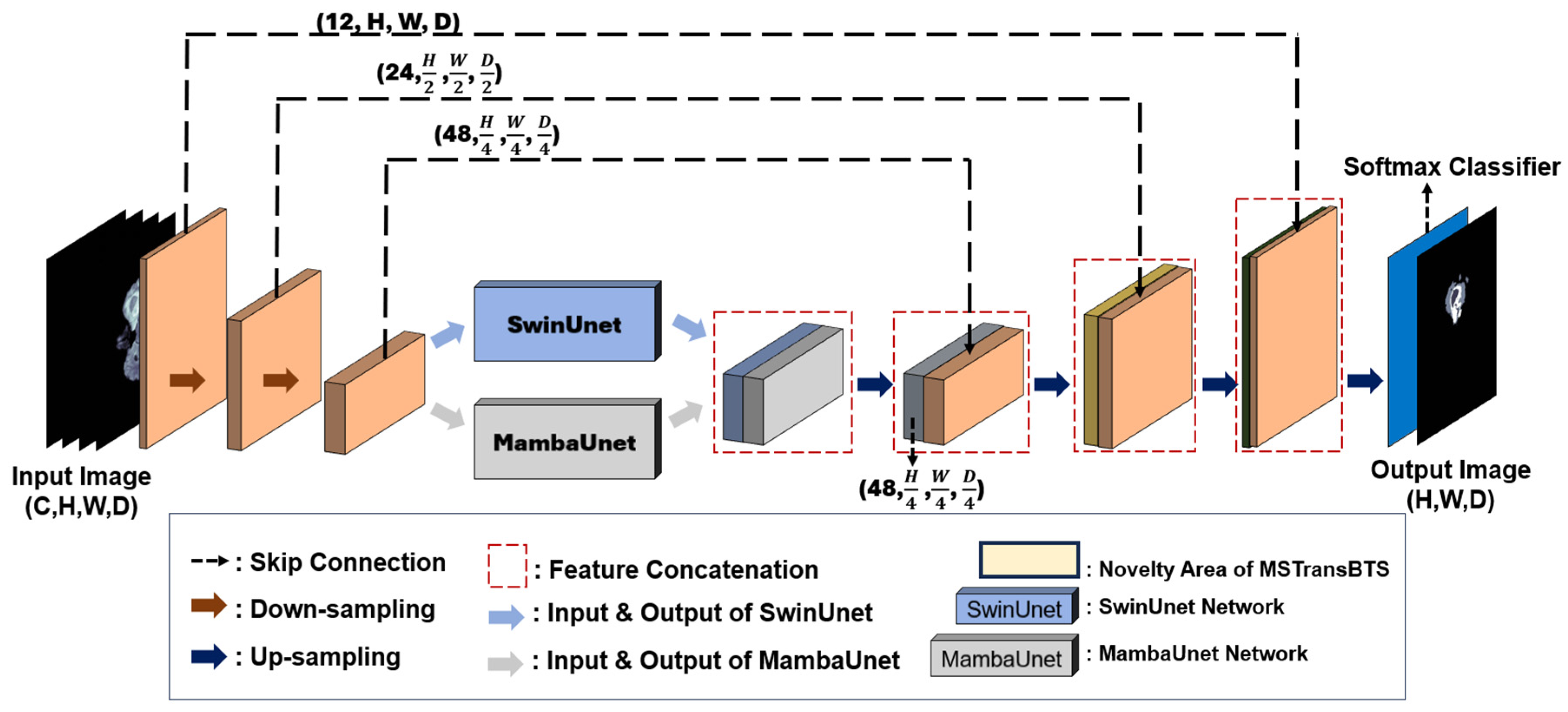
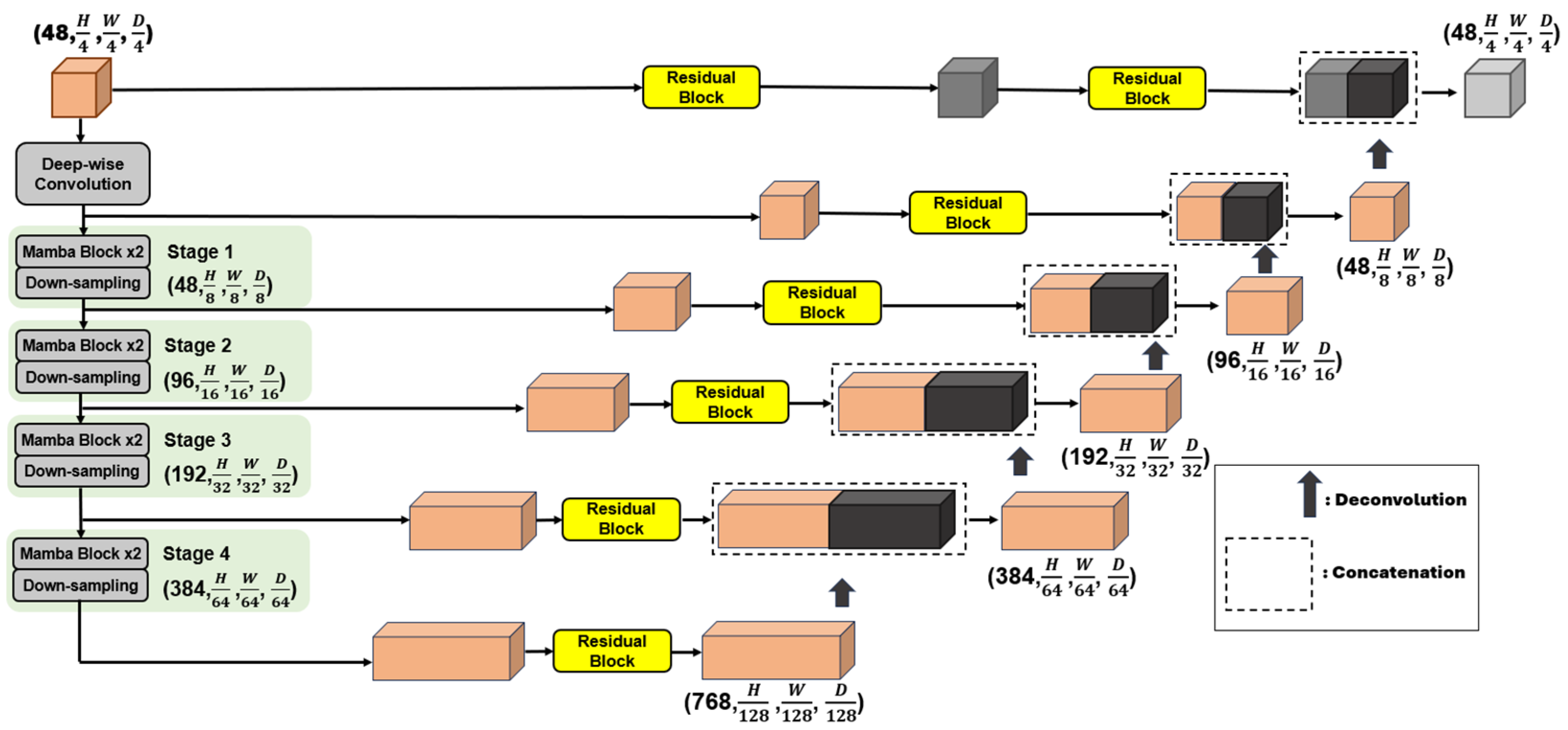
This section provides an overview of the various state-of-the-art DL architectures employed in this work, which were integral to developing our method.
