A Lightweight YOLOv8-Based Network for Efficient Corn Disease Detection
Abstract
1. Introduction
- C2f-GhostNetv2 hybrid backbone: Combines multi-branch feature extraction of C2f with efficient GhostNetV2, incorporating lesion-aware calibration to preserve fine-grained lesion details while remaining lightweight.
- SimAM-guided feature enhancement: Embeds SimAM into GhostNetV2 to emphasize lesion-relevant features and suppress background noise, improving detection of subtle and early-stage lesions.
- Hierarchical feature fusion with BiFPN: Integrates multi-scale features at the neck, enabling robust detection across variable lesion sizes and enhancing overall performance.
- Optimized detection head with SCConv: Uses lesion-adaptive kernel selection guided by BiFPN features, reducing computational overhead while capturing fine-grained and irregular lesion patterns.
2. Related Work
2.1. Crop Disease Detection
2.2. Intelligent Computing for Disease Detection
2.3. Challenges in Disease Detection
3. Previous Work
3.1. Baseline YOLOv8 Architecture
3.2. Related Lightweight Modules
4. Research Method
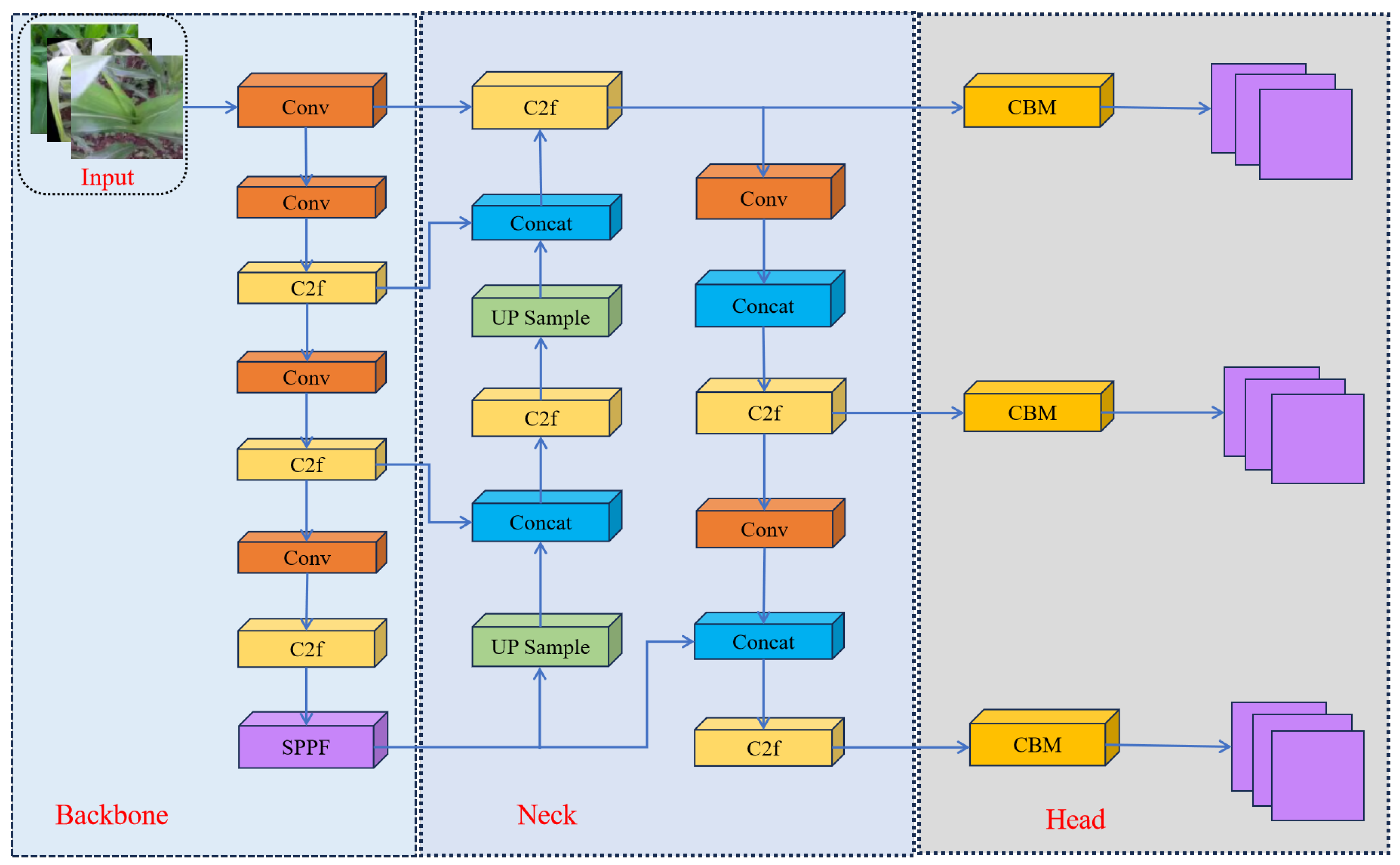
4.1. Improved Structural Framework of YOLOv8
4.2. GhostNetV2 Module
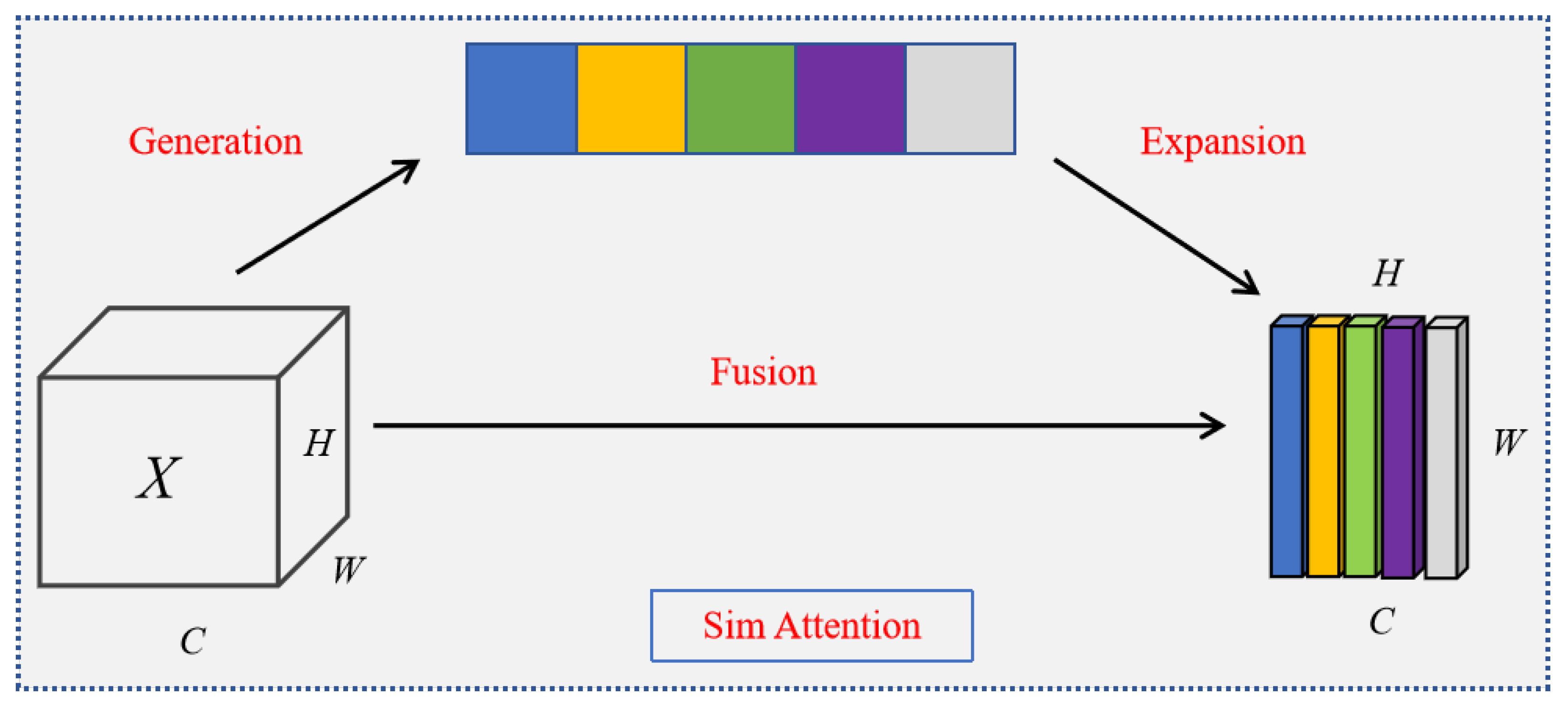
4.3. SimAM Attention Mechanism
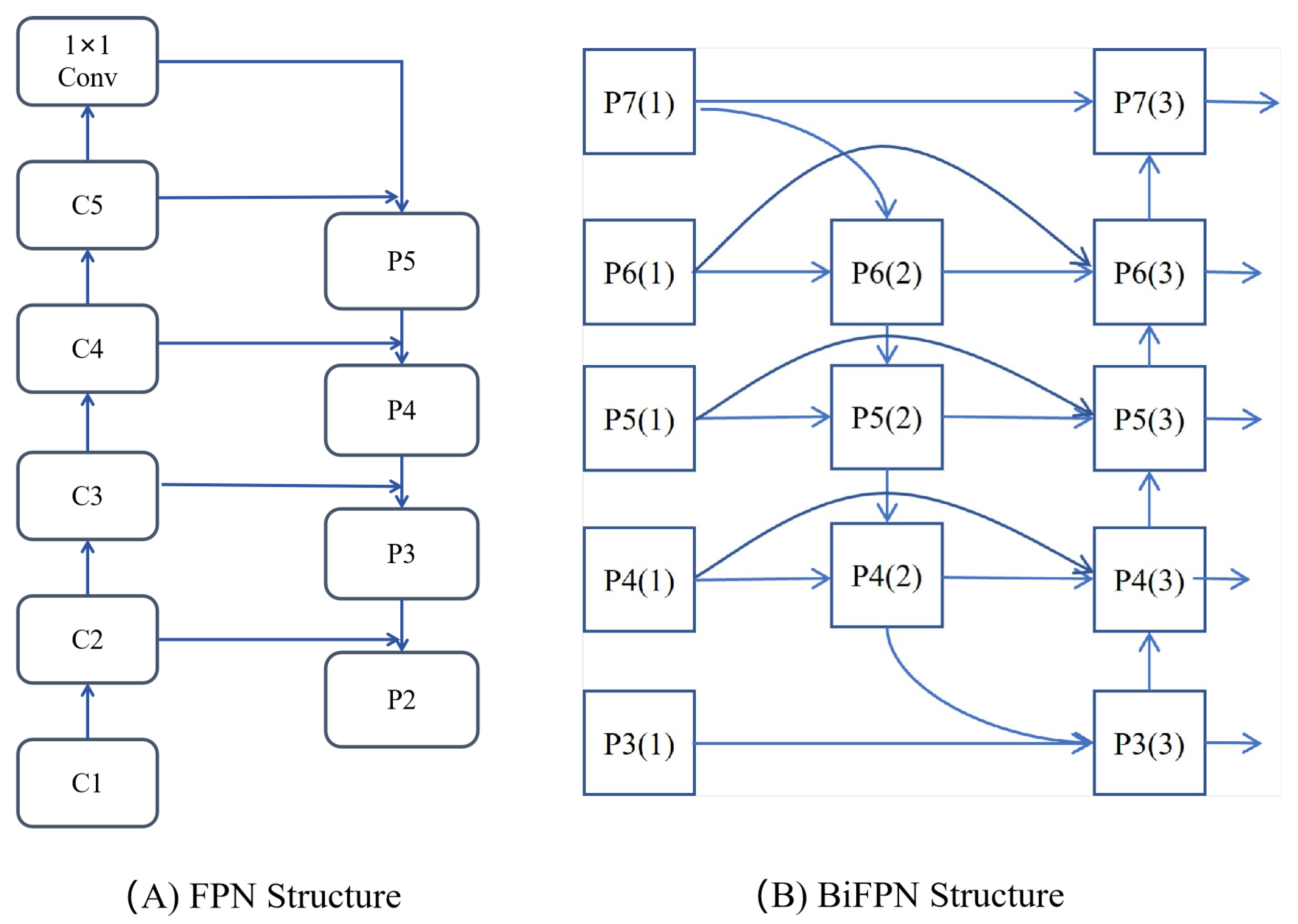
4.4. BiFPN Structure
4.5. Synergistic Mechanism Analysis
5. Experimental Setup
5.1. Dataset
