1. Introduction
Deep learning (DL) has revolutionized medical image analysis, offering robust techniques for disease diagnosis, segmentation, and classification. Among DL methodologies, Convolutional Neural Networks (CNNs) have demonstrated outstanding capability across various imaging applications, primarily due to their proficiency in autonomously extracting salient features from complex datasets. Two important areas of application include the classification of phenotypes in products of conception (PoC) and the identification of brain tumors using magnetic resonance imaging (MRI) [
1]. PoC samples, which are derived from spontaneous abortions, possess critical genetic data that can facilitate the determination of underlying causes of pregnancy loss. Accurate phenotyping of these samples is essential for recognizing genetic disorders and chromosomal anomalies [
2]. Similarly, precise and prompt classification of brain tumors via MRI is imperative, as tumor characterization and grading have a direct impact on clinical decision-making and patient outcomes [
3]. In response to these complexities, transfer learning using established CNNs such as VGG16, ResNet18, InceptionV3, DenseNet121, and EfficientNetB5 has become standard practice, employing pretraining on substantial datasets like ImageNet to enhance effectiveness in medical image analysis.
This research explores phenotype classification in two prominent medical imaging datasets: a brain tumor MRI dataset and a PoC dataset consisting of specimens from spontaneous abortions. In both contexts, achieving high-accuracy classification yields valuable understanding for timely treatment planning, accurate diagnosis, and elucidation of underlying pathological mechanisms. This work systematically assesses the improvements in performance attributed to integrating attention mechanisms within standard CNN models by conducting four distinct experimental scenarios.
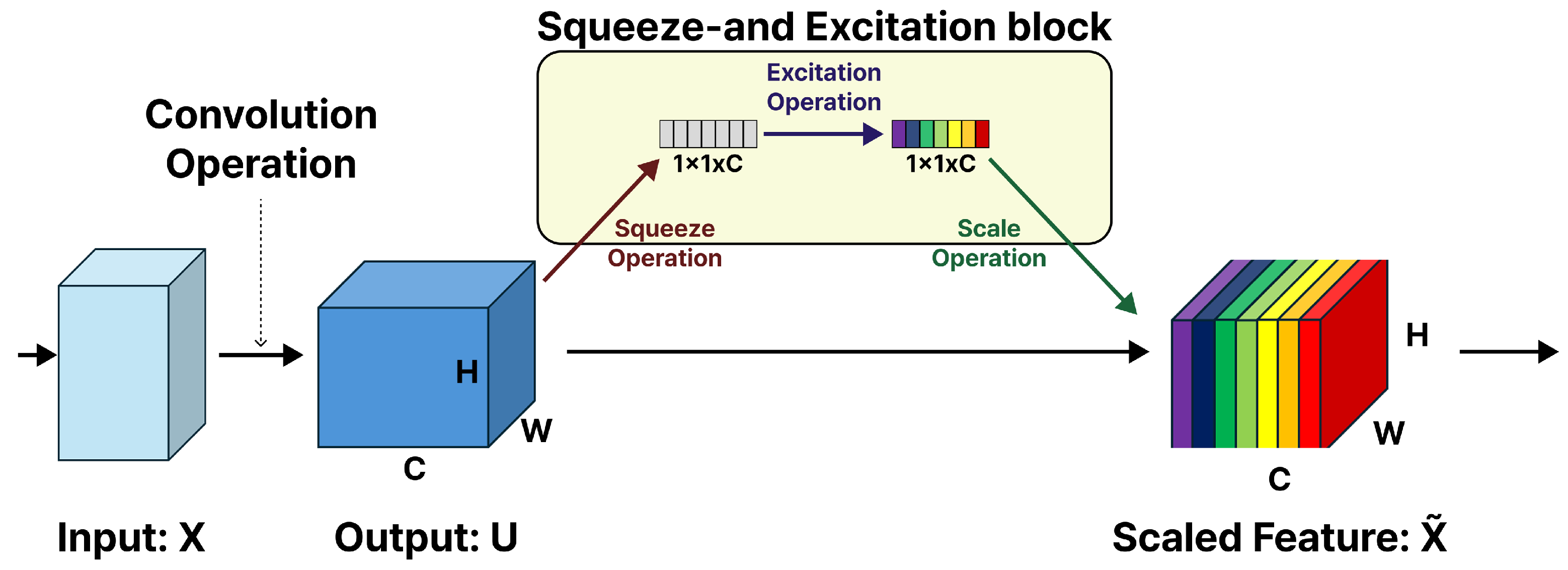
Firstly, we employ pretrained CNN models (VGG16, ResNet18, InceptionV3, DenseNet121, and EfficientNetB5) as benchmarks, retaining their original architectures without modifications. In the second phase, we systematically augment each model by incorporating Squeeze-and-Excitation (SE) modules into every convolutional block. As depicted in
Figure 1, the SE block, initially proposed by Hu et al. [
4], serves as a lightweight addition that strengthens CNN performance by modeling the dependencies between feature map channels. The principal idea behind this module is that feature channels exhibit varying relative significance based on the input, and adaptive recalibration of these channel responses enhances the network’s classification ability. The SE block executes two functional steps: the squeeze operation compresses spatial information into a succinct channel descriptor using global average pooling, capturing comprehensive context while reducing dimensionality; subsequently, with ReLU and sigmoid activations, the excitation operation processes this descriptor using a compact two-layer fully connected (FC) network to yield channel-specific weights. These weights are then used to rescale the corresponding feature maps, amplifying more salient channels and attenuating less relevant ones.
