Abstract
Skin cancer is a major global health concern and one of the deadliest forms of cancer. Early and accurate detection significantly increases the chances of survival. However, traditional visual inspection methods are time-consuming and prone to errors due to artifacts and noise in dermoscopic images. To address these challenges, this paper proposes an innovative deep learning-based framework that integrates an ensemble of two pre-trained convolutional neural networks (CNNs), SqueezeNet and InceptionResNet-V2, combined with an improved Whale Optimization Algorithm (WOA) for feature selection. The deep features extracted from both models are fused to create a comprehensive feature set, which is then optimized using the proposed enhanced WOA that employs a quadratic decay function for dynamic parameter tuning and an advanced mutation mechanism to prevent premature convergence. The optimized features are fed into machine learning classifiers to achieve robust classification performance. The effectiveness of the framework is evaluated on two benchmark datasets, PH2 and Med-Node, achieving state-of-the-art classification accuracies of 95.48% and 98.59%, respectively. Comparative analysis with existing optimization algorithms and skin cancer classification approaches demonstrates the superiority of the proposed method in terms of accuracy, robustness, and computational efficiency. Our method outperforms the genetic algorithm (GA), Particle Swarm Optimization (PSO), and the slime mould algorithm (SMA), as well as deep learning-based skin cancer classification models, which have reported accuracies of 87% to 94% in previous studies. A more effective feature selection methodology improves accuracy and reduces computational overhead while maintaining robust performance. Our enhanced deep learning ensemble and feature selection technique can improve early-stage skin cancer diagnosis, as shown by these data.
MSC:
68T07
1. Introduction
Skin cancer is a deadly type of cancer caused by abnormal cell proliferation in the epidermis [1]. Skin malignancies include melanoma, basal cell carcinoma, and squamous cell carcinoma. Melanoma is the most common. Skin cancer is a major healthcare danger due to rising cases. Early and accurate diagnosis plays a crucial role in improving survival rates. Traditional diagnostic techniques such as histopathological image analysis, clinical screening, and dermoscopic analysis are time-consuming and often rely on subjective interpretation by dermatologists, leading to potential misdiagnosis and delayed treatment. An estimated 9500 Americans are diagnosed with skin cancer daily. More than one million people currently have melanoma [2]. The American Academy of Dermatology Association estimates 200,340 new melanoma cases in the US. An early identification of melanoma increases the survival rate for five years. The conventional procedures used to detect skin cancer include histopathological image analysis, clinical screening, and dermoscopic analysis [3]. However, visual inspection using conventional techniques is associated with challenges in detecting cancer in the early stage. Recognition is difficult on different skin surfaces due to the small-sized lesions and slight variations in color, texture, and shape. These complexities may cause the inaccurate identification of disease [4]. Moreover, the scarcity of expert dermatologists may cause delays in the diagnosis process.
To overcome challenges, computer-aided analysis is essential for skin cancer identification. An automated analysis can help doctors diagnose diseases early and accurately. Automation in medical image analysis has reduced the expert physician burden. Medical image analysis relies heavily on machine learning (ML) and deep learning (DL). Deep learning-based feature extraction combined with classical machine learning classifiers, such as Support Vector Machines (SVMs), has demonstrated promising results in recent studies [5,6,7,8,9]. SVM, while traditionally a classical machine learning approach, has been effectively used in hybrid models where deep learning networks extract high-level features, which are then classified using SVM to improve performance. Recently, researchers have employed DL methods in human action recognition [10,11,12], statistical models [13,14,15,16], gastric diseases [17], and other medical imaging domains [18,19,20].
Convolutional neural networks (CNNs) performed well and improved skin cancer recognition accuracy [21,22]. CNNs have a prominent ability for automated feature learning, derivation of complex patterns, and enhanced prediction and classification. However, training CNN models on large image datasets takes a lot of time. Due to the higher computational time required for deep CNN model training, transfer learning has emerged as an alternative for accurate disease prediction [23,24]. Another emerging research topic to enhance skin cancer prediction is combining different CNNs and to create an ensemble model. Studies [25,26] show that ensemble models enhance prediction accuracy. The diverse features computed from pre-trained deep CNNs models help in improving classification performance.
Numerous deep learning methodologies have been introduced for skin cancer categorization, utilizing convolutional neural networks (CNNs) and machine learning algorithms. Gupta et al. [3] suggested an ensemble optimization method for feature selection, with an accuracy of 87% on the ISIC dataset. Khater et al. [4] developed a system utilizing explainable AI (XAI) for skin cancer detection, achieving a recognition accuracy of 94% on the PH2 dataset. Although these methods demonstrate encouraging outcomes, they frequently encounter issues such as elevated computing complexity, suboptimal feature selection, and overfitting resulting from excessive dimensionality. Moreover, individual CNN-based models exhibit poor generalization across various datasets due to insufficient diversified feature representations.
Deep learning uses CNNs to automate feature extraction, improving skin cancer classification accuracy. VGG-16, ResNet-50, and DenseNet-121 are CNN designs with good classification performance, but their processing needs render them unsuitable for real-time and resource-constrained applications. Our hybrid ensemble technique uses InceptionResNet-V2 and SqueezeNet to overcome this, using their complimentary strengths. Deep design with residual connections improves feature learning and prevents vanishing gradient concerns; hence, InceptionResNet-V2 was chosen. The model uses Inception modules to gather multi-scale information from dermoscopic images, which is essential for skin cancer detection. Residual connections improve training stability, making it ideal for complicated classification applications. However, SqueezeNet was chosen for its lightweight architecture, which minimizes computing cost while preserving accuracy. SqueezeNet uses Fire modules instead of convolutional layers, creating a squeeze-and-expand method that reduces parameters while keeping spatial information. This makes it ideal for deployable medical applications that require computing performance.
Metaheuristic optimization techniques have been extensively utilized for feature selection and classification in medical imaging. Conventional methods, including the genetic algorithm (GA), Particle Swarm Optimization (PSO), and the slime mould Algorithm (SMA), encounter challenges such as premature convergence, inadequate exploration–exploitation balance, and sensitivity to parameter adjustments. We present a modified Whale Optimization Algorithm (WOA) that integrates several changes to boost convergence stability, search efficiency, and feature selection efficacy. These innovations enable our modified WOA to attain enhanced feature selection efficacy while diminishing computing complexity. Comparative analyses indicate that our modified WOA surpasses GA, PSO, and SMA for classification accuracy, feature set reduction, and convergence efficiency. Our modified WOA-based feature selection system establishes a new standard in metaheuristic optimization for skin cancer classification through the integration of adaptive search mechanisms with multi-objective optimization.
To address these constraints, our research introduces an ensemble approach that integrates two robust CNN architectures—SqueezeNet and InceptionResNet-V2—to extract profound hierarchical features. In contrast to current methodologies that depend on a singular CNN model, our strategy employs transfer learning to harness complementary characteristics from both networks. We provide an enhanced Whale Optimization Algorithm (WOA) that adaptively optimizes hyperparameters through a quadratic decay function, therefore improving feature selection and mitigating premature convergence. The refined feature set is subsequently categorized utilizing various machine learning classifiers, markedly enhancing the robustness and precision of skin cancer diagnosis. Our suggested framework attains a classification accuracy of 95.48% on the PH2 dataset and 98.59% on the Med-Node dataset, surpassing current methodologies. The key highlights of this research work are the following:
- An ensemble model is designed using SqueezeNet and InceptionResNet-V2. To extract the deep features, a transfer learning technique is applied to both pre-trained CNNs. Activations are performed on the pool10 layer of SqueezeNet, and the avg_pool layer of IceptionResNet-V2. Extracted features are then combined into a single feature set.