5.2. Experimental Configuration
5.3. Evaluation Metrics
5.4. Comparative Analysis of Algorithms
6. Results
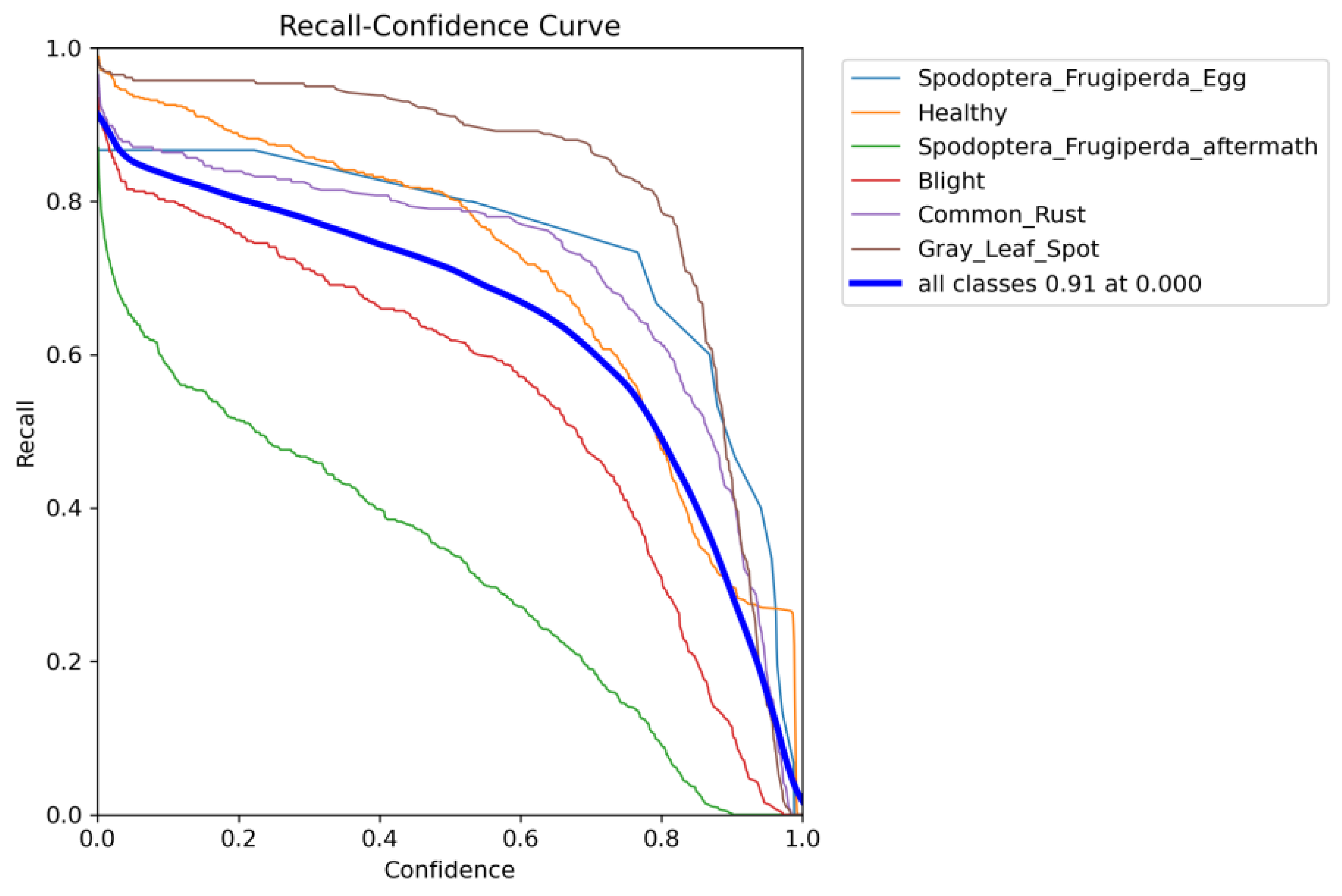
6.1. Data-Enhanced Evaluation
6.2. Ablation Experiment
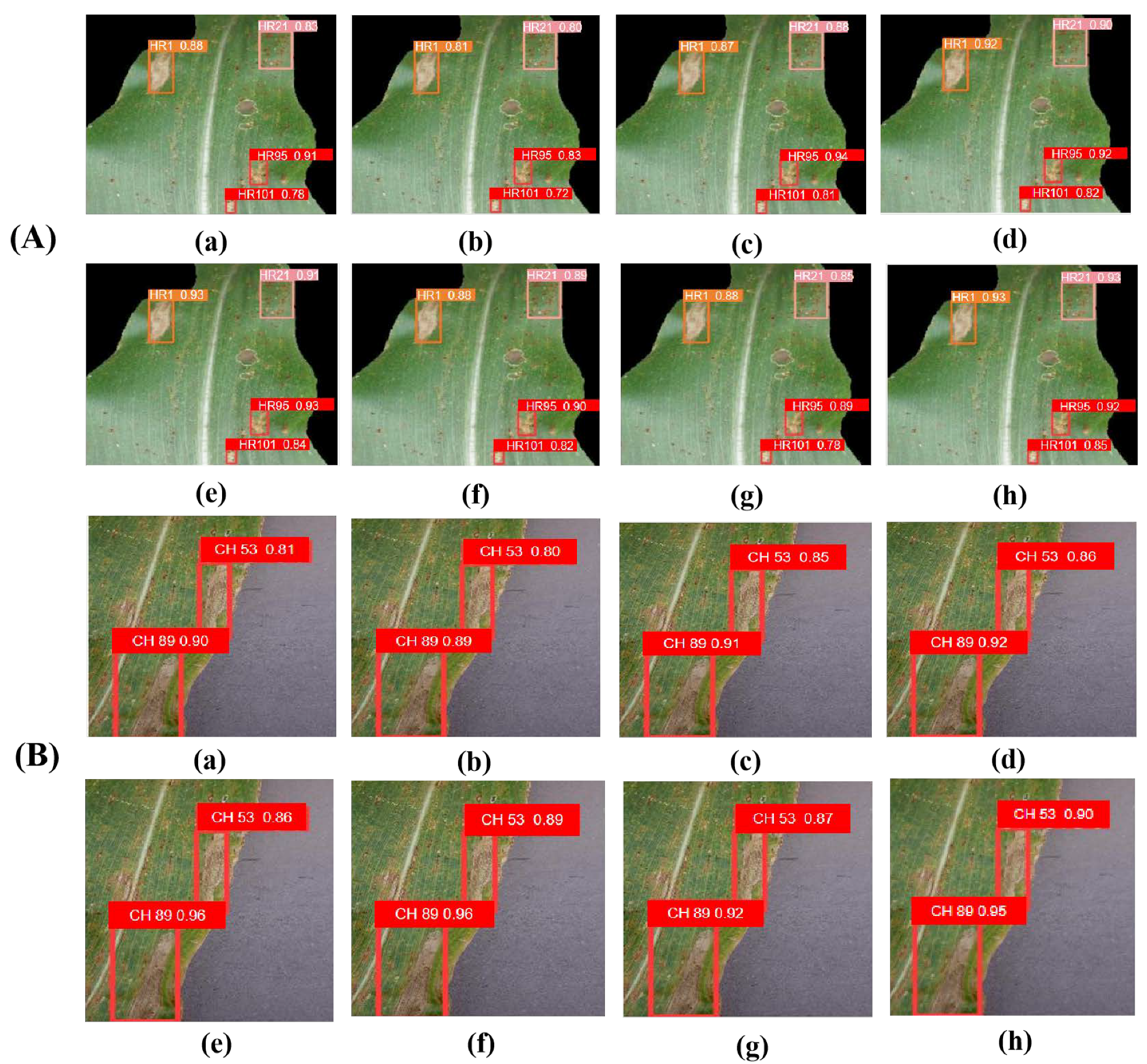
6.3. Comparison and Analysis
6.4. Comparative Experiment
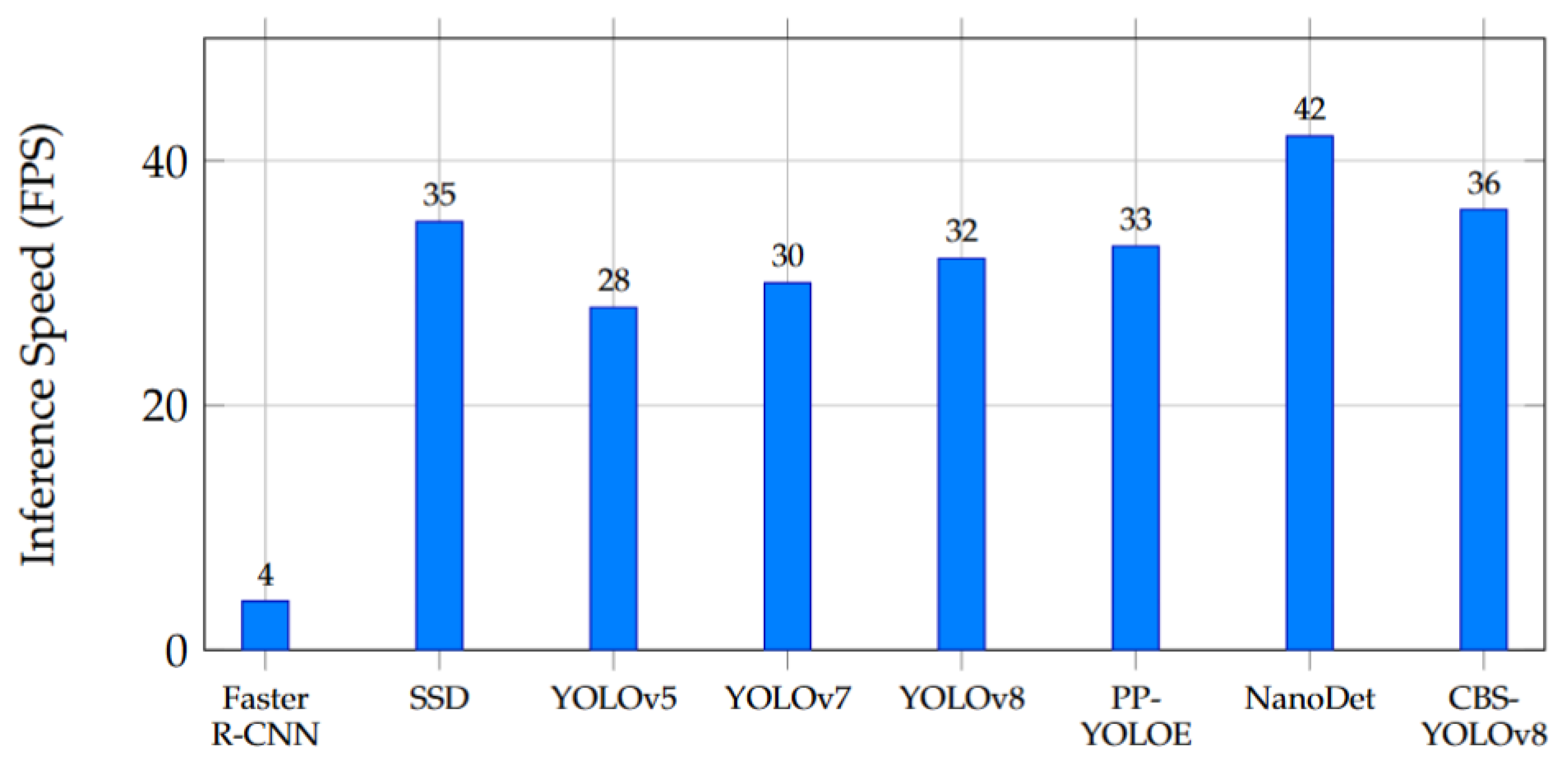
6.5. Inference Speed and Real-Time Performance
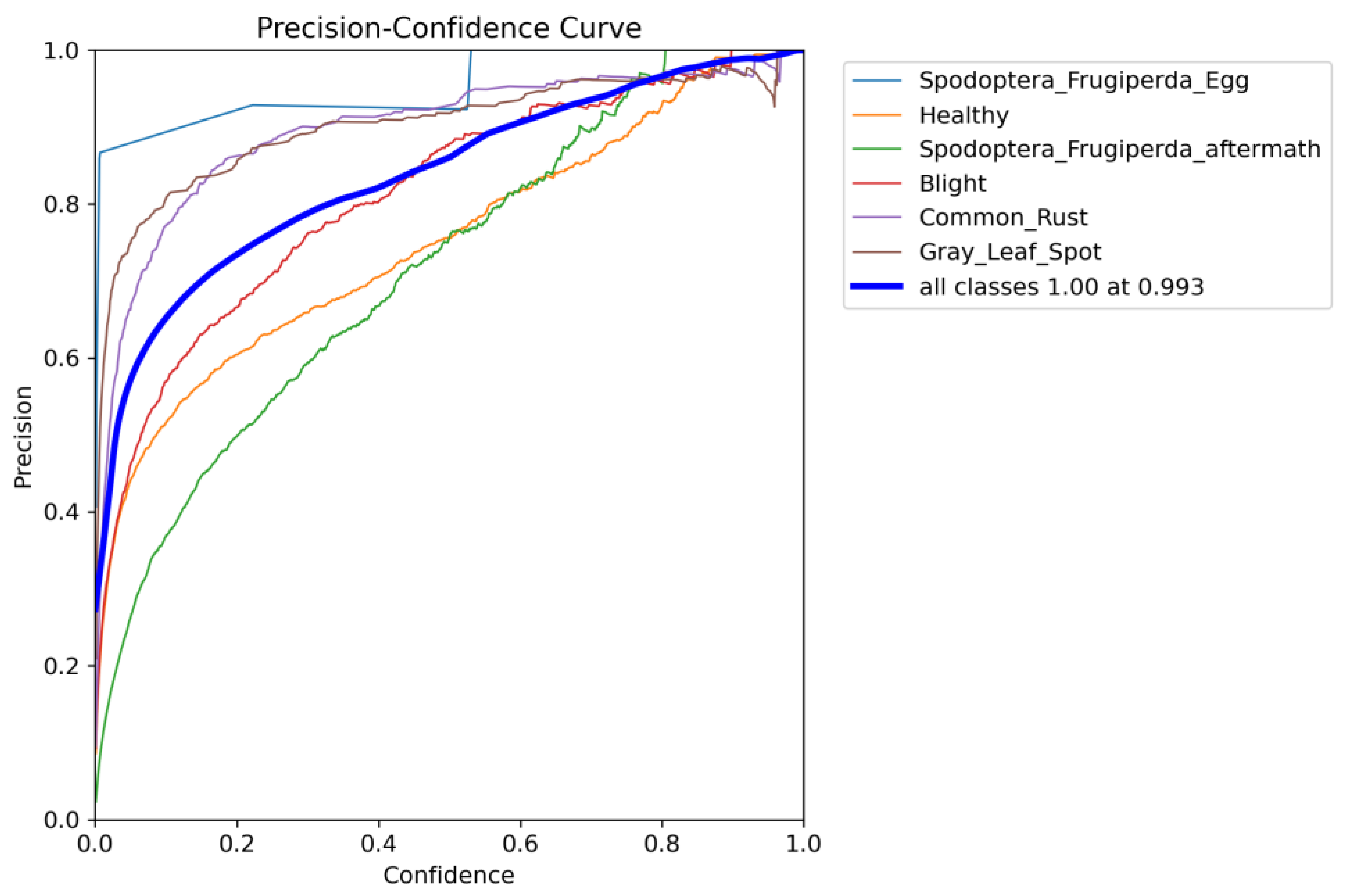
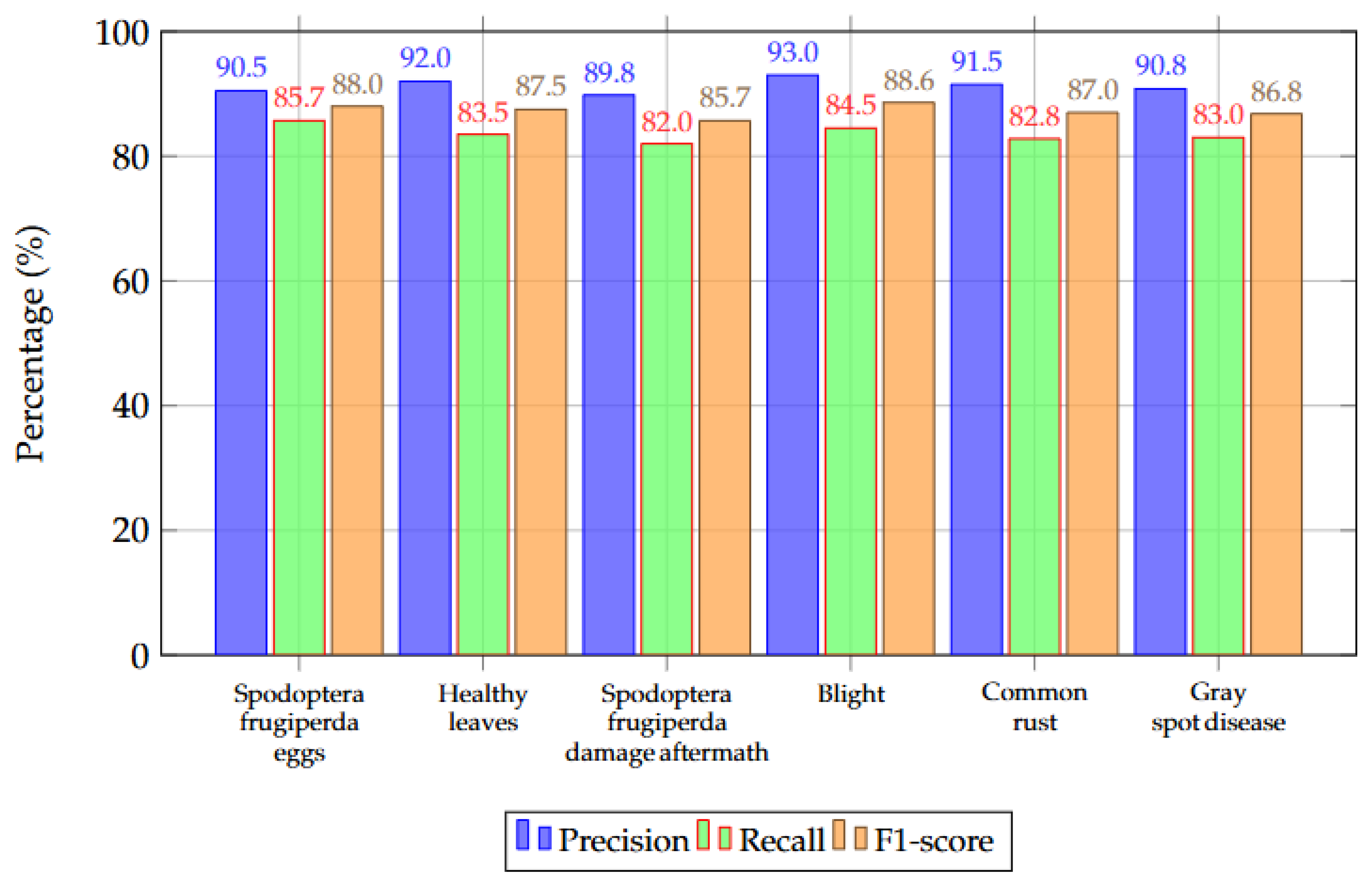
6.6. Class-Wise Detection Performance
6.7. Summary of Results
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shehu, H.A.; Ackley, A.; Mark, M.; Eteng, O.E.; Sharif, M.H.; Kusetogullari, H. YOLO for Early Detection and Management of Tuta absoluta-Induced Tomato Leaf Diseases. Front. Plant Sci. 2025, 16, 1524630. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yao, C.; Yang, R.; Ma, M.; Wu, X.; Xu, Z.; Lu, M.; Zhang, J.; Gong, G. A high-availability segmentation algorithm for corn leaves and leaf spot disease based on feature fusion. Crop Prot. 2024, 187, 106957. [Google Scholar] [CrossRef]
- Zhang, K.; Luo, W.; Zhong, Y.; Ma, L.; Liu, W.; Li, H. Adversarial Spatio-Temporal Learning for Video Deblurring. IEEE Trans. Image Process. 2018, 28, 291–301. [Google Scholar] [CrossRef]
