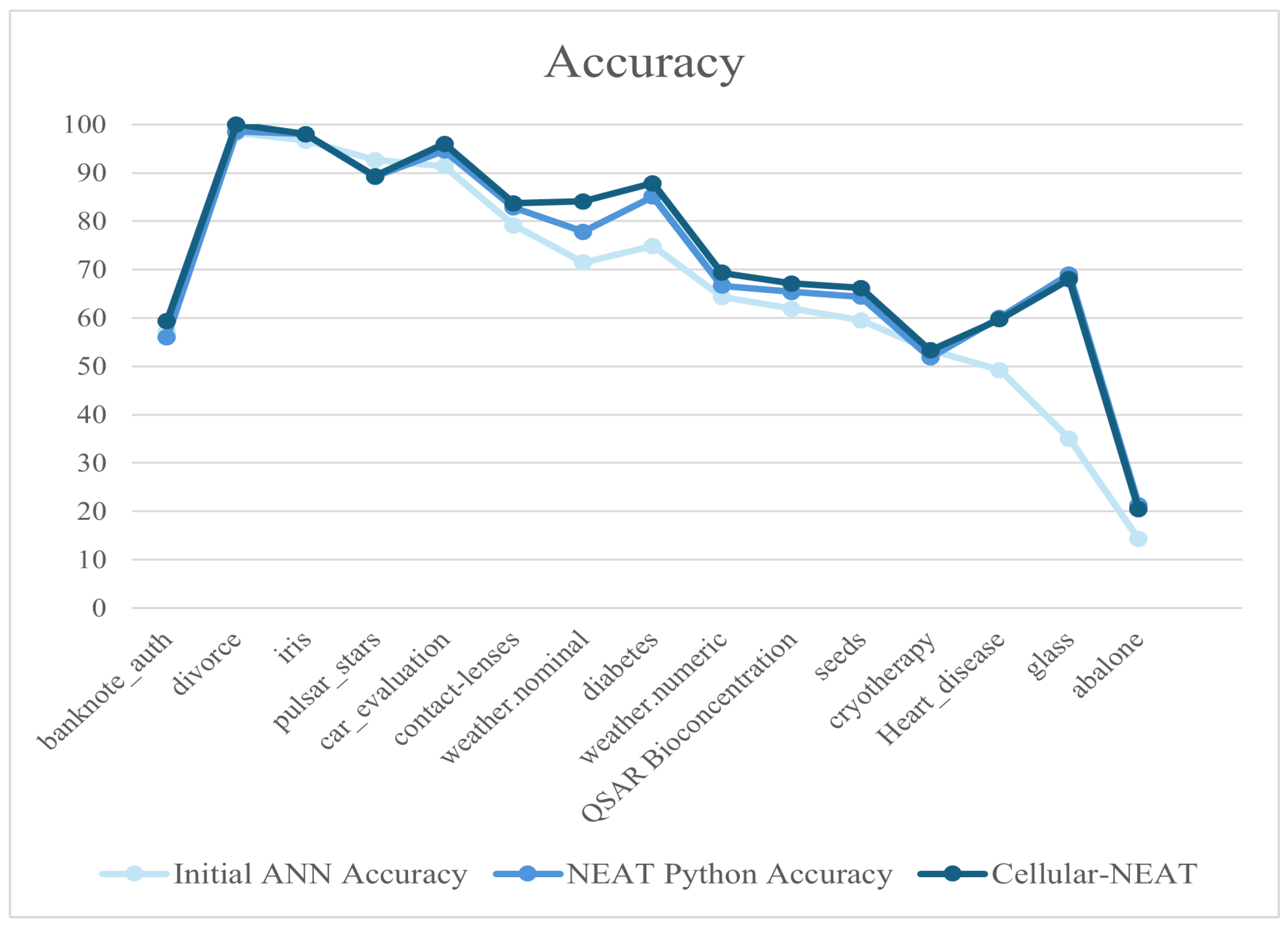
To assess the algorithm robustness, for this work, we consider a set of independent instances with varying characteristics, such as the number of registers and dataset dimensionality. The selection criterion included datasets for classification with at least two evaluation classes and no missing values. The chosen datasets were obtained from publicly available machine learning repositories, specifically Kaggle [
23] and the UCI Machine Learning Repository [
24], providing diverse and well-structured data for a comprehensive performance analysis.