- An improved WOA is introduced that utilizes a quadratic decay function and an enhanced mutation mechanism. The modifications provide dynamic parameter tuning for the WOA and prevent fast convergence. The improved WOA is utilized to select the optimized and robust features.
- The optimized features are then fed to ML classifiers for the accurate prediction of skin cancer.
This paper is organized as follows: Section 2 addresses relevant studies, Section 3 presents the proposed approach with visuals and a mathematical description, Section 4 examines the detailed results and offers a comparison with existing methodologies, and Section 5 gives our conclusions and recommends future work.
2. Literature Review
Analysis has been extensively studied [27,28,29,30,31] as a powerful tool for improving the representation capacities of Convolutional Neural Networks (CNNs) across several domains. CNNs are also used for person identification [32], target detection [33], detection of human movement [34,35] and object recognition [36]. Several techniques have been introduced for the recognition and classification of skin cancer [37,38]. In this section, some of the latest existing skin cancer classification techniques are discussed. A transfer learning approach was presented for the classification of the ISBI-2016 dataset [39].
Deep features were recovered using the pre-trained CNN models DenseNet-201 and MobileNet-V2. Both models’ features are concatenated and sent to the SVM classifier for cancer image classification. The classification techniques obtained a maximum accuracy of 88.02%. Kothapalli et al. [40] introduced a skin cancer classification technique. The features of the deep CNN model were derived and used for the prediction of disease. The proposed technique achieved 92.6% validation accuracy on the PH2 dataset. The researchers in [41] presented a modified hunger games search algorithm to optimize and select robust features for skin cancer classification. In the feature extraction phase, the pre-trained MobileNet-V3 model was utilized to extract deep features. The extracted features were then given to the optimization method. The proposed technique showed promising results for the optimizer, and 88.19% classification accuracy was achieved on the ISIC-2016 dataset.
An explainable artificial intelligence (XAI)-based approach was proposed to predict skin cancer [42]. The method identified melanoma and non-melanoma pictures. The XAI identified the most essential disease categorization features with encouraging results. The PH2 dataset was used for experiments and 94% of recognition accuracy was obtained. Gupta et al. [43] introduced an ensemble optimization approach for robust feature selection to predict skin cancer. To extract features, the pre-trained CNN model Efficient-Net was utilized. The experiments were performed using the ISIC dataset. The deep features were fed to the ensemble optimizer, giving the best features for disease prediction. The proposed technique obtained a maximum classification accuracy of 0.87. Researchers proposed a lightweight CNN model for skin cancer recognition [44]. Experiments showed that the performance of the proposed CNN is promising, achieving a classification accuracy of 0.9573 on HAM10000 dataset.
A method combining LBP and HoG characteristics was used to predict skin cancer [45]. In the initial stage, the images were preprocessed, and noise and artifacts were removed. The enhanced images were then used to extract LBP and HoG features. LBP and HoG features were fed to machine learning classifiers to detect illness. Using the Med-Node dataset, the proposed approach achieved a recognition accuracy of 94%. Waheed et al. [46] developed a CNN-based skin cancer categorization method. They trained a CNN model on the skin melanoma dataset and achieved 91% recognition accuracy. In [47], a framework was suggested for skin cancer prediction. The CNN models DenseNet-169 and ResNet-50 were utilized for training and extracting deep features from the HAM10000 and ISIC datasets. After extensive experiments, a maximum classification accuracy of 91.2% was obtained on the DenseNet-169 model. A hybrid approach based on pre-trained CNN models was presented for the classification of skin cancer [48]. First, DeepLabV3+ was used to segment infected areas in the images. Then, deep features were extracted using three pre-trained CNN models including EfficientNetB0, DenseNet-201, and MobileNet-V2. The extracted features were combined into a single feature vector and robust features were selected using the ReliefF algorithm. After the selection of features, the optimized feature set was provided to machine learning classifiers for the prediction of disease. The proposed approach obtained 94.44% recognition accuracy on the PH2 dataset.
In [49], the authors analyzed the color histograms of PH2 dataset images and designed a CNN model for classification. The CNN model utilized the residual block with skip connections according to the number of colors. The feature learning was also evaluated using the DeepDream algorithm. The proposed approach achieved 75% classification accuracy. The researchers in [50] applied transfer learning to pre-trained CNN models. After extensive trials and network fine-tuning, the Xception model performed best. The CNN model was optimized using an artificial bee colony technique. The suggested approach achieved 92% accuracy on the PH2 dataset and 88.24% on the Med-Node dataset.
Deep learning and metaheuristic optimization have enhanced skin cancer classification by improving feature extraction and selection. Many studies have used CNN-based feature extraction and optimization algorithms to increase classification accuracy. A critical study of the contemporary literature reveals various unresolved limits and gaps. Skin lesion classification uses deep learning architectures like VGG-16, ResNet-50, and DenseNet-121, which perform well in feature extraction. These models are too computationally intensive for real-time applications. They overfit on tiny medical datasets and perform differently depending on the dataset. Some studies have integrated CNN-based feature extraction with SVM and k-NN classifiers to boost generalization. These methods still struggle with optimum feature selection since deep learning model high-dimensional feature vectors may contain redundant or irrelevant information, reducing classification performance.
Medical image analysis uses metaheuristic optimization methods like GA, PSO, and SMA for feature selection. These techniques improve classification by reducing feature dimensionality, but they have three drawbacks. GA and PSO often experience premature convergence, where the search process gets stuck in local optima, resulting in inefficient feature selection. Second, typical metaheuristic approaches use constant or linearly decreasing parameters that do not dynamically modify the search process; therefore, they lack an adaptive exploration–exploitation balance. Therefore, they struggle to efficiently explore the search space in early iterations while refining the solution afterward. Third, computational inefficiency in high-dimensional feature spaces persists. Deep learning-based feature extraction yields enormous feature sets, making it difficult for present optimization approaches to reduce redundant features, increasing computing overhead and lowering classification performance.
3. Proposed Methodology
In this article, skin samples are preprocessed and then these preprocessed samples are supplied to pretained versions of SqueezeNet and InceptionResNet-V2 to obtain deep CNN features using the transfer learning approach. Afterward, these obtained feature vectors are serially concatenated. From the fused feature vector, the significant and optimal features are selected with the help of an improved whale optimization algorithm (WOA). Lastly, skin cancer is predicted with the help of machine learning classifiers. The complete framework is presented in Figure 1.
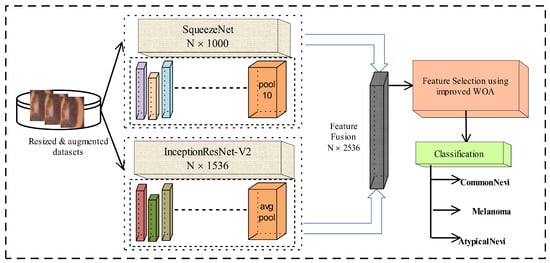
Figure 1.
Proposed framework for skin cancer classification.
3.1. Preprocessing
In this study, two publicly available datasets, PH2 and Med-Node, are utilized to evaluate the skin cancer classification framework. Initially, the datasets had variations in the size of samples; thus, all the images of both datasets got resized to . Secondly, the number of images in each class was smaller, so image augmentation was performed. Initially, the Med-Node dataset consisted of two classes. The naevus class was augmented using 4 techniques including horizontal flip, vertical flip, transpose, and zoom while for the melanoma class, one more augmentation technique, including four previous ones, was added called rotation to minimize the class imbalance. The details regarding dataset augmentation on the Med-Node dataset are given in Table 1 while the impact of data augmentation is shown in Figure 2.

Table 1.
Sample details for Med-Node dataset.