- Chen, N.; Li, B.; Wang, Y.; Ying, X.; Wang, L.; Zhang, C.; Guo, Y.; Li, M.; An, W. Motion and Appearance Decoupling Representation for Event Cameras. IEEE Trans. Image Process. 2025, 34, 5964–5977. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Peng, B.; Gao, M.; Hao, H.; Guo, J.; Liu, X.; Liu, W. Detecting Small Damage on Wind Turbine Surfaces Using an Improved YOLO in Drone-Captured Scenes. J. Fail. Anal. Prev. 2025, 25, 725–740. [Google Scholar] [CrossRef]
- Pan, W.; Yang, Z. A lightweight enhanced YOLOv8 algorithm for detecting small objects in UAV aerial photography. Vis. Comput. 2025, 2, 269. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Z.; Xiao, G.; Han, C.; Fang, C. YOLOv7-DWS: Tea Bud Recognition and Detection Network in Multi-Density Environment via Improved YOLOv7. Front. Plant Sci. 2025, 15, 1503033. [Google Scholar] [CrossRef]
- Diao, Z.; Ma, S.; Li, J.; Zhang, J.; Li, X.; Zhao, S.; He, Y.; Zhang, B.; Jiang, L. Navigation Line Detection Algorithm for Corn Spraying Robot Based on Improved LT-YOLOv10s. Precis. Agric. 2025, 26, 46. [Google Scholar] [CrossRef]
- Feng, Z.; Shi, R.; Jiang, Y.; Han, Y.; Ma, Z.; Ren, Y. SPD-YOLO: A Method for Detecting Maize Disease Pests Using Improved YOLOv7. Comput. Mater. Contin. 2025, 84, 3559–3575. [Google Scholar] [CrossRef]
- Chen, X.; Jiao, Z.; Liu, Y. Improved YOLOv8n based helmet wearing inspection method. Sci. Rep. 2025, 15, 1945. [Google Scholar] [CrossRef]