In the third phase, we investigate a targeted integration approach, whereby SE modules are incorporated solely within the deeper layers of each network architecture. For example, in VGG16, SE blocks are positioned following Blocks 3, 4, and 5. This strategy is designed to concentrate the refinement of attention on higher-level, semantically enriched features, while simultaneously reducing computational demands relative to comprehensive SE integration.
In the fourth and final phase, we expand on selective SE integration by adding Spatial Attention (SA) mechanisms [
5]. This hybrid framework enables the model to focus on both key feature channels and critical spatial regions within the feature maps, thereby enhancing its capacity for localization and classification. The SA module identifies and highlights the regions of a feature map that provide the most valuable spatial information. Unlike channel attention, which distinguishes the most important feature channels, SA assigns significance to precise spatial positions (pixels). This is accomplished by performing global average pooling and max pooling along the channel axis, which generates two spatial descriptors with dimensions
. These descriptors are subsequently concatenated to form an
feature map, which is then processed by a convolutional layer followed by a sigmoid activation to produce the resulting SA map
of size
Recent methods such as Improved EATFormer [
6] have investigated Vision Transformer-based models that utilize advanced self-attention mechanisms for medical image classification. Nevertheless, these approaches often demand significant computational resources and access to large-scale datasets to attain high levels of performance. Furthermore, Vision Transformers usually introduce only one novel architecture, which confines their adaptability to a range of CNN backbones. By comparison, our study introduces a systematic comparative framework that incorporates lightweight yet effective attention modules (SE and Convolutional Block Attention Module (CBAM)) into five widely adopted CNN architectures. This methodology retains computational efficiency and supports deployment in resource-constrained environments while also offering comprehensive insights into the behavior of attention mechanisms across various architectures and imaging modalities. By balancing high accuracy with practical deployment considerations, our research both extends Transformer-based studies and improves the generalizability and clinical relevance of these methods.
Figure 2 presents the steps involved in generating the spatial attention map.
This study examines whether embedding attention mechanisms, specifically SE and CBAM, within established CNN architectures can meaningfully enhance feature representation and boost classification accuracy within medical image analysis. Therefore, the primary research question is as follows: To what degree does the methodical integration of SE and CBAM attention modules within CNN frameworks elevate the accuracy, precision, recall, and F1-score in medical image classification, and how does this performance differ when applied to varying network depths and separate datasets?
Additionally, the goal of this work is to deliver not only a performance benchmarking but also a reproducible and systematic approach for integrating attention modules, specifically SE and CBAM, across several CNN models under uniform experimental protocols. In contrast to previous research that has focused only on single models or datasets, our approach makes possible an equitable, cross-model, and cross-task assessment encompassing both classification and segmentation. Moreover, we investigate the impact of different layer-wise placements of attention modules, providing actionable insights regarding the operation of attention mechanisms at multiple levels of feature abstraction. Through this rigorous examination, we underscore the enhancement in performance as well as the nuanced behavior and generalization capabilities of attention integration in medical image analysis.
The principal objective of this research is to evaluate how distinct attention strategies, global, selective, and hybrid, affect the classification outcomes of pretrained CNN architectures in difficult medical imaging tasks. Our results indicate that the use of attention modules, particularly when applied in a targeted and spatially guided manner, substantially improves the discriminative performance of CNNs while ensuring computational efficiency. The code is available at:
https://github.com/Zahid672/Brain-Tumor-and-POC-Classification (accessed on 20 October 2025).
The main contributions of this study are outlined below:
We integrate lightweight attention modules (SE and CBAM) into five well-established CNN backbones, generating multiple model variants to systematically evaluate the roles of channel and spatial-level attention.
In contrast to prior studies that typically concentrate on a single backbone or Transformer-based architecture, our research establishes a unified comparative platform to investigate attention integration across several CNN architectures, providing meaningful perspectives for medical image classification.
We conduct extensive validation on two separate medical imaging datasets, such as brain tumor MRI (including multiple subtypes) and POC histopathology (encompassing various tissue classes), thereby demonstrating the robustness of our approach within both radiological and pathological contexts.
Experimental results demonstrate that attention-augmented CNNs consistently surpass their respective baselines, with EfficientNetB5 combined with hybrid attention achieving the highest accuracy. Moreover, the introduction of attention facilitates enhanced feature localization, which supports improved model interpretability and greater clinical value.
The remainder of this manuscript is organized as follows:
Section 2 reviews foundational literature.
Section 3 details the methodologies and datasets employed.
Section 4 presents outcomes and discussion.
Section 5 discusses limitations and suggests future research directions. Finally,
Section 6 concludes the study.