Table 1 shows the characteristics of each selected dataset, where columns show the name of the dataset, the repository from which the dataset was extracted, the data types of the corresponding attributes of each dataset, and the number of registers, attributes, and classes.
2.1. Preprocessing
Once the instances were defined, we applied a normalization method. For this work, we decided on the normalization method to be used to transform each dataset. We used a normalization; this process rescales data to a specific range, typically between 0 and 1 or −1 and 1, ensuring a consistent scale across feature. Moreover, standardization adjusts data by centering them around a mean of 0 and scaling them to have a standard deviation of 1. This technique preserves the original distribution and is particularly effective when working with normally distributed data.
For the selection of the normalization method, each attribute of each dataset was reviewed to evaluate which type of standardization would be applied, considering if the attributes presented a normal distribution after we applied standardization, and, in cases where the data needed rescaling, we applied normalization.
Table 2 shows a sample of the results of the analysis performed for the selection of the normalization technique for all datasets, where columns show the attribute number of the dataset, the statistical information for each attribute (minimum value, maximum value, mean, and standard deviation), and the decision taken to normalize each attribute, where the norm is obtained from Equation (
1), std is calculated with Equation (
2), and the − symbol indicates that no normalization method was required for this attribute.
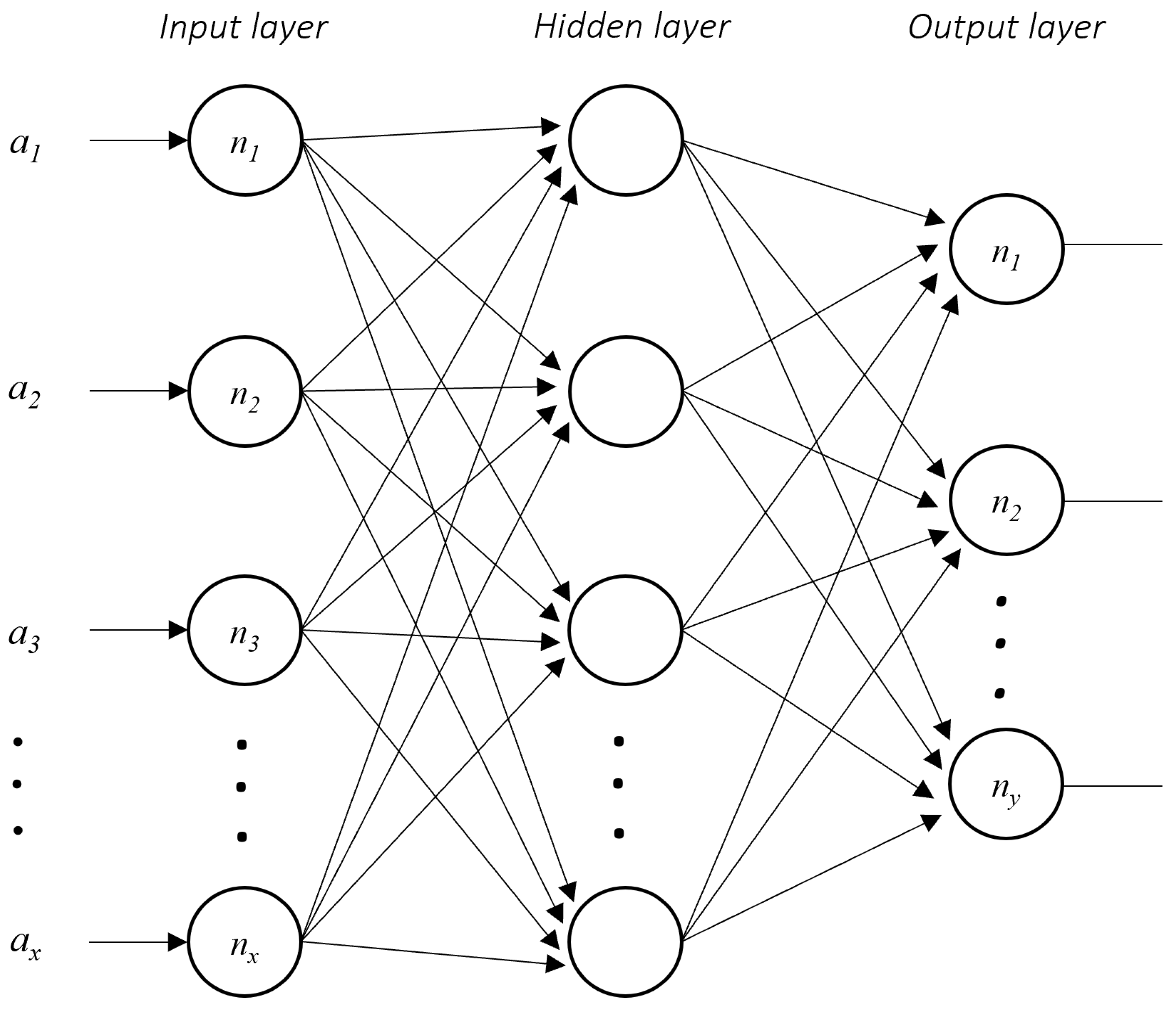
In addition, we perform an Artificial Neural Network with the TensorFlow library in Python to have an accuracy reference value. The Artificial Neural Network structure for this experiment was an input layer with the number of neurons equal to the number of attributes, a hidden layer with the same number of neurons as the input layer, and an output layer with one neuron. Each dataset was divided into training and testing. The production percentages of each set in the experimentation were 70% training and 30% test.
2.2. NEAT Algorithm
For the neuroevolution of neural networks, we apply the NeuroEvolution of Augmenting Topologies (NEAT) algorithm [
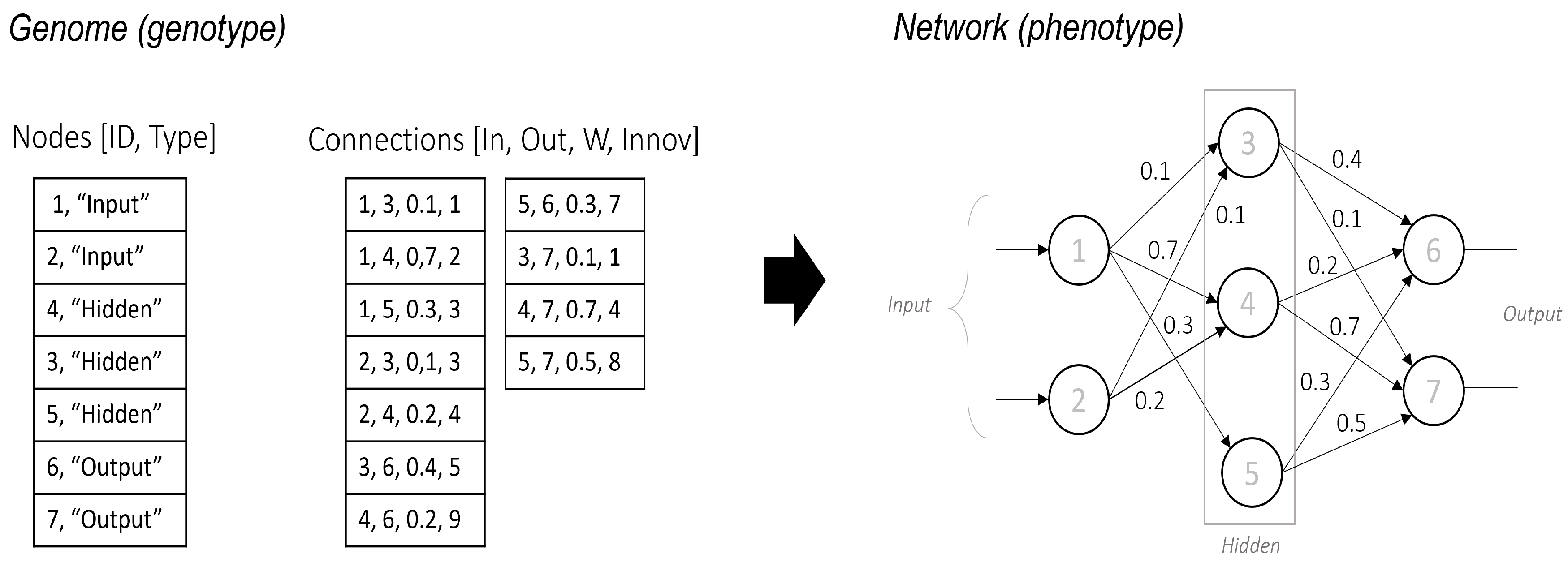
25], which evolves Artificial Neural Networks through mutation and selection. The genetic coding scheme represents the neural network in two phases: genotype and phenotype.