- Xu, X.; Zhou, B.; Li, W.; Wang, F. A Method for Detecting Persimmon Leaf Diseases Using the Lightweight YOLOv5 Model. Expert Syst. Appl. 2025, 284, 127567. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, M.; Wang, Y. YOLO-BS: A traffic sign detection algorithm based on YOLOv8. Sci. Rep. 2025, 15, 7558. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Xie, G.; Cui, J.; Guo, M. Surface Mine Personnel Object Video Tracking Method Based on YOLOv5-Deepsort Algorithm. Sci. Rep. 2025, 15, 17123. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kaur, J.; Singh, K.; Singh, M.L. Deep transfer learning-based automated detection of blast disease in paddy crop. Signal Image Video Process. 2024, 18, 569–577. [Google Scholar] [CrossRef]
- Feng, Z.R.; Li, Y.H.; Chen, W.Z.; Su, X.P.; Chen, J.N.; Li, J.P.; Liu, H.; Li, S.B. Infrared and Visible Image Fusion Based on Improved Latent Low-Rank and Unsharp Masks. Spectrosc. Spectr. Anal. 2025, 45, 2034–2044. [Google Scholar]
- Radočaj, D.; Radočaj, P.; Plaščak, I.; Jurišić, M. Evolution of Deep Learning Approaches in UAV-Based Crop Leaf Disease Detection: A Web of Science Review. Appl. Sci. 2025, 15, 10778. [Google Scholar] [CrossRef]
- Daphal, D.; Koli, S.M. Enhanced deep learning technique for sugarcane leaf disease classification and mobile application integration. Heliyon 2024, 10, e29438. [Google Scholar] [CrossRef]
- Nuanmeesri, S. Enhanced hybrid attention deep learning for avocado ripeness classification on resource-constrained devices. Sci. Rep. 2025, 15, 3719. [Google Scholar] [CrossRef]