2. Related Work
DL techniques, particularly CNNs, have revolutionized medical image analysis by facilitating automated processing and improving diagnosis for numerous medical conditions [
7]. CNNs have demonstrated significant impact in computer-aided diagnosis, marking substantial advances within the discipline [
8]. For instance, CNN architectures are capable of autonomously extracting essential features from brain MRI scans, which enhances the effectiveness of cancer detection relative to traditional methods [
9,
10]. Additionally, CNN-based approaches have yielded improvements in diagnostic accuracy for neurodegenerative disorders through multi-class classification tasks [
11]. In brain tumor analysis, CNNs remain extensively explored, with several publications reporting high performance in identifying and distinguishing various tumor types [
12,
13].
A key factor underlying these achievements is transfer learning [
14], which utilizes pretrained models built on large-scale datasets such as ImageNet. Transfer learning has demonstrated particular significance in medical imaging, where available datasets are frequently small in size [
15]. Pretrained CNNs, like those used in this study, offer a solid groundwork by applying knowledge obtained through extensive training on natural images [
16]. Fine-tuning these models on domain-specific medical datasets typically leads to considerable gains in performance in comparison to initializing models from scratch. To enhance model effectiveness further, attention mechanisms, such as SE modules and SA, have been integrated into CNN architectures. These mechanisms enable networks to focus on the most relevant features or areas within medical images, thereby increasing diagnostic accuracy [
17].
Continuing this research trajectory, Rongjun et al. [
18] proposed an approach where, in 1D residual CNNs, SE blocks are primarily incorporated to detect ECG arrhythmia. They merged SE modules with temporal 1D convolutions to dynamically enhance channel features associated with arrhythmia, eliminate redundant features, and eliminate the need for preprocessing (i.e., denoising) steps. Residual connections contribute to stabilizing model training and optimizing efficiency. Their experimental findings reveal that this model achieved high performance, underscoring its potential for robust automated ECG analysis.
Li et al. [
19] proposed a unified temporal–spectral SE framework that extracts multi-scale temporal patterns and multi-level spectral features simultaneously from EEG signals. Their model utilizes convolutional blocks to capture nonstationary patterns, while parallel spectral convolutional blocks are employed to extract frequency-band characteristics. These extracted features are integrated via a novel SE module, which adaptively prioritizes the most salient channel-wise representations. To address overfitting resulting from the limited number of seizure events, the authors implemented an information-maximizing loss function. Their approach achieved more accurate results than prior methods, demonstrating the advantage of jointly modeling temporal and spectral domains for seizure detection. In a comparable manner, Kitada et al. [
20] use both labeled and unlabeled data to refine an ensemble of pretrained SENets through a mean-teacher semi-supervised learning framework. They further augmented the training process with dermatology-specific data augmentations to expand the dataset. This augmentation strategy led to a substantial improvement in balanced accuracy, increasing it from 79.2% to 87.2% on the ISIC 2018 validation set.
Zheng et al. [
21] developed a Multi-Attention CNN aimed at fine-grained image recognition. The model simultaneously learns both attention localization and feature extraction by identifying multiple discriminative parts without the need for part annotations. This demonstrates that attention mechanisms facilitate enhanced class separability and interpretability, paving the way for subsequent attention-based models in medical and visual recognition. Gu et al. [
22] presented CA-Net (Comprehensive Attention Network), a framework incorporating spatial, channel, and scale attention mechanisms to improve feature representation in medical image segmentation. The model increases segmentation accuracy and interpretability by selectively emphasizing key anatomical regions. This study underscores the value of multi-dimensional attention fusion, supporting our study’s focus on integrating channel and SA within CNN architectures.
Although prior studies have demonstrated the utility of SE networks [
23] and attention mechanisms for specialized applications such as ECG analysis [
18], EEG-based seizure detection [
19], and skin lesion classification [
20], these approaches largely remain limited to task-specific and single-modality settings. In contrast, the present study systematically evaluates attention integration across multiple widely adopted CNN architectures, VGG16, ResNet18, InceptionV3, DenseNet121, and EfficientNetB5, applied to two distinct medical imaging modalities: brain tumor MRI and histopathological images of PoC. Unlike earlier work that concentrates on a single signal dataset, our analysis includes both channel-oriented (SE) and hybrid channel-spatial (CBAM) modules, thereby presenting a more comprehensive perspective on feature recalibration and localization. The findings indicate that CNNs enhanced with attention modules achieve not only higher classification accuracy but also improved generalizability across heterogeneous datasets, with the combination of EfficientNetB5 and CBAM yielding the highest overall performance. This broad, modality-independent comparative assessment distinguishes our research from previous task-specific studies and offers actionable insights for developing robust, generalizable attention-based models for clinical decision support.
3. Materials and Methods
The methodology of this study consisted of a sequence of experiments designed to evaluate the performance of pretrained CNN architectures and their versions augmented with attention modules for phenotype classification in PoC and brain tumor MRI datasets.