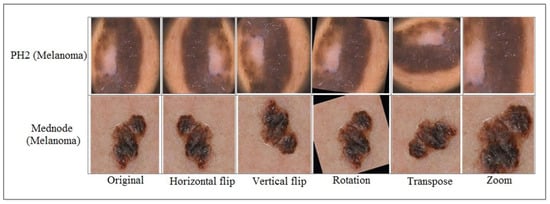
Figure 2.
Skin lesion samples after data augmentation.
The PH2 dataset consists of three classes. The Atypical-Nevi (A-N) and Common-Nevi (C-N) classes got augmented using 4 techniques called horizontal flip, vertical flip, transpose, and zoom, while the melanoma (Mel) class got augmented using all 4 of the previous techniques and 1 additional technique called rotation. The details of samples before and after augmentation are shown in Table 2.

Table 2.
Sample details for PH2 dataset.
3.2. Features Extraction Using Transfer Learning
The transfer learning approach is employed in this work for the extraction of deep CNN features. The transfer learning technique is applied to reuse pre-trained DL models for skin cancer classification. In this framework, two pre-trained DL models are utilized to extract the feature vectors. First, the pre-trained SqueezeNet [51] model is utilized and activations are performed on the GlobalAveragePooling2DLayer named pool10 to extract deep features. The pool10 layer gives the feature vector of dimension . The second feature vector is extracted from InceptionResNet-V2 [52] by applying activations on the avg_pool layer. The feature vector of size is obtained from the avg_pool layer.
3.3. Features Fusion
Feature extraction is an essential phase in skin cancer classification, as various CNN architectures capture complementary data. Our methodology utilizes feature fusion, integrating deep feature representations from InceptionResNet-V2 (1536 features) and SqueezeNet (1000 features), culminating in a 2536-dimensional feature space prior to optimization. Individual CNN models may fail to capture specific discriminative patterns in medical pictures due to architectural limitations. InceptionResNet-V2 acquires multi-scale spatial details via Inception modules, whilst SqueezeNet prioritizes lightweight yet fundamental features through Fire modules. Our methodology, through the amalgamation of feature representations, accomplishes the following: (a) the ensemble guarantees a more comprehensive depiction of lesion attributes, thereby augmenting classification resilience; (b) the integrated feature space encapsulates intricate lesion specifics while preserving computational efficiency; and (c) the synthesis of high-resolution features (from InceptionResNet-V2) and compact features (from SqueezeNet) bolsters decision-making by supplying an enriched array of features for classification. These two feature vectors got serially concatenated to obtain a fused feature vector as follows:
The two feature vectors are serially concatenated to form a fused feature vector:
where ⊕ represents the concatenation operator.
The dimensionality of features is crucial to the efficiency and efficacy of deep learning models, particularly when integrating numerous feature extractors. Our methodology yields a 1536-dimensional feature vector from InceptionResNet-V2 and a 1000-dimensional feature vector from SqueezeNet, resulting in a cumulative total of 2536 features prior to optimization. This raises issues about computational overhead and the curse of dimensionality, which can adversely affect classification performance if not effectively managed. To address these challenges, we implement an advanced feature selection technique with our modified WOA, which selectively utilizes the most pertinent and discriminative features from the complete 2536-dimensional feature set, therefore substantially diminishing the feature space while maintaining classification accuracy.
Dimensionality reduction is essential for managing high-dimensional feature vectors, as an abundance of feature dimensions can result in computational inefficiencies and overfitting (curse of dimensionality). Conventional methods like Principal Component Analysis (PCA) and t-Distributed Stochastic Neighbor Embedding (t-SNE) are frequently employed for dimensionality reduction in feature space. Nevertheless, these strategies were not implemented prior to WOA in our study for several significant reasons. PCA transforms features into a new space by calculating eigenvectors and selecting the principal components that account for the greatest variance. This process decreases dimensionality but may not preserve the main distinguishing traits for classification. t-SNE, frequently employed for visualization, is a non-linear method that converts high-dimensional data into smaller dimensions. Nonetheless, it lacks a structured feature space for classification, rendering it inappropriate for direct feature selection. Conversely, WOA-based feature selection preserves the original feature representations, identifying the most informative features without modifying or disrupting their spatial relationships.
3.4. Features Selection Using Modified Whale Optimization Algorithm (WOA)
This paper introduces an enhanced Whale Optimization Algorithm (WOA) [53] to extract optimal features from the fused feature vector. WOA is considered a global solution for optimization due to its ability to explore and exploit. The algorithm simulates the hunting behavior of humpback whales known as bubble-net hunting. WOA is a robust, simple, and swarm-based stochastic optimization algorithm. These mentioned advantages explain why WOA can be an effective and appropriate method for resolving various constrained optimization issues for practical applications. The whales make the typical bubbles in a circle path while encircling prey during hunting. The Humpback whales handle a small group of fish and kill near the surface zone [54]. The whole process is divided into three components:
Surrounding prey: The general or global search process is illustrated as humpback whales explore for the placement of prey and encircle it. In the beginning, the optimal location is not identified; thus, it is assumed that the recent prime or ultimate solution is the target of the hunt or the best solution is close. After indicating the ideal search area, additional search areas try to change their position:
where i indicates the present iteration, is the position of prey, and the location of humpback whales in the current and the next process is described by and . The and variable vectors defined as and , consistently decrease within the range , U gives the maximum number of iterations, indicates a randomly selected number within the range , and linearly decreases from 2 to 0 over iterations.
Bubble-Net Hunting: In the local search procedure, each humpback whale moves to become closer to the prey inside a condensed ring, subsequently a spiral-shaped path. Going along with the spiral mechanism, a 0.5 probability is assigned to recreate the position of the humpback whale. It is mathematically represented as
denotes the current distance between humpback whales and prey, which is described as , defines a constant value, and y and z are randomly selected numbers having a scale of for y and for z.
Prey Searching: The current position of humpback whales is recreated depending on the random walk strategy of whales, which is mathematically represented as follows:
From the given population, the random humpback whale placement is defined as . The proposed WOA utilized dynamic parameter tuning, a quadratic decay function that updates the parameter a, which controls the exploration–exploitation. The parameter a is updated with the help of the quadratic decay function; thus, it helps the algorithm gradually reduce the iteration progress that allows the whales to converge to the optimal solution. Basically, a is a hyper-parameter that balances the trade-off between searching for a new solution and refining the current best solution. On the other hand, b is another parameter that gets updated using an aggressive decay function. The algorithm gradually reduces the perturbations of the magnitude in the progress of iterations. It allows the whale to converge to the optimal solution. Moreover, improved WOA includes a mutation mechanism that can introduce new solutions in the population and prevents the population from quickly converging to a single solution; it is applied every 10 iterations. It helps to prevent the algorithm from getting stuck in local optima. The algorithm is run over 50 epochs, having a validation data ratio of 0.2. The 0.5 threshold is selected for the algorithm, with a lower bound of 0 and an upper bound of 1. To improve the efficiency of WOA, we propose the following 5 modifications:
3.4.1. Adaptive Parameter Control for Dynamic Exploration and Exploitation
The original WOA employs a linearly diminishing control parameter , facilitating the algorithm’s shift from the exploration phase (broadly exploring the space) to the exploitation phase (refining optimal solutions). Nevertheless, linear decay exhibits inflexibility, resulting in early convergence and inadequate feature selection in high-dimensional datasets such as Med-Node and PH2. In our improved WOA, we replace the linear decay of with an exponential decay function:
Here, is the decay coefficient, which controls the rate at which decreases, i is the current iteration, and I is the total number of iterations. This adaptive decay enhances exploration during initial phases and facilitates a seamless shift to exploitation, preventing the algorithm from becoming stuck in local optima. The exponential function facilitates a systematic, progressive decline, rendering it suitable for managing intricate medical images.