- Li, Z.; Wu, W.; Wei, B.; Li, H.; Zhan, J.; Deng, S.; Wang, J. Rice disease detection: TLI-YOLO innovative approach for enhanced detection and mobile compatibility. Sensors 2025, 25, 2494. [Google Scholar] [CrossRef]
- Shafay, M.; Hassan, T.; Owais, M.; Hussain, I.; Khawaja, S.G.; Seneviratne, L.; Werghi, N. Recent advances in plant disease detection: Challenges and opportunities. Plant Methods 2025, 21, 140. [Google Scholar] [CrossRef]
- Tan, T.; Cao, X.; Liu, H.; Chen, L.; Wang, J.; Chen, X.; Wang, G. Characteristic analysis and model predictive-improved active disturbance rejection control of direct-drive electro-hydrostatic actuators. Expert Syst. Appl. 2026, 301, 130565. [Google Scholar] [CrossRef]
- Wang, N.; Liu, H.; Li, Y.; Zhou, W.; Ding, M. Segmentation and Phenotype Calculation of Rapeseed Pods Based on YOLO v8 and Mask R-Convolution Neural Networks. Plants 2023, 12, 3328. [Google Scholar] [CrossRef]
- Mu, C.; Zhang, F.; Feng, J.; Haidarh, M.; Liu, Y. GhostNet and pair-wise similarity module for cross-domain few-shot classification of hyperspectral images. Appl. Soft Comput. 2025, 183, 113717. [Google Scholar] [CrossRef]
- DeRose, J.F.; Wang, J.; Berger, M. Attention Flows: Analyzing and Comparing Attention Mechanisms in Language Models. IEEE Trans. Vis. Comput. Graph. 2021, 27, 1160–1170. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Computer Vision—ECCV 2016; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2016; Volume 9905, pp. 21–37. [Google Scholar]