2.3. Transformer
Driven by the success of attention mechanisms in natural language processing (NLP), attention mechanisms were introduced into CNN models to capture long-range dependencies. Initially developed for sequence-to-sequence tasks, the Transformer framework is notable for its ability to model relationships between points within a sequence. In contrast to conventional CNN-based methods, the Transformer utilizes self-attention processes and completely removes the necessity for convolutions. This architecture exhibits exceptional proficiency in capturing the whole context and remarkable effectiveness in subsequent tasks, especially when pre-trained on extensive datasets [
12].
Transformers process inputs as 1D sequences and focus on global context modelling, which results in low-resolution features. The conventional up-sampling of these features is insufficient to fully recover the lost degree of detail. On the other hand, a hybrid technique that merges CNN and Transformer encoders has greater potential. This approach effectively enhances the high-resolution spatial information captured by CNNs while simultaneously taking advantage of the global context offered by Transformers [
13]. Therefore, the Transformer in 3D CNN for 3D MRI Brain Tumour Segmentation (TransBTS) model is proposed by Wang et al. [
12].
The TransBTS model improves the existing encoder–decoder architecture to deal with volumetric data. The network’s encoder initially uses 3D CNNs to extract spatial features from the input 3D images while simultaneously down-sampling them. This approach delivers condensed volumetric feature maps that effectively localizes 3D contextual information; subsequently, every volume is reshaped into a vector (known as a token) and fed into a Transformer for global feature modelling. On the decoder side, a 3D CNN decoder utilizes the feature embedding acquired from the Transformer to gradually up-sample the resolution and anticipate the whole segmentation map. This hybrid methodology integrates the advantages of both CNNs and Transformers, facilitating the effective processing of volumetric data while including local and global contexts for precise segmentation outcomes.
TransBTS has remarkable average Dice scores of 78.73% for enhancing tumours (ETs), 90.09% for whole tumours (WTs), and 81.73% for tumour cores (TCs) for the BraTS2020 dataset. These scores indicate the model’s exceptional ability to efficiently segment various tumour locations, proving its efficacy in 3D BT segmentation.
2.4. Swin Transformer
Transferring strong performance from language tasks to the visual domain poses major challenges due to differences between the two modalities. One notable distinction is the scale. Unlike word tokens, which serve as the fundamental processing units in language Transformers, visual components exhibit significant variations in scale. This issue is especially apparent in object detection. Since tokens have a fixed scale, current Transformer-based models are not suited for vision applications. In addition, images have a superior pixel resolution compared to words in text passages. Tasks involving vision, such as semantic segmentation, need precise predictions at the pixel level. However, Transformers struggle with performing this task on high-resolution images as the computational complexity of self-attention increases quadratically with image size, which makes it impractical.
A flexible framework called the Swin Transformer is implemented to solve the above issues. This methodology creates hierarchical feature maps with linear computational complexity compared to the image size.
By using these hierarchical feature maps, the Swin Transformer model may effectively integrate techniques for dense prediction, such as U-Net. The Swin Transformer achieves linear computational complexity by performing self-attention calculations locally within non-overlapping windows that partition an image. The quantity of patches within each window remains consistent, leading to a complexity that increases proportionally with the size of the image. The features of Swin Transformer make it very suitable as an adaptable framework for many visual tasks. In contrast to previous Transformer-based designs, which produce feature maps with a single resolution and have a quadratic complexity, this approach is unique and distinct. Additionally, an important aspect of the Swin Transformer’s architecture is its shifting of the window partition between successive self-attention levels
Beginning with the first layer, a standard window partitioning approach is implemented, and self-attention is calculated within each window. In the next layer, the window partitioning is varied, resulting in additional windows. The self-attention computation in these new windows extends beyond the bounds of the preceding windows in the first layer, thereby establishing connections between them [
14].
In general, transferring high-performance Transformer features from language tasks to the visual domain presents major problems, arising from the discrepancies in scale and resolution. The Swin Transformer framework is proposed to solve these issues, providing hierarchical feature maps with a linear computational complexity. Furthermore, the Swin Transformer has a dynamic window partitioning mechanism that establishes connections between feature maps at different scales. In the next subsection, a specific model constructed with the Swin Transformer is introduced to justify the effectiveness of the Swin Transformer in vision tasks.