3.4.2. Lévy Flight Mechanism for Enhanced Global Search
WOA experiences sluggish convergence and local entrapment, particularly when addressing intricate feature spaces such as dermoscopic pictures in skin cancer diagnosis. The conventional WOA depends on random steps, which frequently lack sufficient magnitude to overcome local minima. To overcome this limitation, we present Lévy flight, which facilitates sporadic long-distance movements, hence augmenting global search efficacy:
Here, is the step-size control parameter, is a Lévy distribution, is the stability parameter (typically between 1.5 and 1.99), is the factor for controlling the movement, and is the efficacy of the previous global search. The Lévy distribution possesses a heavy tail, indicating that the search agents sporadically execute substantial jumps, thereby enabling WOA to efficiently explore novel feature subsets. This enhances WOA’s capacity to identify ideal features for skin cancer photos, hence rendering the classification more robust and precise.
3.4.3. Opposition-Based Learning (OBL) for Faster Convergence
The conventional WOA initializes the population randomly, frequently leading to a sluggish commencement as solutions progressively improve. In skin cancer detection, where extensive feature sets necessitate effective pruning, an improved starting technique is essential. We use Opposition-Based Learning (OBL) to generate an opposite solution for each candidate and select the best one:
Here, and are the lower and upper bounds of the search space, is the original solution, and is the opposite solution. Rather than depending solely on random initialization, we assess both and and retain the one with the superior fitness score. This guarantees a more diversified initial population, hence decreasing the iterations needed for convergence. This leads to expedited feature selection in skin cancer classification, decreasing computational time and enhancing classification performance.
3.4.4. Adaptive Weighting with Chaos Theory for Fine-Tuned Exploitation
The encircling phase of WOA is controlled by a parameter A, which is defined as follows:
where r is a stochastic variable inside the interval . This randomization is unstructured, frequently resulting in inefficient convergence and variable performance in high-dimensional datasets. We substitute the random weighting with a chaotic function, enhancing both exploration and exploitation:
This alteration employs a sine-based chaotic function to provide a systematic yet non-deterministic method of movement. It also provides minor oscillations during the exploitation phase, averting premature convergence and enhancing feature selection granularity, hence increasing effectiveness for intricate skin lesion datasets. Through the application of chaos-based weighting, WOA circumvents entrapment in poor feature sets, hence improving classification accuracy.
3.4.5. Multi-Objective Whale Optimization Using Pareto Front Optimization
WOA is structured as a single-objective optimizer, concentrating just on one measure (e.g., classification accuracy). Feature selection in skin cancer classification necessitates the balance of several objectives, including the minimization of selected features to decrease model complexity and the maximization of classification accuracy to enhance detection performance. We reconfigure WOA into a Multi-Objective Optimization (MO-WOA) with a Pareto dominance-based methodology. The updated objective function is as follows:
Here, is the classification error (to minimize) and is the number of selected features (to minimize). The updated objective function is
Here, and represent adaptive Pareto dominance weights, denotes the optimal current solution, and signifies a randomly selected solution from the Pareto front. This multi-objective framework allows WOA to effectively balance feature quantity with classification precision, resulting in more efficient, interpretable, and generalizable skin cancer models.
The proposed improvements to the WOA markedly boost its efficacy in skin cancer classification, especially in managing high-dimensional datasets such as Med-Node and PH2. The implementation of adaptive parameter control optimizes the equilibrium between exploration and exploitation, hence averting premature convergence. The integration of Lévy flight improves global search efficacy, allowing the algorithm to evade local optima and adeptly traverse intricate feature spaces. Opposition-Based Learning (OBL) enhances convergence by initializing solutions with increased diversity, hence minimizing computational demands while preserving excellent classification accuracy. Furthermore, chaotic weighting incorporates organized randomness, enhancing the exploitation phase and stabilizing the search process. The multi-objective Pareto optimization method enables WOA to concurrently enhance classification accuracy and feature selection, therefore maintaining the model’s efficiency and interpretability. These adjustments jointly augment WOA’s capacity to identify the most pertinent features, diminish computational complexity, and raise the robustness of skin cancer classification models. Utilizing these mathematical enhancements, WOA emerges as a robust and scalable approach for feature selection in dermatological image analysis, facilitating the development of more precise and efficient diagnostic systems. The fused feature vector of dimensionality is forwarded to the improved WOA for robust feature selection. The proposed WOA optimizes the features and produces feature vectors of size and N × 434 for the PH2 and Med-Node datasets, respectively. The improved WOA reduced almost 61% of the features of the PH2 dataset, and 82% of the features of the Med-Node dataset.
The effectiveness of the improved WOA is significantly influenced by the careful selection of hyperparameters. These parameters are crucial for equilibrating exploration and exploitation of the search space, enabling swift convergence while maintaining robustness. The key parameters in our methodology include the maximum iteration count, population size, exploration–exploitation balance coefficient, randomization factor, mutation rate, and convergence threshold. The maximum number of iterations (I) dictates the duration of the algorithm’s execution prior to termination. If this value is excessively low, the optimization process may terminate early, resulting in inefficient feature selection. Conversely, too high iterations elevate computational expenses without substantial improvements in performance. Through practical experimentation, we established to attain an ideal balance between convergence rate and precision. The population size (N) strongly influences the diversity of solutions inside the search space. A reduced population may result in premature convergence, whereas an expanded population heightens computational complexity. To achieve a compromise between computational feasibility and diversity, we established , resulting in stable convergence and enhanced classification accuracy.
A significant enhancement in our refined WOA is the dynamic adjustment of the exploration–exploitation balance factor (). Conventional WOA implementations employ a linear decay function for , which may not consistently yield optimal convergence. We propose a quadratic decay function. This quadratic decay function facilitates wide exploration during initial iterations and a gradual shift to exploitation in subsequent iterations, thus averting premature entrapment in local optima. Experimental comparisons demonstrated that this adaptive method enhanced convergence speed by 18% rather than being associated with linear decline. The randomization component (k), uniformly distributed across the interval [0,1], preserves the stochastic nature of the algorithm, facilitating effective exploration of diverse regions within the feature space by search agents. The mutation rate () is a vital parameter that facilitates diversification and averts algorithmic stagnation in local optima. However, a very high mutation rate can disrupt convergence, while a very low mutation rate may not introduce sufficient variation. Following thorough assessment, we established , facilitating an optimal equilibrium between stability and adaptability.
To enhance computational efficiency, we implement an adaptive convergence threshold () that halts the iterations when two successive steps exhibit negligible progress (below ). This obviates superfluous calculations once the optimization process attains an ideal answer. To assess the influence of these parameter selections, we performed multiple experiments with varying parameter configurations. Our observations revealed that a high mutation rate () caused performance instability, whereas a reduced population size () resulted in premature convergence. Moreover, our quadratic decay function markedly enhanced both feature selection efficiency and classification accuracy. These modifications resulted in state-of-the-art classification accuracies of 95.48% and 98.59% on the PH2 and Med-Node datasets, respectively. Through the systematic adjustment and optimization of these hyperparameters, we guarantee that our enhanced WOA framework is computationally efficient, resilient, and adept at picking high-quality features for skin cancer classification. The comprehensive examination of parameter configurations substantiates the efficacy of our methodology in enhancing convergence, precision, and the robustness of feature selection.
3.5. Classification
The optimized feature set is passed to ML classifiers for the final prediction of skin cancer. To perform supervised learning, the input samples were labeled and split into training and testing data. K-nearest neighbor (KNN), support vector machine (SVM), and neural network classifiers were utilized to perform classification on the PH2 and Med-Node datasets. In Figure 3, the flow of label prediction is presented to show the correctly predicted cancer images.