- Yaseen, M. YOLOv5, YOLOv8 and YOLOv10: The Go-To Detectors for Real-Time Vision. arXiv 2024, arXiv:2407.02988. [Google Scholar]
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. arXiv 2023, arXiv:2207.02696. [Google Scholar]
- Yaseen, M. What is YOLOv8: An In-Depth Exploration of the Internal Architecture, Training, and Performance. arXiv 2024, arXiv:2408.15857. [Google Scholar]
- Xu, S.; Wang, X.; Lv, W.; Chang, Q.; Cui, C.; Deng, K.; Wang, G.; Dang, Q.; Wei, S.; Du, Y.; et al. PP-YOLOE: An Evolved Version of YOLO. arXiv 2022, arXiv:2203.16250. [Google Scholar] [CrossRef]
- Miao, D.; Wang, Y.; Yang, L.; Wei, S. Foreign Object Detection Method of Conveyor Belt Based on Improved NanoDet. IEEE Access 2023, 11, 23046–23052. [Google Scholar] [CrossRef]




| Disease Category | Training | Validation | Test | Total |
|---|---|---|---|---|
| Healthy Leaves | 856 | 245 | 122 | 1223 |
| Spodoptera frugiperda eggs | 576 | 164 | 82 | 822 |
| Spodoptera frugiperda damage aftermath | 932 | 266 | 133 | 1331 |
| Blight | 752 | 215 | 107 | 1074 |
| Rust Disease | 698 | 199 | 99 | 996 |
| Gray Leaf Spot Disease | 453 | 130 | 65 | 648 |
| Name | Parameter |
|---|---|
| Operating System | Windows 11 |
| CPU | AMD Ryzen 7 7735H |
| GPU | NVIDIA RTX 4060 |
| GPU Memory | 16GB |
| Training Epochs | 120 |