To encode the genome, NEAT utilizes a list of nodes and a list of connections. The Nodes list defines neurons within the network, each uniquely identified by an and categorized as input, hidden, or output. The Connections list specifies synaptic links between neurons, encoding key attributes such as input and output nodes, connection weight w, and innovation number , which ensures the historical tracking of mutations.
Figure 1 also shows the phenotype, where the neural network structure emerges from the encoded genome. Input neurons receive data, hidden neurons process information, and output neurons generate final predictions. The structural complexity of the network increases as mutations introduce new nodes and connections, enabling adaptive evolution.
This encoding strategy ensures a direct mapping between genotype and phenotype, supporting both structural and parametric evolution while preserving innovation through historical markings. As a result, NEAT effectively optimizes neural network architectures by balancing the exploration of new topologies and the inheritance of beneficial traits.
2.3. Evolution Process
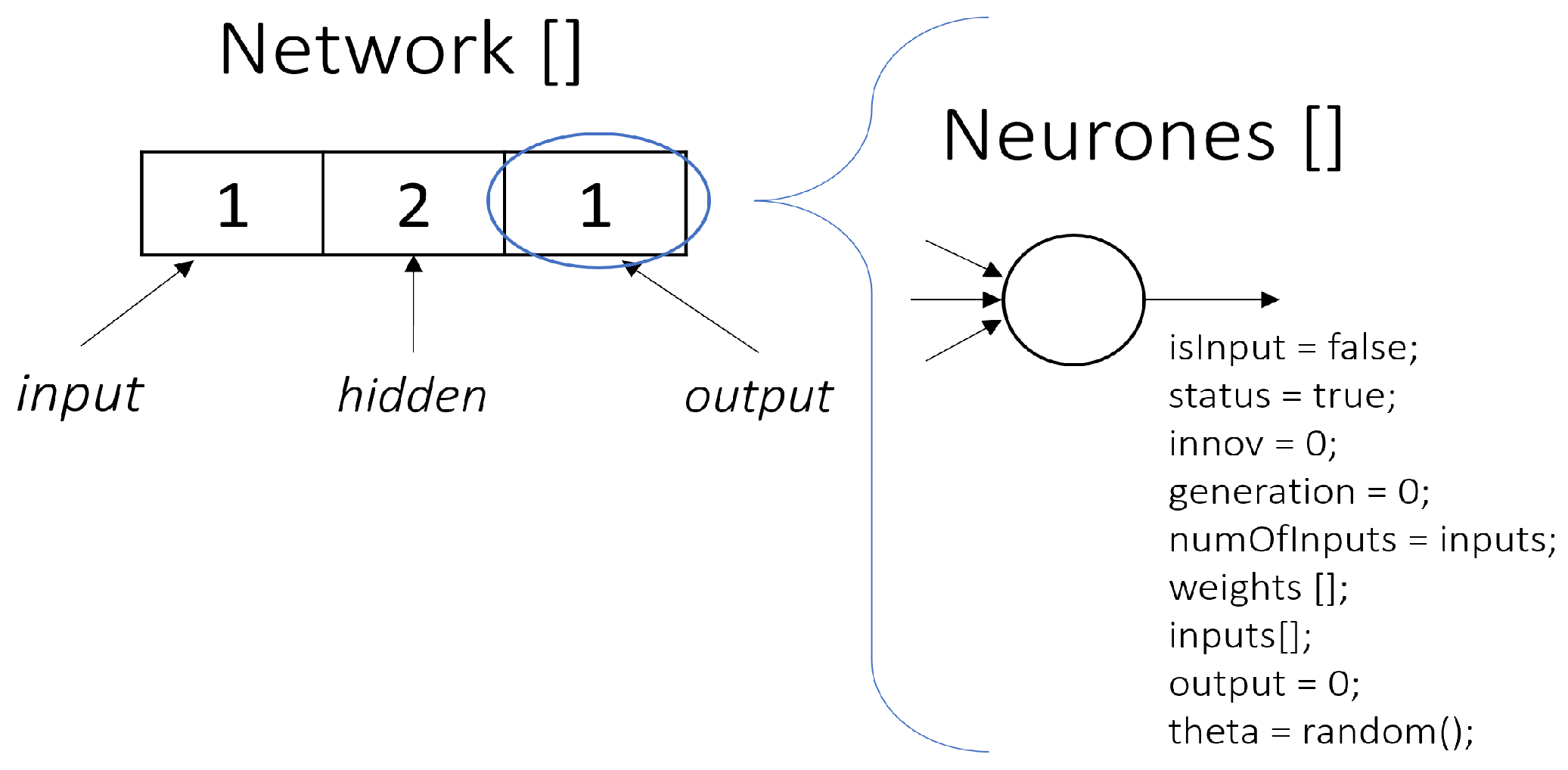
In this paper, we propose a modified genetic encoding scheme for a neural network within the NeuroEvolution of Augmenting Topologies (NEAT) algorithm. This adaptation incorporates structural modifications to the genotype by integrating additional connection parameters and representing the phenotype as a structured list format as shown in
Figure 2.
At the genotypic level, the encoding includes a set of connection attributes, specifying details such as activation states (e.g., true/false flags), numerical weights (float values), and additional properties that refine the mutation and selection mechanisms. These modifications enhance the expressiveness of the genome, allowing for greater control over the evolutionary process.