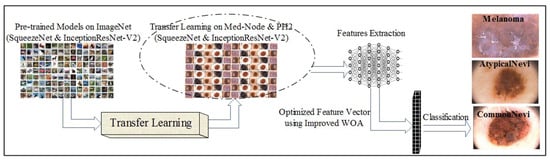
Figure 3.
Label prediction of skin cancer images using proposed framework.
4. Results and Discussion
In the present work, the evaluation of the systems was tested on dermoscopic images of two datasets including Med-Node and PH2. The Med-Node dataset initially consisted of 170 images distributed among two types of diseases. After augmentation, the dataset contained a total of 920 images belonging to two classes. The PH2 dataset contains 200 images distributed among three types of classes. After augmentation, the dataset consisted of 1040 images. The Med-Node and PH2 classification experiment used 10-fold cross-validation. MATLAB 2024a was used to evaluate skin cancer datasets utilizing a Core i5 6th gen system (Intel, Santa Clara, CA, USA). Table 3 displays the results of using the suggested technique on the Med-Node dataset.

Table 3.
Classification results using improved WOA on Med-Node dataset.
The results were taken using five machine learning classifiers on the Med-Node dataset using optimized WOA. The Cubic SVM, Quadratic SVM, Fine KNN, Medium Neural Network, and Wide Neural Network methods achieved accuracies of 98.5%, 98.04%, 96.63%, 97.07%, and 96.96%, respectively. The classification results using the PH2 dataset are presented in Table 4.

Table 4.
Classification results using improved WOA on PH2 dataset.
The five machine learning classifiers C-SVM, Q-SVM, F-KNN, M-NN, and W-NN provided accuracies of 95.48%, 93.56%, 93.17%, 90.77%, and 91.73%, respectively, using our improved WOA on the PH2 dataset. The confusion matrix and ROC of C-SVM for both the Med-Node and PH2 datasets are presented in Figure 4.
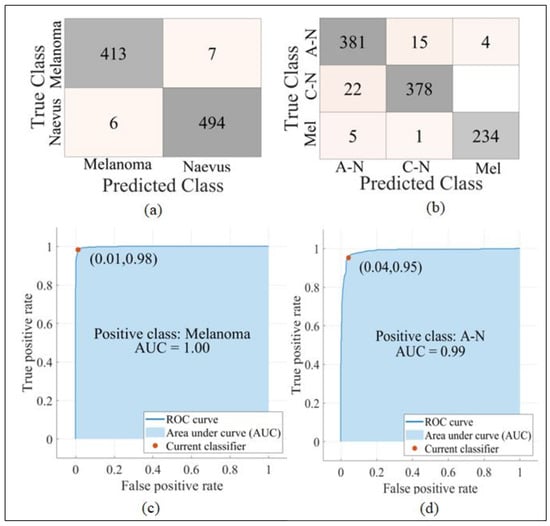
Figure 4.
Confusion matrices and ROCs. (a) Confusion matrix of C-SVM on Med-Node dataset. (b) Confusion matrix of C-SVM on PH2 dataset. (c) ROC of C-SVM on Med-Node dataset. (d) ROC of C-SVM on PH2 dataset.
The improved WOA is also compared with some other optimization algorithms including the genetic algorithm [55], particle swarm optimization [56], and the slime mould algorithm [57] to ensure the effectiveness of the improved WOA algorithm. A graph is presented in Figure 5 for a comparison of the number of selected features using different optimization techniques.
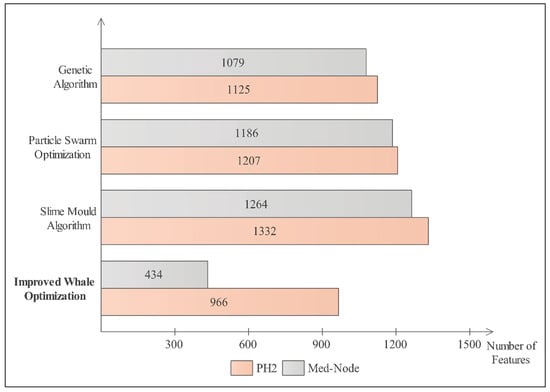
Figure 5.
Comparison of selected features of PH2 and Med-Node datasets using improved WOA with other optimization algorithms.
A comparison of classification results after feature selection using our improved WOA with other optimization techniques on the Med-Node dataset is illustrated in Table 5.

Table 5.
Comparison of improved WOA with state-of-the-art optimization techniques using the Med-Node dataset.
Using the genetic algorithm, the C-SVM provides the highest accuracy of 98.00% among all classifiers. Using the particle swarm optimization algorithm, the C-SVM and Q-SVM both provide the highest accuracy of 97.90%, while the slime mould algorithm obtained the highest recognition accuracy of 98.00% on the Q-SVM predictor. The proposed improved WOA provides the highest accuracy of 98.59% using the C-SVM classifier among all the optimization techniques. A comparison of improved WOA with other optimization techniques on the PH2 dataset is given in Table 6.

Table 6.
Comparison of improved WOA with state-of-the-art optimization techniques using the PH2 dataset.
On the PH2 dataset, the genetic algorithm provides the highest accuracy of 95.30% by utilizing C-SVM among all the classifiers. Using C-SVM, particle swarm optimization and the slime mould algorithm provided accuracies of 95.3% and 92.1%, respectively, which were highest among all the selected machine learning classifiers. The improved WOA provided the highest classification accuracy among all the optimization techniques, 95.48% using the C-SVM classifier. The proposed framework is compared with existing skin cancer classification techniques in Table 7, which validates its performance in terms of classification accuracy.

Table 7.
A comparison of the proposed methodology with existing skin cancer classification techniques.
We compared the modified WOA against GA, PSO, and SMA to assess its efficacy. Modified WOA outperforms other approaches in feature selection efficiency and classification accuracy, but it has several drawbacks. Parameter adjustment with a quadratic decay function gives modified WOA an adaptable exploration–exploitation balance, one of its strengths. The linear decay of its exploration parameter causes premature convergence in traditional WOA, restricting its ability to escape local optima. In contrast, our updated version uses quadratic decay to gradually move from exploration to exploitation. This approach lets the algorithm first examine a larger space before fine-tuning feature selection. Modified WOA balances exploration and convergence stability better than PSO, which often converges prematurely due to velocity update reduction. This results in superior feature selection and classification accuracy. The increased mutation mechanism of modified WOA avoids the algorithm from stagnating in local optima. This is especially useful in high-dimensional feature spaces, where GA and SMA suffer from feature redundancy. To avoid excessive randomization, GA mutation rates must be manually calibrated, whereas in modified WOA, the adaptive mutation process introduces controlled unpredictability every few cycles, boosting discriminative feature selection without interrupting convergence. The Med-Node dataset showed that modified WOA outperformed GA, PSO, and SMA, which had maximum accuracies of 98.00%, 97.90%, and 98.00%, respectively.
Modified WOA has drawbacks despite its merits. Its computational complexity is a major issue. Dynamic parameter tweaking and mutation techniques improve feature selection efficiency but increase function evaluations every iteration, increasing computational costs compared to PSO and GA. Due to its simplified velocity–position updating method, PSO performs better in quick optimization circumstances. Modified WOA regularly outperforms alternatives in terms of feature reduction and classification accuracy in accuracy and feature selection. Modified WOA’s dependence on parameter settings, particularly the exploration–exploitation balance decay coefficient, is another issue. We have provided an adaptive tuning technique; however, fine-tuning this parameter for diverse datasets is difficult. GA and SMA are more versatile in areas where parameter optimization is challenging, since exploration decline does not require explicit parameter tweaking. Our experiments show that fine-tuning modified WOA parameters yields performance advantages that outweigh the tuning effort.