| Training/Validation Split | 8:2 |
| Batch Size | 16 |
| Optimizer | Adam |
| Initial Learning Rate | Adam: 0.001 |
| Learning Rate Schedule | Cosine decay |
| Weight Decay | 0.0005 |
| Momentum | 0.9 |
| IoU Threshold (mAP) | 0.5 for full evaluation |
| Image Size | 640 × 640 |
| Data Augmentation | Random flip, scale, color jitter, Mosaic, MixUp |
| Mirror | Scale | Precision (%) | Recall (%) | mAP@0.5 (%) |
|---|---|---|---|---|
| × | × | 90.3 | 81.8 | 86.1 |
| × | √ | 91.5 | 82.1 | 87.2 |
| √ | × | 91.8 | 82.9 | 87.9 |
| √ | √ | 92.3 | 82.9 | 88.6 |
| No. | Model | Precision | Recall | mAP@0.5 |
|---|---|---|---|---|
| 1 | YOLOv8 | 0.896 | 0.875 | 0.865 |
| 2 | YOLOv8+GhostNetV2 | 0.899 | 0.861 | 0.873 |
| 3 | YOLOv8+SimAM | 0.908 | 0.876 | 0.869 |
| 4 | YOLOv8+BIFPN | 0.910 | 0.885 | 0.875 |
| 5 | CBS-YOLOv8 | 0.913 | 0.889 | 0.882 |
| Model | Precision (%) | Recall (%) | mAP@0.5 (%) | Error (%) | Time (ms) |
|---|---|---|---|---|---|
| Faster R-CNN [25] | 92.1 | 81.5 | 88.3 | 11.7 | 215 |
| SSD [26] | 85.7 | 78.4 | 81.9 | 18.1 | 30.4 |
| YOLOv5 [27] | 90.2 | 82.7 | 87.5 | 12.5 | 33.1 |
| YOLOv7 [28] | 91.1 | 83.5 | 88.2 | 11.8 | 34.2 |
| YOLOv8 [29] | 91.5 | 83.0 | 88.5 | 11.5 | 32.5 |
| PP-YOLOE [30] | 90.8 | 82.9 | 87.8 | 12.2 | 29.8 |
| NanoDet [31] | 89.5 | 81.7 | 86.8 | 13.2 | 28.5 |
| CBS-YOLOv8 | 92.3 | 84.2 | 88.9 | 11.1 | 31.0 |
| Model | FPS | Params (M) | GFLOPs |
|---|---|---|---|