The phenotype is represented as a list (network), where the first element corresponds to the number of neurons in the input layer, followed by elements representing the number of neurons in each hidden layer. The final element in the list denotes the number of neurons in the output layer. This structured format provides an intuitive representation of the neural network architecture, enabling the efficient manipulation and analysis of evolving topologies.
The proposed genome was used in conjunction with a genetic algorithm to develop a genetic algorithm aimed at enhancing neural network development.
Genetic algorithms (GAs) [
26] are optimization techniques inspired by the principles of natural selection and genetics. GAs operate by evolving a population of candidate solutions through iterative processes of selection, crossover, and mutation, aiming to find optimal or near-optimal solutions to complex problems. In the context of neural networks, GAs have been employed to optimize various aspects, including network architecture [
27,
28], weights [
29,
30], and hyperparameters [
31,
32].
The genome structures of the neural network were developed for the initial population of the genetic algorithm. The algorithm began by selecting candidates for the generation through a binary tournament.
For genetic operators, mutation and crossover methods were designed based on the NEAT genome. The mutation method introduced changes at the genotype level, specifically modifying the weight on a specific connection. Algorithm 1 modifies the structure of neural networks by adjusting the weights and innovation numbers within the hidden layers. The procedure begins by selecting a random hidden layer and a neuron within that layer. A specific weight from the selected neuron is then chosen and modified. Then, the algorithm increments the innovation numbers for the hidden layer and the neuron to reflect the changes and updates the chosen weight with a new random value.