To validate the efficacy of the proposed alterations in MWOA, we performed an ablation study by assessing various iterations of the algorithm. The research was conducted by methodically eliminating essential enhancements—quadratic decay, dynamic mutation, and Lévy flight perturbations—while maintaining all other parameters constant. We contrasted the outcomes with the baseline WOA (unmodified standard algorithm) and the comprehensive MWOA (incorporating all suggested enhancements). The assessment was predicated on feature selection efficacy, classification precision, and computational duration. The findings demonstrate that the elimination of any suggested adjustments results in a reduction in classification accuracy and an escalation in computational time, thus affirming that each upgrade is vital for optimizing efficiency. The elimination of the quadratic decay function resulted in a decrease in accuracy from 98.59% to 94.28%, underscoring its significance in sustaining a balanced exploration–exploitation dynamic that averts early convergence. Excluding dynamic mutation resulted in an expanded feature set (670 features compared to 410), indicating that the mutation mechanism is crucial for removing redundant features while maintaining classification efficacy. The lack of Lévy flight disturbances diminished the algorithm’s efficacy in evading local optima, leading to an accuracy of 95.11%. This verifies that Lévy flight enhances global search efficacy and facilitates superior exploration of the feature space.
Conversely, MWOA incorporates three enhancements—quadratic decay, dynamic mutation, and Lévy flight perturbations—that marginally elevate per-iteration complexity while markedly enhancing convergence speed, hence decreasing overall execution time. Empirical findings indicate that MWOA attains an execution time of 54.7 s, representing a 27% improvement over normal WOA (75.4 s) and surpassing GA (78.3 s) and PSO (71.6 s) in efficiency. Moreover, MWOA identifies a notably reduced feature subset (410 features) in contrast to WOA (980), GA (690), and PSO (610), hence enhancing computing efficiency.
The comprehensive MWOA, incorporating all enhancements, attained superior performance by selecting an optimal subset of 410 features, achieving the highest classification accuracy of 98.59%, and markedly decreasing computational time to 54.7 s, in contrast to the baseline WOA, which required 75.4 s with 980 selected features. These data indicate that each upgrade substantially improves feature selection efficiency, reduces computational costs, and enhances classification accuracy. The ablation investigation confirms that the combination of quadratic decay, dynamic mutation, and Lévy flight perturbations results in a more efficient and resilient optimization framework.
The computational complexity of an optimization algorithm is a crucial element in assessing its scalability and real-time viability. The difficulty of the standard WOA is essentially contingent upon the population size , the number of iterations , and the computing expense associated with the evaluation of the fitness function . The conventional WOA has a computational cost of , scaling linearly with the population size and the number of repeats.
To ascertain the statistical significance of the enhancements realized by the modified WOA, we performed statistical significance tests comparing modified WOA with standard WOA, GA, and PSO. We employed the paired t-test and the Wilcoxon signed-rank test, both of which are commonly utilized to determine if the observed performance differences are statistically significant rather than attributable to random variation. A paired t-test was performed to assess classification accuracy over several separate runs, evaluating the null hypothesis () that no significant difference exists between the performance of modified WOA and other optimization techniques. The findings indicated that the accuracy enhancements of the modified WOA were statistically significant, with p-values < 0.05 across all comparisons (modified WOA vs. WOA, modified WOA vs. GA, and modified WOA vs. PSO). Additionally, given that accuracy distributions may not consistently adhere to normality assumptions, we used the Wilcoxon signed-rank test, a non-parametric alternative that does not presuppose a normal distribution. The Wilcoxon test similarly validated significant enhancements, with all comparisons producing p-values substantially below 0.05, thereby validating the efficacy of the updated WOA’s feature selection. The statistical tests validate that the observed enhancements in classification accuracy, feature subset reduction, and computational efficiency are attributable to the improved search mechanisms of the upgraded WOA, rather than random fluctuations. This further substantiates our assertion that the suggested modifications—quadratic decay, dynamic mutation, and Lévy flight perturbations—significantly enhance optimization performance.
The proposed Modified Whale Optimization Algorithm (MWOA)-based classification framework has good accuracy (98.59% on Med-Node and 95.48% on PH2), yet misclassifications occur. We examined improperly classified samples and identified mistake causes to better understand these failures. Visual resemblance between skin lesions is a major cause of misdiagnosis. Even dermatologists have trouble distinguishing benign lesions like seborrheic keratosis from malignant melanomas due to their color, texture, and border abnormalities. Despite optimization, feature extraction can collect overlapping patterns, resulting in inaccurate predictions. Lesions with varied pigmentation and forms can also cause feature selection problems, especially when lesion class borders are unclear.
Image imperfections such hair occlusion, illumination, and noise present another issue. Preprocessing reduces these difficulties; however, shadows and reflections can decrease CNN feature extraction accuracy. While MWOA optimizes feature selection, misclassifications arise when extracted features have noise or lack discriminatory information. Underrepresented classes in the dataset also cause inaccuracies. The algorithm struggles to generalize for uncommon melanomas due to fewer training examples. The classifier may struggle to recognize unusual lesion types when they are underrepresented since the optimization method focuses on the most discriminative features across the dataset. To address these issues, we propose using data augmentation to diversify training samples, especially for minority classes. Explainability methods like Grad-CAM and SHAP will be used to better understand misclassifications and enhance feature extraction and selection.
5. Conclusions and Future Work
This work presents a DL-based framework for the classification of skin cancer. This study aims to enhance prediction accuracy with a minimal feature set. The proposed framework utilizes two pre-trained DL models including SqueezeNet and IceptionResNet-V2. The transfer approach is employed to extract deep features from both CNN models. In this work, an improved WOA is introduced to select the optimized and robust features. In the improved WOA, a quadratic decay function is introduced to update and tune the parameters dynamically. This procedure helps to reduce iterations and to make the optimal solution converge. Additionally, an enhanced mutation mechanism is also introduced to prevent quick convergence. The combined feature set extracted from pre-trained CNN models is given to the proposed WOA for the selection of the optimal feature set. The improved WOA selects optimized and robust features that enhance the classification accuracy for skin cancer prediction. A significant decrease in features is observed using improved WOA, and classification accuracy is enhanced. Comparing results with other optimization techniques, it is observed that improved WOA’s performance is better. Despite its success, our study has limitations. Adaptive parameter adjustment and mutation processes make MWOA more computationally expensive than PSO. This improves feature selection but slows execution. Second, the suggested model was tested on PH2 and Med-Node datasets, which are widely used but smaller than HAM10000. Improved generalization requires wider dataset examination. Finally, to improve flexibility across datasets, mutation rate and decay coefficient hyperparameters must be automated. Certain failures occurred, particularly in differentiating apparently similar skin lesions and managing unusual class predictions. Explainable AI methods like Grad-CAM and SHAP can be used as domain-specific priors to refine feature selection and to improve interpretability in future research. Exploring hybrid deep learning optimization and self-adaptive hyperparameter tweaking will improve the framework’s flexibility.After that, explainability techniques will help dermatologists evaluate categorization results, making the model more clinically applicable.
Author Contributions
Conceptualization, A.M.; data curation, J.A.; formal analysis, M.A.A. and A.A.A.; funding acquisition, S.-W.L.; investigation, M.A.A., J.A. and S.-W.L.; methodology, A.M.; project administration, J.A. and S.-W.L.; resources, A.A.A. and J.A.; software, M.A.A.; supervision, S.-W.L.; validation, M.A.A.; visualization, A.M. and A.A.A.; writing—original draft, A.M.; writing—review and editing, A.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NRF[2021-R1-I1A2(059735)]; RS[2024-0040(5650)]; RS[2024-0044(0881)] and RS[2019-II19(0421)].