We initially established benchmark results by employing the pretrained models, VGG16 [
24], ResNet18 [
25], InceptionV3 [
26], EfficientNetB5 [
27], and DenseNet121 [
28]. Each network was fine-tuned for the target classification tasks without altering its architecture, enabling us to assess its baseline performance and set reference points for evaluating subsequent modifications. In the following phase, we augmented the pretrained CNNs with SE modules. These additions enhance the representational power of the networks by adaptively modulating channel-wise feature responses, which amplify salient features while diminishing those deemed less informative. To investigate the potential benefit of a more targeted SE application, the third experimental phase implemented selective integration. Specifically, for VGG16, SE modules were inserted only after Blocks 3, 4, and 5, based on the rationale that deeper layers capture progressively more abstract and semantically meaningful features. Finally, in the fourth phase, we incorporated SE modules alongside SA. While SE enhances feature interdependencies between channels, SA emphasizes the most discriminative spatial regions within feature maps. By integrating these two complementary approaches, our aim was to develop models that are receptive to feature channels as well as spatial information, thereby enhancing both classification accuracy and interpretability.
3.1. Datasets
Two publicly available datasets were utilized in this study, specifically the POC dataset and the BT-Large-4C dataset, as illustrated in
Figure 3. An overview of these datasets can be found in
Table 1.
3.1.1. Products of Conception Dataset
The POC dataset [
29] used in this work is publicly available for research purposes. It contains histopathological image samples categorized into 4 distinct tissue types: chorionic villi, decidual tissue, hemorrhage, and trophoblastic tissue. The training set consists of 4155 samples, specifically 1138 hemorrhage, 1391 chorionic villi, 926 decidual, and 700 trophoblastic tissue images. The testing set comprises 421 hemorrhage images out of a total of 1511 samples, including 390 chorionic villi, 349 decidual, and 351 trophoblastic tissue images.
3.1.2. Brain Tumor Dataset
The second dataset used in this research is the publicly available brain tumor MRI dataset obtained from the Kaggle repository (
https://www.kaggle.com/sartajbhuvaji/brain-tumor-classification-mri (accessed on 20 October 2025)). The dataset comprises 3064 T1-weighted, contrast-enhanced brain MR images corresponding to three primary tumor types: gliomas, meningiomas, and pituitary tumors. To facilitate a more in-depth evaluation, we expanded the dataset to 4 categories by adding normal brain MR images, referring to this version as BT-Large-4c. The BT-Large-4c dataset, therefore, encompasses four classes: Normal, Glioma tumor, Meningioma tumor, and Pituitary tumor. Given the limited number of MRI images, we increased dataset diversity using image augmentation techniques, particularly two methods: horizontal flipping and rotation. In the first step, input images were randomly rotated by 90 degrees one or more times, after which each rotated image was horizontally flipped, thereby generating additional training data.
3.2. Baseline CNN Architectures
3.2.1. VGG16
VGG16, as introduced by Simonyan and Zisserman [
24], achieved prominence due to its strong results in the ImageNet Large-Scale Visual Recognition Challenge (ILSVRC) 2014. Its architecture is characterized by a consistent application of small
convolutional filters with a stride of 1 and padding, which serves to preserve spatial dimensions. The sequential stacking of these layers enables the network to capture hierarchical representations of increasing abstraction while controlling the parameter count relative to architectures using larger filters. VGG16 consists of 13 convolutional layers and 3 FC layers, yielding a total of 16 weight-bearing layers and thus providing the rationale for the model’s name.
In VGG16, the convolutional layers are divided into five consecutive blocks, with each block succeeded by a max-pooling layer with stride 2, progressively reducing spatial resolution while efficiently maintaining essential feature information. The output feature maps from the final block are then flattened and passed through 3 FC layers, with the last layer typically utilizing a softmax activation function to produce class probabilities. Despite its substantial depth, the architecture’s consistent and straightforward design has contributed to VGG16’s broad adoption in computer vision applications. The baseline, which focused on medical image analysis, was selected for its strong feature extraction capabilities and the availability of pretrained weights on large-scale datasets such as ImageNet.
To explore how attention mechanisms affect convolutional networks, we constructed three VGG16-based variants with attention modules, as depicted in
Figure 4. The first variant, VGG16-SE v1, extends the baseline by inserting SE blocks following every convolutional layer. This approach enables channel recalibration at all depths, ensuring that informative channels are persistently prioritized whereas redundant channels are diminished. The second model, VGG16-SE v2, adopts a more targeted approach by integrating SE blocks only after the final convolutional layers of Blocks 3, 4, and 5. Focusing recalibration on higher-level semantic features in this manner reduces computational demands and enhances the expressive power of deeper layers where abstraction is paramount. In the third version, VGG16-SE-SA, both channel and SA are employed; SA modules are added after the final convolutions of Blocks 1 and 2 to capture key spatial regions, while SE blocks are inserted after the final convolutions of Blocks 3, 4, and 5 for refined channel adjustments. This configuration is intended to leverage the complementary properties of spatial and channel attention, allowing the network to effectively determine both the location and the nature of feature importance. This strategy supports selective emphasis within feature maps.
3.2.2. ResNet18
ResNet18, introduced by He et al. [
25], marked a key innovation in deep learning by addressing the vanishing gradient challenge via residual learning. Instead of requiring the network to learn direct input–output mappings, ResNet introduces shortcut (skip) connections that allow it to focus on learning residual functions. This design permits the successful training of deeper architectures, circumventing the degradation of accuracy commonly observed in very deep networks.