| Faster R-CNN | 4 | 42.3 | 215 |
| SSD | 35 | 26.5 | 30.4 |
| YOLOv5 | 28 | 9.4 | 26.4 |
| YOLOv7 | 30 | 10.9 | 27.5 |
| YOLOv8 | 32 | 11.2 | 28.6 |
| PP-YOLOE | 33 | 8.8 | 22.1 |
| NanoDet | 42 | 4.1 | 12.3 |
| CBS-YOLOv8 | 36 | 8.1 | 21.0 |
| Category | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|
| Spodoptera frugiperda eggs | 90.5 | 85.7 | 88.0 |
| Healthy leaves | 92.0 | 83.5 | 87.5 |
| Spodoptera frugiperda damage aftermath | 89.8 | 82.0 | 85.7 |
| Blight | 93.0 | 84.5 | 88.6 |
| Common rust | 91.5 | 82.8 | 87.0 |
| Gray spot disease | 90.8 | 83.0 | 86.8 |
| Overall | 91.3 | 83.6 | 87.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, D.; Peng, Y.; Gu, X.; U, K. A Lightweight YOLOv8-Based Network for Efficient Corn Disease Detection. Mathematics 2025, 13, 4002. https://doi.org/10.3390/math13244002
Song D, Peng Y, Gu X, U K. A Lightweight YOLOv8-Based Network for Efficient Corn Disease Detection. Mathematics. 2025; 13(24):4002. https://doi.org/10.3390/math13244002
Chicago/Turabian StyleSong, Deao, Yiran Peng, Xinyuan Gu, and KinTak U. 2025. "A Lightweight YOLOv8-Based Network for Efficient Corn Disease Detection" Mathematics 13, no. 24: 4002. https://doi.org/10.3390/math13244002
APA StyleSong, D., Peng, Y., Gu, X., & U, K. (2025). A Lightweight YOLOv8-Based Network for Efficient Corn Disease Detection. Mathematics, 13(24), 4002. https://doi.org/10.3390/math13244002