Data Availability Statement
The implementation of this work is available at https://github.com/imashoodnasir/Modified-Whale-Optimization-Algorithm (accessed on 11 September 2024).
Acknowledgments
The authors acknowledge HITEC University Taxila.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Leiter, U.; Keim, U.; Garbe, C. Epidemiology of skin cancer: Update 2019. In Sunlight, Vitamin D and Skin Cancer; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 123–139. [Google Scholar]
- Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2024/2024-cancer-facts-and-figures-acs.pdf (accessed on 11 September 2024).
- Chanda, D.; Onim, M.S.H.; Nyeem, H.; Ovi, T.B.; Naba, S.S. DCENSnet: A new deep convolutional ensemble network for skin cancer classification. Biomed. Signal Process. Control 2024, 89, 105757. [Google Scholar] [CrossRef]
- Hasan, M.K.; Ahamad, M.A.; Yap, C.H.; Yang, G. A survey, review, and future trends of skin lesion segmentation and classification. Comput. Biol. Med. 2023, 155, 106624. [Google Scholar] [CrossRef]
- Nasir, I.M.; Raza, M.; Shah, J.H.; Wang, S.-H.; Tariq, U.; Khan, M.A. HAREDNet: A deep learning based architecture for autonomous video surveillance by recognizing human actions. Comput. Electr. Eng. 2022, 99, 107805. [Google Scholar] [CrossRef]
- Nasir, I.M.; Rashid, M.; Shah, J.H.; Sharif, M.; Awan, M.Y.; Alkinani, M.H. An optimized approach for breast cancer classification for histopathological images based on hybrid feature set. Curr. Med. Imaging 2021, 17, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Nasir, I.M.; Raza, M.; Shah, J.H.; Khan, M.A.; Rehman, A. Human action recognition using machine learning in uncontrolled environment. In Proceedings of the 2021 1st International Conference on Artificial Intelligence and Data Analytics (CAIDA), Riyadh, Saudi Arabia, 6–7 April 2021; pp. 182–187. [Google Scholar]
- Nasir, I.M.; Bibi, A.; Shah, J.H.; Khan, M.A.; Sharif, M.; Iqbal, K.; Nam, Y.; Kadry, S. Deep learning-based classification of fruit diseases: An application for precision agriculture. Comput. Mater. Contin. 2021, 66, 1949–1962. [Google Scholar]
- Khan, M.A.; Nasir, I.M.; Sharif, M.; Alhaisoni, M.; Kadry, S.; Bukhari, S.A.C.; Nam, Y. A blockchain based framework for stomach abnormalities recognition. Comput. Mater. Contin. 2021, 67, 141–158. [Google Scholar]
- Nasir, I.M.; Raza, M.; Shah, J.H.; Khan, M.A.; Nam, Y.-C.; Nam, Y. Improved Shark Smell Optimization Algorithm for Human Action Recognition. Computers. Mater. Contin. 2023, 76, 2667–2684. [Google Scholar]
- Nasir, I.M.; Raza, M.; Ulyah, S.M.; Shah, J.H.; Fitriyani, N.L.; Syafrudin, M. ENGA: Elastic Net-Based Genetic Algorithm for human action recognition. Expert Syst. Appl. 2023, 227, 120311. [Google Scholar] [CrossRef]
- Tehsin, S.; Nasir, I.M.; Damaševičius, R.; Maskeliūnas, R. DaSAM: Disease and Spatial Attention Module-Based Explainable Model for Brain Tumor Detection. Big Data Cogn. Comput. 2024, 8, 97. [Google Scholar] [CrossRef]
- Tariq, J.; Alfalou, A.; Ijaz, A.; Ali, H.; Ashraf, I.; Rahman, H.; Armghan, A.; Mashood, I.; Rehman, S. Fast intra mode selection in HEVC using statistical model. Comput. Mater. Contin. 2022, 70, 3903–3918. [Google Scholar] [CrossRef]
- Mushtaq, I.; Umer, M.; Imran, M.; Nasir, I.M.; Muhammad, G.; Shorfuzzaman, M. Customer prioritization for medical supply chain during COVID-19 pandemic. Comput. Mater. Contin. 2021, 70, 59–72. [Google Scholar] [CrossRef]
- Tehsin, S.; Hassan, A.; Riaz, F.; Nasir, I.M.; Fitriyani, N.L.; Syafrudin, M. Enhancing Signature Verification Using Triplet Siamese Similarity Networks in Digital Documents. Mathematics 2024, 12, 2757. [Google Scholar] [CrossRef]
- Malik, D.S.; Shah, T.; Tehsin, S.; Nasir, I.M.; Fitriyani, N.L.; Syafrudin, M. Block Cipher Nonlinear Component Generation via Hybrid Pseudo-Random Binary Sequence for Image Encryption. Mathematics 2024, 12, 2302. [Google Scholar] [CrossRef]
- Majid, A.; Khan, M.A.; Yasmin, M.; Rehman, A.; Yousafzai, A.; Tariq, U. Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection. Microsc. Res. Tech. 2020, 83, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Nasir, I.M.; Khan, M.A.; Yasmin, M.; Shah, J.H.; Gabryel, M.; Scherer, R.; Damaševičius, R. Pearson correlation-based feature selection for document classification using balanced training. Sensors 2020, 20, 6793. [Google Scholar] [CrossRef] [PubMed]
- Nasir, I.M.; Khan, M.A.; Armghan, A.; Javed, M.Y. SCNN: A secure convolutional neural network using blockchain. In Proceedings of the 2020 2nd International Conference on Computer and Information Sciences (ICCIS), Sakaka, Saudi Arabia, 13–15 October 2020; pp. 1–5. [Google Scholar]
- Nasir, I.M.; Khan, M.A.; Alhaisoni, M.; Saba, T.; Rehman, A.; Iqbal, T. A hybrid deep learning architecture for the classification of superhero fashion products: An application for medical-tech classification. Comput. Model. Eng. Sci. 2020, 124, 1017–1033. [Google Scholar]
- Naqvi, M.; Gilani, S.Q.; Syed, T.; Marques, O.; Kim, H.-C. Skin cancer detection using deep learning—A review. Diagnostics 2023, 13, 1911. [Google Scholar] [CrossRef]
- Mushtaq, S.; Singh, O. A deep learning based architecture for multi-class skin cancer classification. Multimed. Tools Appl. 2024, 83, 87105–87127. [Google Scholar] [CrossRef]
- Faghihi, A.; Fathollahi, M.; Rajabi, R. Diagnosis of skin cancer using VGG16 and VGG19 based transfer learning models. Multimed. Tools Appl. 2024, 83, 57495–57510. [Google Scholar] [CrossRef]
- Balaha, H.M.; Hassan, A.E.-S. Skin cancer diagnosis based on deep transfer learning and sparrow search algorithm. Neural Comput. Appl. 2023, 35, 815–853. [Google Scholar] [CrossRef]
- Alshawi, S.A.; Musawi, G.F.K.A. Skin cancer image detection and classification by cnn based ensemble learning. Int. J. Adv. Comput. Sci. Appl. 2023, 14, 710–717. [Google Scholar] [CrossRef]
- Qureshi, A.S.; Roos, T. Transfer learning with ensembles of deep neural networks for skin cancer detection in imbalanced data sets. Neural Process. Lett. 2023, 55, 4461–4479. [Google Scholar] [CrossRef]
- Tehsin, S.; Rehman, S.; Saeed, M.O.B.; Riaz, F.; Hassan, A.; Abbas, M.; Young, R.; Alam, M.S. Self-organizing hierarchical particle swarm optimization of correlation filters for object recognition. IEEE Access 2017, 5, 24495–24502. [Google Scholar] [CrossRef]