The architecture of Swin Transformers is highly suitable for a wide range of downstream activities as it allows for the extraction of multi-scale features that can be used for further processing [
15]. To utilize this capability, an innovative design known as Swin UNETR (Swin U-Net Transformers) is proposed. Swin UNETR employs a U-shaped network structure and a Swin Transformer, serving as the encoder and connected to a CNN-based decoder at distinct resolutions through skip connections.
The model’s input consists of 3D multi-modal MRI scans with four channels. The Swin UNETR model first splits the input data into non-overlapping patches and leverages a patch partition layer to establish windows of the necessary size to carry out self-attention calculations. Subsequently, the encoded feature representations generated by the Swin Transformer are fed to a CNN-based decoder through skip connections at different resolutions. Ultimately, the segmentation outputs are computed through a convolutional layer followed by a sigmoid activation function. The resulting segmentation output consists of three output channels, each representing the ET, WT, and TC sub-regions.
According to the findings reported by Hatamizadeh and colleagues (2022) [
16], the SwinUNETR framework exhibits exceptional performance on the BraTS2021 training dataset. Notably, it attains average Dice scores of 85.30% for ET, 92.70% for WT, and 87.60% for TC segmentation. These outcomes highlight the efficacy of SwinUNETR in precisely identifying tumour areas in multi-modal MRI imaging. In summary, the outstanding performance demonstrated by SwinUNETR highlights the potential of integrating the Swin Transformer with U-net as a promising approach for medical image segmentation. In the following year, He and his teammates [
7] enhanced the SwinUNETR model to a greater extent and named it as SwinUNETR-V2. The improved version incorporates the stagewise convolutions with window-based self-attention in each stage, which provides a stronger backbone for the feature extraction. It is also the main component in our proposed model, and the integration method is elucidated in the Materials and Methods Section.
A 3D convolutional patch embedding layer (with a stride = 2,2,2 and kernel size = 2,2,2) is implemented in SwinUNETR-V2 to convert patches into tokens. Subsequently, the outputs undergo four stages of Swin Transformer blocks and patch merging to encode the input patches. At Swin block j, given an input tensor of size
, the Swin Transformer block partitions the tensor into windows of size
. The mechanism of two successive Swin Transformer blocks is depicted and shown in
Figure 2.
Based on
Figure 2, it is clear to state that four computations are conducted as Equations (1)–(4) [
14].
where
= output elements of the SW-MSA module;
= output of the MLP module; W-MSA = window-based multi-head self-attention partitioning function; SW-MSA = shifted-window based multi-head self-attention partitioning function; MLP = multilayer perceptron function; LN = layer normalization function,
; and
= number of Swin Transformer blocks in each stage.
To halve each spatial dimension, a patch merging layer is placed after the Swin Transformer blocks in each stage. In addition, at the beginning of each stage, the input tokens are reshaped to restore their original 3D volume form. Next, a Residual Convolution block is used, which consists of two sets of
convolutions, instance normalization, and leaky rectified linear unit (ReLU) activations. It has the same architecture as the residual connection in
Figure 1, except for the ReLU having been changed to leaky ReLU and layer normalization to instance normalization. The resulting output proceeds to a series of subsequent Swin Transformer blocks with depths of
. Notably, three Residual Convolution blocks are incorporated across the three stages [
7].
2.5. Mamba
CNNs are extensively used in image processing, especially techniques such as Fully Convolutional Networks (FCNs), which are exceptionally effective at extracting hierarchical features. Transformers, initially developed for NLP and subsequently extended for visual tasks via architectures such as Vision Transformers (ViT) and Swin Transformer, are highly effective at capturing global information. Their incorporation into CNN frameworks has resulted in the development of hybrid models such as SwinUNETR, which greatly enhances the representation of long-range connections [
15].