| Algorithm 1 Genoytpe Mutation Operator |
- 1:
procedure () - 2:
for each do - 3:
(0, ) - 4:
(0, ) - 5:
(0, ) - 6:
- 7:
- 8:
- 9:
- 10:
end for - 11:
end procedure
|
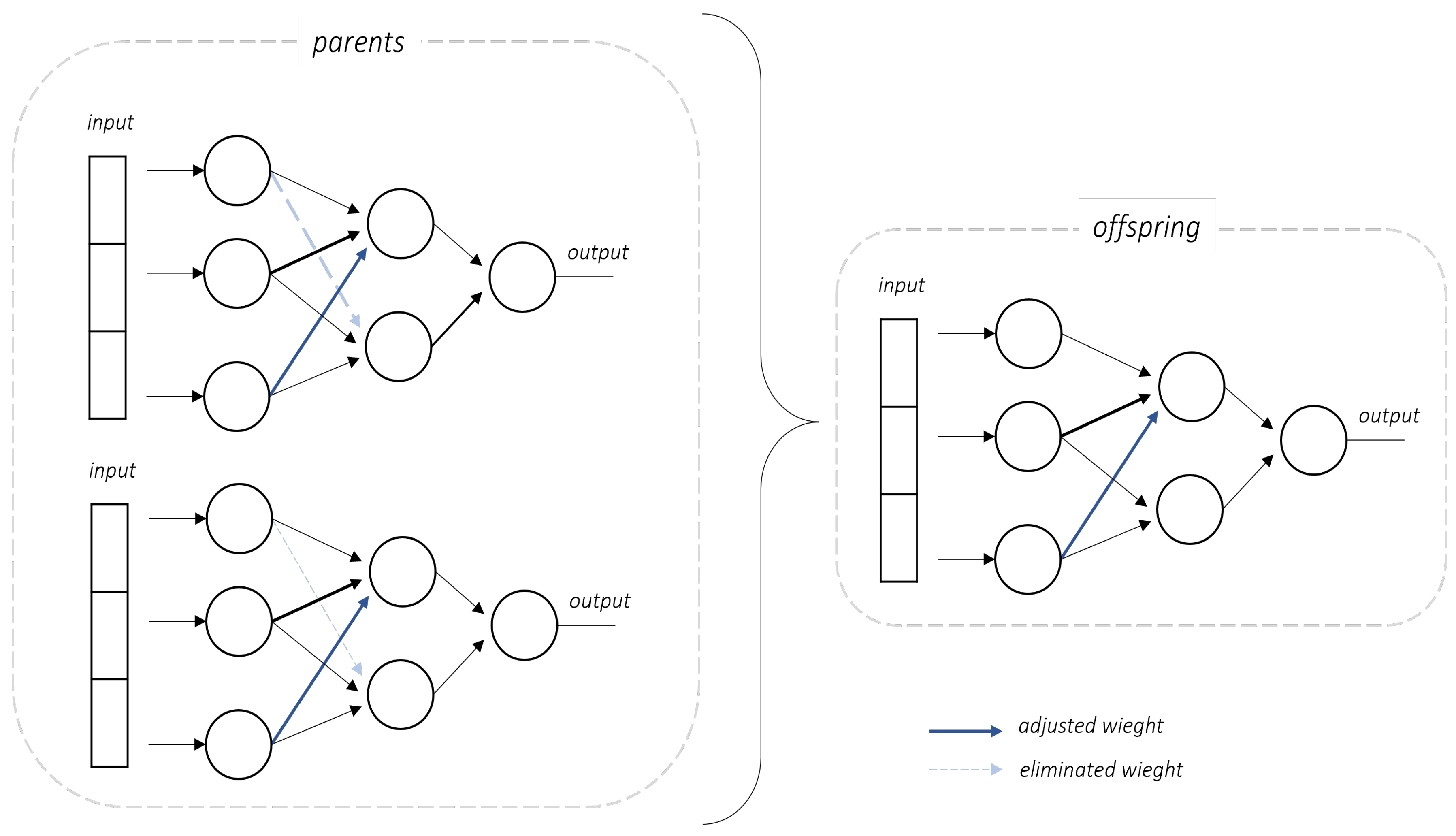
For crossover, modifications were applied to the genome’s information regarding the connections within the neural network. The crossover operator consists of a differential evolution strategy within a neural network framework. It adjusts the network weights based on differences computed from two-parent networks and introduces stochastic mutations while tracking modifications via innovation counters, as illustrated in
Figure 3.
Algorithm 2 modifies the neural network structure based on evolutionary principles. It processes three neural networks, denoted as
,
, and
, along with an integer parameter numEdges, which represents the total number of edges in the network.
| Algorithm 2 Differential Crossover Operator |
- 1:
procedure gendc_operator() - 2:
- 3:
for to do - 4:
- 5:
- 6:
- 7:
- 8:
- 9:
- 10:
- 11:
if then - 12:
- 13:
- 14:
- 15:
- 16:
- 17:
- 18:
else - 19:
- 20:
- 21:
- 22:
- 23:
end if - 24:
end for - 25:
return - 26:
end procedure
|
The number of modifications applied is computed as
The algorithm iterates for
, and, in each iteration, it performs the following steps. A hidden layer index HL is selected randomly, followed by a neuron index N within that layer and a weight index
from the neuron’s weight vector. These indices define a specific weight to modify. The corresponding weights from
and
are retrieved:
The new weight is computed as
where
v is a predefined scalar multiplier defined by experimentation.