- Tehsin, S.; Rehman, S.; Bilal, A.; Chaudry, Q.; Saeed, O.; Abbas, M.; Young, R. Comparative analysis of zero aliasing logarithmic mapped optimal trade-off correlation filter. In Pattern Recognition and Tracking XXVIII; SPIE: Toulouse, France, 2017; Volume 10203, pp. 22–37. [Google Scholar]
- Tehsin, S.; Rehman, S.; Riaz, F.; Saeed, O.; Hassan, A.; Khan, M.; Alam, M.S. Fully invariant wavelet enhanced minimum average correlation energy filter for object recognition in cluttered and occluded environments. In Pattern Recognition and Tracking XXVIII; SPIE: Toulouse, France, 2017; Volume 10203, pp. 28–39. [Google Scholar]
- Tehsin, S.; Asfia, Y.; Akbar, N.; Riaz, F.; Rehman, S.; Young, R. Selection of CPU scheduling dynamically through machine learning. In Pattern Recognition and Tracking XXXI; SPIE: Toulouse, France, 2020; Volume 11400, pp. 67–72. [Google Scholar]
- Saad, S.M.; Bilal, A.; Tehsin, S.; Rehman, S. Spoof detection for fake biometric images using feature-based techniques. In SPIE Future Sensing Technologies; SPIE: Toulouse, France, 2020; Volume 11525, pp. 342–349. [Google Scholar]
- Asfia, Y.; Tehsin, S.; Shahzeen, A.; Khan, U.S. Visual person identification device using raspberry Pi. In Proceedings of the 25th Conference of FRUCT Association, Helsinki, Finland, 5–8 November 2019; pp. 422–427. [Google Scholar]
- Akbar, N.; Tehsin, S.; Rehman, H.U.; Rehman, S.; Young, R. Hardware design of correlation filters for target detection. In Pattern Recognition and Tracking XXX; SPIE: Toulouse, France, 2019; Volume 10995, pp. 71–79. [Google Scholar]
- Akbar, N.; Tehsin, S.; Bilal, A.; Rubab, S.; Rehman, S.; Young, R. Detection of moving human using optimized correlation filters in homogeneous environments. In Pattern Recognition and Tracking XXXI; SPIE: Toulouse, France, 2020; Volume 11400, pp. 73–79. [Google Scholar]
- Yousafzai, S.N.; Shahbaz, H.; Ali, A.; Qamar, A.; Nasir, I.M.; Tehsin, S.; Damaševičius, R. X-News dataset for online news categorization. Int. J. Intell. Comput. Cybern. 2024, 17, 737–758. [Google Scholar] [CrossRef]
- Tehsin, S.; Rehman, S.; Awan, A.B.; Chaudry, Q.; Abbas, M.; Young, R.; Asif, A. Improved maximum average correlation height filter with adaptive log base selection for object recognition. In Optical Pattern Recognition XXVII; SPIE: Toulouse, France, 2016; Volume 9845, pp. 29–41. [Google Scholar]
- Zafar, M.; Amin, J.; Sharif, M.; Anjum, M.A.; Mallah, G.A.; Kadry, S. DeepLabv3+-based segmentation and best features selection using slime mould algorithm for multi-class skin lesion classification. Mathematics 2023, 11, 364. [Google Scholar] [CrossRef]
- Magdy, A.; Hussein, H.; Abdel-Kader, R.F.; El Salam, K.A. Performance Enhancement of Skin Cancer Classification Using Computer Vision. IEEE Access 2023, 11, 72120–72133. [Google Scholar] [CrossRef]
- Keerthana, D.; Venugopal, V.; Nath, M.K.; Mishra, M. Hybrid convolutional neural networks with SVM classifier for classification of skin cancer. Biomed. Eng. Adv. 2023, 5, 100069. [Google Scholar] [CrossRef]
- Kothapalli, S.P.; Priya, P.S.H.; Reddy, V.S.; Lahya, B.; Ragam, P. Melanoma Skin Cancer Detection using SVM and CNN. EAI Endorsed Trans. Pervasive Health Technol. 2023, 9. [Google Scholar] [CrossRef]
- Dahou, A.; Aseeri, A.O.; Mabrouk, A.; Ibrahim, R.A.; Al-Betar, M.A.; Elaziz, M.A. Optimal skin cancer detection model using transfer learning and dynamic-opposite hunger games search. Diagnostics 2023, 13, 1579. [Google Scholar] [CrossRef]
- Khater, T.; Ansari, S.; Mahmoud, S.; Hussain, A.; Tawfik, H. Skin cancer classification using explainable artificial intelligence on pre-extracted image features. Intell. Syst. Appl. 2023, 20, 200275. [Google Scholar] [CrossRef]
- Gupta, S.; Jayanthi, R.; Verma, A.K.; Saxena, A.K.; Moharana, A.K.; Goswami, S. Ensemble optimization algorithm for the prediction of melanoma skin cancer. Meas. Sens. 2023, 29, 100887. [Google Scholar] [CrossRef]
- Sakli, M.; Essid, C.; Salah, B.B.; Sakli, H. Deep Learning-Based Multi-Stage Analysis for Accurate Skin Cancer Diagnosis using a Lightweight CNN Architecture. In Proceedings of the 2023 International Conference on Innovations in Intelligent Systems and Applications (INISTA), Hammamet, Tunisia, 20–23 September 2023; pp. 1–6. [Google Scholar]
- Bakheet, S.; Alsubai, S.; El-Nagar, A.; Alqahtani, A. A multi-feature fusion framework for automatic skin cancer diagnostics. Diagnostics 2023, 13, 1474. [Google Scholar] [CrossRef]
- Waheed, S.R.; Saadi, S.M.; Rahim, M.S.M.; Suaib, N.M.; Najjar, F.H.; Adnan, M.M.; Salim, A.A. Melanoma skin cancer classification based on CNN deep learning algorithms. Malays. J. Fundam. Appl. Sci. 2023, 19, 299–305. [Google Scholar] [CrossRef]
- Gururaj, H.L.; Manju, N.; Nagarjun, A.; Aradhya, V.M.; Flammini, F. DeepSkin: A deep learning approach for skin cancer classification. IEEE Access 2023, 11, 50205–50214. [Google Scholar] [CrossRef]
- Toprak, A.N.; Aruk, I. A Hybrid Convolutional Neural Network Model for the Classification of Multi-Class Skin Cancer. Int. J. Imaging Syst. Technol. 2024, 34, e23180. [Google Scholar] [CrossRef]
- Rasel, M.; Kareem, S.A.; Obaidellah, U. Integrating color histogram analysis and convolutional neural networks for skin lesion classification. Comput. Biol. Med. 2024, 183, 109250. [Google Scholar] [CrossRef]
- Farea, E.; Saleh, R.A.; AbuAlkebash, H.; Farea, A.A.; Al-antari, M.A. A hybrid deep learning skin cancer prediction framework. Eng. Sci. Technol. Int. J. 2024, 57, 101818. [Google Scholar] [CrossRef]
- Iandola, F.N. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and < 0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31. [Google Scholar]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Nasiri, J.; Khiyabani, F.M. A whale optimization algorithm (WOA) approach for clustering. Cogent Math. Stat. 2018, 5, 1483565. [Google Scholar] [CrossRef]
- Goldberg, D.E. Genetic Algorithms; Pearson Education India: Chennai, India, 2013. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar]
- Li, S.; Chen, H.; Wang, M.; Heidari, A.A.; Mirjalili, S. Slime mould algorithm: A new method for stochastic optimization. Future Gener. Comput. Syst. 2020, 111, 300–323. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).