Transformers, despite being effective at capturing long-range dependencies, encounter challenges because of their high computational cost. This is mainly due to the self-attention mechanism, which scales quadratically with the input size. This impact is particularly obvious in high-resolution biomedical images. Recent advancements in State Space Models (SSMs), notably Structured SSMs (S4), present a promising solution for efficiently processing large sequences. The Mamba model further improves S4 by incorporating selective mechanisms and hardware optimization, resulting in exceptional performance in dense data domains [
17].
Introduce SegMamba, a novel design that integrates the U-shape structure with Mamba to effectively capture global contexts at different scales over the entire volume. SegMamba consists of three primary components: (1) A 3D feature encoder that includes multiple tri-orientated spatial Mamba blocks to capture global information at various scales. (2) A 3D decoder that leverages convolution layers to predict segmentation outcomes. (3) Skip connections are established between the global multi-scale features and the decoder. These connections allow for the reuse of features. The SegMamba framework is utilized in our proposed model, and the integration process is described in the Materials and Methods Section.
The encoder in SegMamba consists of a Depth-Wise Convolution layer and multiple Mamba blocks. The Depth-Wise convolution is built with a 7 × 7 × 7 kernel size, 3 × 3 × 3 padding, and 2 × 2 × 2 stride. Given a 3D input image, the Depth-Wise Convolution layer further extracts the input features into a size of
. Subsequently,
proceeds through each Mamba block and its corresponding down-sampling layers. The computation process for the
Mamba block is as shown in Equations (5)–(7) [
8]:
where
= Gated Spatial Convolution function;
= Tri-orientated Mamba function;
= multilayer perceptron function;
= layer normalization function;
; and
= number of Mamba blocks in each stage.
The GSC mechanism is depicted in
Figure 3. The GSC module is integrated before Mamba layer processing to enhance feature extraction and capture spatial relationship. This module first passes the input 3D features through two convolution blocks, each of which contains a convolution operation, instance normalization, and a non-linear layer. The convolution blocks contain kernel sizes of 3 × 3 × 3 and 1 × 1 × 1. Subsequently, the outputs of these blocks are element-wise multiplied to modulate information flow which is identical to a gate function. An additional convolution block is used to perform further feature fusion, which is enriched by a residual connection that recovers the initial features. This design is essential for efficiently extracting spatial dependencies before proceeding to the Mamba layer.
In the Mamba block, the importance lies in its ability to comprehensively capture global information from high-dimensional features. This is accomplished by implementing a Tri-orientated Mamba (ToM) module that computes feature dependencies from three different directions. As illustrated in
Figure 4, the process involves flattening the 3D input features into three sequences, allowing the operation of feature interaction from distinct directions [
8]. As a result, this approach facilitates the generation of fused 3D features, which strengthens the model’s capability to comprehend and employ intricate spatial relationships within the data.
In 3D medical image segmentation, the inclusion of global features and multi-scale features is of utmost importance. Transformer architectures are highly effective in extracting global information, but they encounter difficulties in terms of computational burden when dealing with extremely long feature sequences. To handle this issue, techniques such as SwinUNETR employ the direct down-sampling of the 3D input to effectively decrease the length of the sequence. However, this method undermines the encoding of multi-scale characteristics that are essential for precise segmentation predictions. To address this constraint, the TSMamba block is designed to enable the concurrent modelling of multi-scale and global features while ensuring efficient performance during both training and inference.
Based on the findings presented by Xing et al. [
8], the SegMamba framework demonstrates exceptional performance on the BraTS2023 training dataset. It obtains remarkable average Dice scores of 87.71% for ET, 93.61% for WT, and 92.65% for TC segmentation. The results highlight the effectiveness of SegMamba in precisely identifying tumour areas in multi-modal MRI imaging. The results of SegMamba show that combining Mamba with U-net may give excellent results for medical image segmentation.