ResNet18 consists of 18 trainable layers, comprising 17 convolutional layers and a final FC layer. The model structure is organized into five primary stages: an initial convolution with stride 2, followed by a max-pooling layer, and four distinct groups of residual blocks. Within each residual block are two convolutional layers, each succeeded by batch normalization and ReLU activation, with an accompanying shortcut connection that bypasses one or more layers. Depending on dimensional consistency, these shortcut connections can function as identity mappings or employ convolutions to align dimensions.
Residual connections in ResNet18 facilitate the learning of more distinctive feature representations and help mitigate the vanishing gradient issue, offering an effective trade-off between accuracy and efficiency. The architecture’s lightweight nature and access to pretrained weights on large-scale datasets such as ImageNet have established ResNet18 as a prominent backbone in computer vision, supporting applications such as image classification, segmentation, and medical image analysis.
To systematically analyze the impact of attention mechanisms within residual networks, three ResNet18-based variants were developed as depicted in
Figure 5. The first variant, SEResNet18 v1, alters the baseline by substituting every BasicBlock with an SE-enhanced version, thus enabling adaptive channel recalibration at each residual unit throughout the network. The second variant, SEResNet18 v2, incorporates SE blocks specifically after layers 2, 3, and 4, focusing on mid and high-level feature representations where greater semantic abstraction is observed, while also minimizing additional computational demands. The third variant, SEResNet18-SA, sequentially integrates both SE and SA modules after layers 2, 3, and 4. In this variant, SE modules adaptively reweight channel responses, while SA modules highlight the most relevant spatial regions. Through the combination of these complementary mechanisms, the model enhances its capacity to capture both channel dependencies and spatial relationships at the same time.
3.2.3. InceptionV3
InceptionV3, proposed by Szegedy et al. [
30], is a deep CNN designed to achieve high accuracy with computational efficiency. As an advanced version of the initial Inception approach, it incorporates factorization methods, dimensionality reduction, and optimized resource utilization. The defining principle of the Inception module lies in its ability to capture spatial information at multiple scales by using convolutional kernels of various sizes (
,
, and
) in parallel, in addition to pooling layers. These outputs are merged by concatenation, which enables the network to extract both detailed and global features concurrently.
InceptionV3 introduces several significant improvements over its predecessors. Large convolutions, such as , are decomposed into two sequential convolutions, which reduces the parameter count substantially without sacrificing representational capacity. In addition, asymmetric convolutions, such as a followed by a , are utilized to further optimize computational efficiency. Extensive application of batch normalization stabilizes and expedites the training process, while auxiliary classifiers at intermediate layers enhance gradient propagation and reduce the risk of overfitting. The final network configuration comprises multiple stacked Inception modules, succeeded by a global average pooling layer and FC layers, with a softmax activation facilitating classification. Owing to its optimal balance between computational efficiency and predictive accuracy, InceptionV3 has achieved widespread adoption in image classification, including medical imaging applications, especially in settings with constrained computational resources.
Due to its modular architecture, InceptionV3 is well-suited for evaluating the integration of attention mechanisms. Accordingly, we introduced three attention-enhanced variants, depicted in
Figure 6. The first variant, InceptionV3-SE v1, incorporates SE blocks after selected stages to recalibrate channel-wise feature responses in intermediate layers, adaptively enhancing the most informative channels while suppressing less significant ones. The second variant, InceptionV3-SE v2, implements SE blocks more selectively, positioning them exclusively after the Inception-C, Inception-D, and Inception-E modules. As these modules correspond to deeper layers with heightened semantic abstraction, this targeted approach strengthens high-level representational power without adding undue complexity. The third variant, InceptionV3-SE-SA, integrates both SE and SA mechanisms. In this configuration, SA modules are inserted following the Inception-B module and again before the global pooling layer, emphasizing salient spatial regions at both mid-level and high-level stages. SE blocks are applied after the Inception-C, D, and E modules to refine channel interdependencies. The combined use of spatial and channel attention in this hybrid structure facilitates more robust and discriminative feature extraction.
3.2.4. EfficientNetB5
EfficientNetB5, introduced by Tan and Le [
27], is part of the EfficientNet series of CNNs designed using a compound scaling methodology. Rather than scaling depth, width, or input resolution independently, EfficientNet utilizes a compound coefficient that consistently scales these three factors together. This results in models that achieve high accuracy with improved computational efficiency.
The EfficientNet architecture originates from a baseline network optimized via neural architecture search (NAS) and is primarily structured around Mobile Inverted Bottleneck Convolution (MBConv) blocks integrated with SE modules. Each MBConv block increases the number of input channels, employs depthwise separable convolutions, and subsequently projects the result back to a lower dimensionality, which reduces both parameter count and floating-point operations (FLOPs). The incorporated SE modules perform adaptive recalibration of channel responses, allowing the model to prioritize the most relevant features. EfficientNetB5 is a scaled-up variant of the baseline EfficientNet, employing a compound coefficient to simultaneously scale network depth, width, and input resolution in a unified manner. Relative to smaller versions (B0–B4), B5 is capable of processing higher-resolution inputs (), which enables it to extract finer structural details. The architecture’s final layers include a global average pooling procedure followed by FC layers ending with a softmax classifier. Because of its efficient architecture and robust feature extraction, EfficientNetB5 has been widely adopted as a backbone in complex vision applications, with notable use in medical image classification and segmentation, domains that demand both high accuracy and computational efficiency.