A probability value
p is sampled from a uniform distribution in
. If
, which consists of the probability of selecting a third parent as a candidate to improve the solution as determined using Grid Search, where the final value selected was 0.30, a crossover occurs by incorporating a weight modification from
:
The temporary weight is then updated:
In both cases, the algorithm updates the innovation counters:
The last stored weight is also updated accordingly:
Finally, the modified weight is assigned:
Once the offspring are generated, the neural network’s accuracy serves as the objective function for the next phase.
The genetic algorithm (GA) used follows a structured approach to optimizing neural network configurations. An individual represents a neural network genotype, and each candidate solution consists of a structured list defining the number of neurons in each layer and their associated connection weights.
The binary tournament technique was used as a selection operator, ensuring diversity while prioritizing high-performing individuals [
33]. For example, given two candidates
A and
B with fitness scores
and
, the selection of the best individual is as follows:
We applied the differential evolution strategy for crossover, where weight adjustments are computed based on differences between two parent networks. If two-parent weights are
and
with a scaling factor
, the new weight is computed as
With a probability of 0.3, an additional modification is introduced using a third parent’s weight
:
The mutation process occurs by introducing weight perturbations in randomly selected connections. For example, if we consider a mutation rate of 0.1 to a weight
, the mutation modifies it as
Thus, if
generates a value of 0.02, the new weight is
The replacement technique adopts an elitist strategy, preserving the best solutions across generations to maintain stable convergence. If the best solution in generation t has and in generation a candidate achieves , the elite strategy ensures retaining the new best solution.
The GA parameters, mutation, and crossover probability were tuned through Grid Search, ensuring an optimal balance between maintaining diversity and refining promising network architectures.
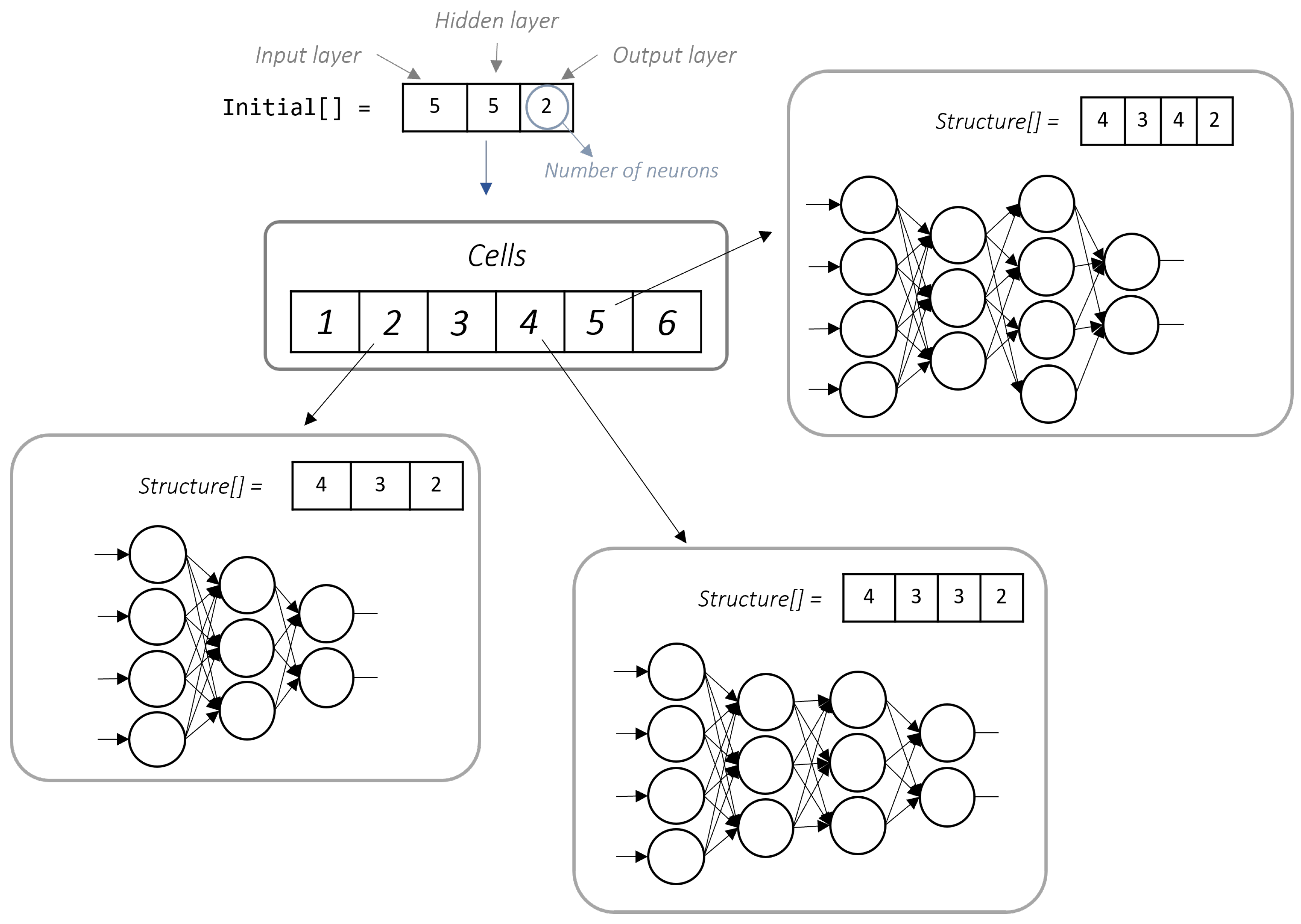
2.4. Cellular Processing
To enhance the quality of the solution obtained by the genetic algorithm, the search space was expanded using a Cellular Processing Algorithm (PCELL). This model simulates the communication and information-processing mechanisms of biological cells.
In this work, PCELL utilized six processing cells, each containing a distinct neural network phenotype derived from the initial solution. Additionally, each cell incorporated the proposed genetic algorithm to explore local solutions effectively.
Algorithm 3 begins by initializing an initial solution
, which serves as the basis for generating a set of