To enhance its representational strength, we introduced three attention-augmented variants of EfficientNetB5, outlined in
Table 2. The first variant, EfficientNet-B5 (MB- Conv + SE), alters the original MBConv blocks to insert SE modules within each block, positioned immediately after the depthwise convolution and before the projection layer. This arrangement provides channel recalibration at every stage, ensuring detailed channel-level attention. The second variant, EfficientNet-B5 (SE after blocks), places SE modules selectively after key architectural stages: after Block 2 (index 4, 176 output channels), Block 3 (index 9, 512 output channels), and Block 4 (index 13, 512 output channels). This selective strategy emphasizes mid-level and high-level semantic features, while minimizing the additional computational cost associated with uniform SE integration. The third variant, EfficientNet-B5 (SE + SA), integrates both channel and SA by applying SE modules after Blocks 2, 3, and 4, and further introducing an SA module after Block 3 directly following SE3. This combined approach enables synergistic refinement, as SE focuses on channel discrimination and SA highlights spatially significant regions, thereby advancing both the location and nature of feature extraction.
3.2.5. DenseNet121
DenseNet121, introduced by Huang et al. [
28], is a CNN that uses dense connectivity to promote feature reuse and improve gradient flow. In this model, each layer receives the feature maps of all preceding layers as input and then passes its own outputs to every subsequent layer in the same block. This structure supports more efficient transmission of both information and gradients through the network, alleviating the vanishing gradient issue and enhancing parameter efficiency.
DenseNet121 consists of 121 layers grouped into four dense blocks, which are separated by transition layers. In each dense block, the feature maps produced by a layer are concatenated, not summed, with those of all earlier layers, resulting in varied and informative data representations. The transition layers, made up of convolutions followed by average pooling, are inserted between dense blocks to control the expansion of feature-map dimensions while reducing computation. The model’s structure starts with a convolution and max-pooling step and ends with global average pooling and an FC softmax classifier.
One of the primary advantages of DenseNet121 is its efficient use of parameters. Due to the extensive reuse of features across layers, the model achieves robust accuracy with fewer parameters than alternative architectures of comparable depth. This efficiency is particularly advantageous for medical image analysis, where datasets are often limited and there is a critical need for both effective feature extraction and minimization of overfitting. To evaluate the impact of various attention mechanisms, we developed three DenseNet121 variants that incorporate SE and SA modules, as depicted in
Figure 7. The first variant, DenseNet121-SE v1, extends the baseline by inserting SE blocks after each of the four dense blocks, thereby providing consistent channel recalibration throughout all levels of representation, from low to high-level features. The second variant, DenseNet121-SE v2, takes a selective approach by introducing SE blocks solely after dense blocks 2, 3, and 4. As these later dense blocks capture increasingly abstract semantic representations, this strategy directs the attention mechanisms toward deeper layers, maintaining a balance between computational cost and performance enhancement. The third variant, DenseNet121-SE-SA, incorporates both SE and SA modules. Specifically, SE modules are implemented after dense blocks 2–4 to strengthen channel-wise feature dependencies, while SA modules are positioned after transition layers 1–3 and prior to the global average pooling layer to focus on salient spatial regions. By integrating both channel and spatial forms of attention, the model achieves complementary refinement, improving its capacity to identify and prioritize relevant features and where they are most pertinent within the spatial domain.
3.3. Evaluation Metrics
To quantitatively evaluate the effectiveness of the model, we utilized four widely recognized classification metrics: accuracy, precision, recall, and F1-score. The definitions of these metrics are as follows:
where
,
,
, and
denote true positives, true negatives, false positives, and false negatives, respectively.
3.4. Mathematical Influence of SE Units on Learning
Let the output of a convolutional block be denoted as
, where
C,
H, and
W represent the number of channels, height, and width, respectively. SE unit first performs a
squeeze operation through global average pooling to generate a channel descriptor:
where
captures the global spatial information of each channel.
Next, an excitation operation learns channel-wise dependencies using two FC layers with a non-linear activation:
where
and
are learnable weight matrices,
denotes the ReLU activation, and
is the sigmoid function. The resulting vector
encodes the relative importance of each feature channel.
The reweighted feature maps are obtained by channel-wise multiplication:
where
acts as a dynamic scaling coefficient for channel
c.
This modulation influences the learning process by adaptively controlling both forward activations and backward gradients. During backpropagation, the gradient of the loss
with respect to
becomes the following:
This relationship demonstrates that channels with greater attention weights have correspondingly higher gradients. As a result, features that are discriminative are intensified, while redundant or noisy features are reduced, which enhances convergence stability and improves generalization. Such adaptive gradient scaling provides a mathematical rationale for the observed performance improvements when SE modules are selectively integrated into CNN architectures trained on the POC dataset.