N processing cells
. Each cell
is assigned a unique phenotype
based on a transformation function applied to
, given by
, where
represents a diversity parameter to explore different regions in the search space. Within each processing cell, a GA iteratively optimizes the assigned solution, leading to an update step defined as
. The best solution found in each cell is evaluated and stored as
(Equation (
13)), where
is the objective function to maximize.
After processing all cells, the global best solution
updates according to
This process continues iteratively until a stopping criterion is met, which is defined when a predefined number of iterations is reached or when the improvement in
global falls below a threshold
. Finally, the algorithm returns the most-optimized solution obtained.
| Algorithm 3 Cellular Processing |
- 1:
Input: Initial solution , number of cells N - 2:
Generate N processing cells - 3:
for each cell do - 4:
- 5:
- 6:
- 7:
end for - 8:
- 9:
Output:
|
Algorithm Complexity
The PCELL algorithm divides the search into
N independent cells, where
N is the number of cells,
G represents the number of generations per cell,
P denotes the population size in each cell,
C stands for the cost of applying genetic operators (mutation, crossover, etc.) to a candidate, and
E indicates the cost of evaluating the objective function for each candidate; in this case, the overall temporal complexity is expressed as
A conventional genetic algorithm that operates on a single population has a temporal complexity of
Comparing Equations (
15) and (
16) shows that the computational cost of the PCELL approach increases by a factor of
N. Although this factor raises the computational load, the parallel exploration of multiple cells enhances the diversity in the search for solutions.
In backpropagation, the training process iterates over the dataset to update the network parameters, where
I represents the number of iterations (epochs) and
M denotes the total number of parameters (weights and biases) in the network; accordingly, the complexity approximates to
Equation (
17) indicates a lower cost per iteration compared to Equation (
15); this method risks becoming trapped in local optima and does not explore the solution space as efficiently.
The PCELL approach exhibits a temporal complexity of , which exceeds that of a traditional () due to the factor N. This extra cost justifies itself by the improvement in the exploration of the solution space. Additionally, the backpropagation method has a complexity of , which generally offers higher efficiency per iteration but may not provide the global search robustness achieved by evolutionary methods.
This comparison clarifies the implications of the increased computational cost of the PCELL approach compared to traditional methods and highlights the trade-off between computational efficiency and the ability to explore diverse solutions.