3.5. Computational Efficiency Analysis
To evaluate the computational demands of attention integration, we compared the parameter count, FLOPs, inference time, and memory consumption for both the baseline and attention-enhanced models.
Table 3 presents the trade-offs in efficiency, indicating that SE and CBAM modules result in negligible additional cost while yielding substantial performance improvements.
3.6. Experimental Framework
3.6.1. Baseline Fine-Tuning of Pretrained CNNs
In the initial phase of the study, baseline models were established using several widely accepted CNN architectures, including VGG16, ResNet18, InceptionV3, DenseNet121, and EfficientNetB5. To maintain experimental consistency and utilize transfer learning, we employed their pretrained versions on the ImageNet dataset. These models were implemented in their default forms, with no changes in architecture, thereby ensuring a fair and robust benchmark. For fine-tuning, the terminal classification layer is substituted with an FC layer that matches the number of categories in our datasets. Convolutional layers utilize pretrained weights to convey generalizable feature representations, while the new layers are learned from randomly initialized parameters to fit the specific classification targets. This approach to fine-tuning provides a robust baseline for directly comparing model performance across architectures and serves as a basis for assessing the impact of subsequent improvements.
3.6.2. Integration of SE Modules
To augment the representational capability of the baseline architectures, we integrated SE modules into the convolutional blocks of each network to enhance the representation power of each model. As shown in
Figure 1, the SE block applies channel-wise attention by dynamically recalibrating the feature maps. This process starts with global average pooling, which summarizes spatial information into a channel descriptor. The descriptor then passes through two FC layers (realized as
convolutions) with a reduction ratio of 16, followed by a sigmoid activation to generate channel-specific weights. These weights are then multiplied by the original feature maps, thereby emphasizing informative channels and suppressing less relevant ones.
In the modified VGG16 architecture, SE modules are introduced after each convolutional layer and placed immediately prior to the corresponding pooling layer, where appropriate. Specifically, SE blocks are incorporated into all five of VGG16’s convolutional blocks. For example, in the initial block, each of the two convolutional layers is directly followed by an SE module and subsequently by a max-pooling operation, with this sequential structure maintained across the remaining blocks. This systematic application of channel attention across all abstraction levels enables dynamic recalibration of feature dependencies throughout the feature extraction process. The classifier segment of VGG16 is retained from the pretrained model, with the exception of the final FC layer, which is substituted to match the number of target classes (four in this context). Thus, the model preserves original ImageNet-pretrained feature representations while incorporating SE-based channel recalibration for finer-grained feature adjustment.
3.6.3. Selective Placement of SE Modules in CNN Models
Instead of positioning SE modules after every convolutional layer, we utilized a selective placement approach to optimize the trade-off between performance gains and computational efficiency. As outlined previously, an SE block enacts channel-wise attention by conducting global average pooling coupled with a bottleneck consisting of a reduction ratio of 16, yielding channel descriptors that guide recalibration of feature maps. Within the modified VGG16 structure, SE modules are placed solely after the final convolutional layers of the 3rd, 4th, and 5th convolutional blocks (i.e., conv_3_3, conv_4_3, and conv_5_3 in the canonical VGG16), which correspond to layers 10, 17, and 24 in the PyTorch implementation. This targeted integration emphasizes deeper layers, which encode higher-order semantic information, thereby augmenting representational capabilities while minimizing computational overhead.
By excluding SE modules from the early layers, the model efficiently preserves low-level representations such as texture and edge information, while the deeper layers are enhanced through adaptive channel recalibration to capture more complex contextual features. This configuration limits the number of added parameters, diminishing the likelihood of overfitting on comparatively small datasets. The classifier portion of VGG16 remains consistent with the pretrained backbone, aside from the final FC layer, which is revised to produce four target class outputs. In this manner, ImageNet-pretrained features are utilized, and critical stages of the network are selectively enhanced via channel-focused attention mechanisms.
We introduce SE modules following the mid-to-late convolutional blocks and immediately before global average pooling. This placement is informed by (i) the development of semantic richness in features, (ii) alignment of the receptive field with lesion scale, and (iii) the maintenance of stable gradient propagation in residual structures when SE follows the residual branch. Early layers extract basic textures and stain patterns; thus, reweighting at this stage is less beneficial. This information is less indicative of class and may increase the presence of noise. In contrast, later layers display greater inter-channel redundancy; SE diminishes this redundancy and enhances class differentiation.
3.6.4. Combining SA with SE Modules
To increase the representational capability of the network, we integrated both SE and SA modules within the VGG16 architecture in a complementary manner. SE blocks enable channel-wise recalibration, whereas SA modules focus on highlighting spatially relevant regions within feature maps. This integrated strategy enables the model to more effectively capture both inter-channel relationships and spatial contextual information. The SA module creates a SA map by applying max pooling and average pooling along the channel dimension, concatenating these results, and processing them with a convolution followed by a sigmoid activation. This operation enhances prominent spatial regions while diminishing the effects of background noise.
In our revised VGG16, SE and SA modules are strategically placed at distinct depths to optimize effectiveness. SA modules are introduced after the last convolutional layers of the first two convolutional blocks (conv1_2 and conv2_2), where low-level features such as edges and textures emerge, making spatial localization more valuable. SE modules are subsequently applied after the final convolutional layers of the third, fourth, and fifth convolutional blocks (conv3_3, conv4_3, and conv5_3), where abstract semantic features are prominent and channel recalibration is most advantageous. This arrangement creates a balance, enabling the network to extract detailed spatial features in the shallow layers and refine complex semantic representations in the deeper layers through channel attention. The classifier component of VGG16 remains consistent with the original pretrained version, except for substituting the last FC layer to accommodate the four classification categories. Through this complementary integration of SE and SA, the model enhances feature expression and classification performance while retaining computational efficiency.
3.7. Implementation Details
All experiments were conducted using PyTorch 2.7.1+cu118 on a system with an NVIDIA GPU. Two open-access medical imaging datasets, the Brain Tumor MRI dataset and the POC dataset, were utilized. Each dataset was partitioned into training and testing subsets, and all images were resized to
. Training was performed for 60 epochs with a batch size of 32. We utilized five pretrained CNN architectures, VGG16, ResNet18, InceptionV3, DenseNet121, and EfficientNetB5, as baseline models. To examine the influence of attention mechanisms, experiments were organized into four phases. The initial phase involved evaluating the baseline models in their unaltered form. In the following phases, SE blocks [
4] were systematically incorporated into all convolutional blocks to capture channel interdependencies and enhance feature representations. We further investigated selective integration, adding SE modules exclusively to upper layers (for example, Blocks 3–5 in VGG16) to reduce computational requirements while targeting improvements in semantic features. In the final phase, SA modules [
5] were integrated alongside SE blocks in the deeper layers to promote improved spatial localization and further enhance classification results.
All models were trained employing cross-entropy loss and the Adam optimizer. Two distinct parameter groups were established: the backbone CNN layers, assigned a learning rate of 0.0001, and the SE block parameters, granted a higher learning rate of 0.0006. A StepLR scheduler was used, decreasing the learning rate by a factor of 0.1 after every 10 epochs. Early stopping, using a 20-epoch patience threshold, was implemented to mitigate overfitting. Model performance assessment was conducted using accuracy, precision, recall, and F1-score, all computed from the confusion matrix of the test set. During training, the model with the highest test accuracy was preserved, and its evaluation metrics were documented in an output file for further analysis. To promote reproducibility, results for all experimental runs were comprehensively logged, including each confusion matrix and per-class values for precision, recall, and F1-score. Experiments were conducted with fixed random seeds to ensure consistency across all settings.
Training Parameters
The models were implemented and trained using the PyTorch framework for a total of 60 epochs with a batch size of 32. All input images were resized to pixels, and normalized according to the default torchvision preprocessing protocol. Both the Brain Tumor and POC datasets were structured as four-class classification tasks, corresponding to their predefined categories. Training utilized the Cross-Entropy Loss function throughout. The Adam optimizer was configured with two parameter groups that separately updated backbone layers and attention mechanisms. For the backbone CNN, a learning rate of 0.0001 and weight decay of 0.0001 were applied. Regarding the attention modules (SE/CBAM), learning rates of 0.0006 and a weight decay of 0.0001 were employed. A StepLR scheduler served to systematically decrease the learning rate, operating with a step size of 10 epochs and a decay factor of . Early stopping was applied, utilizing a patience value of 20 epochs and monitoring test accuracy; training was halted if no further improvement was detected. At the conclusion of each epoch, the model’s performance was assessed using the test set. The model exhibiting the highest test accuracy was designated as the best-performing model and subsequently saved. For each experiment, evaluation metrics, confusion matrix, precision, recall, and F1-score were systematically recorded. All experimental procedures were executed in an NVIDIA GPU-enabled computing environment.
The comprehensive list of training parameters utilized in all experiments is summarized in
Table 4.
4. Results and Discussion
This section details and interprets the classification results of various CNN architectures augmented with channel and spatial attention modules on two datasets: the POC dataset and the Brain Tumor MRI dataset. The evaluation criteria include test accuracy, precision, recall, and F1-score. The experimental findings are systematically organized into four parts: baseline CNN performance, global SE integration, selective SE integration, and hybrid attention integration.
4.1. Baseline Performance of Pretrained CNN Models
Table 5 and
Table 6 display the baseline outcomes of pretrained CNN architectures excluding attention mechanisms. For the POC dataset, EfficientNetB5 achieved the highest test accuracy at 86.05%, while VGG16 demonstrated the lowest performance at 78.22%. In the Brain Tumor dataset, DenseNet121 achieved the highest result, with 81.00% accuracy and an F1-score of 0.7940; EfficientNetB5 and InceptionV3 yielded similar performance levels.
These initial findings highlight the superior generalization abilities found in deeper and compound-scaled architectures such as EfficientNet and DenseNet, positioning them as effective baselines for evaluating the influence of attention mechanisms.
4.2. Global Integration of SE Modules
To investigate the effect of globally applied channel-wise attention, SE blocks were embedded into every convolutional block within each pretrained architecture (
Table 7 and
Table 8). All models exhibited consistent improvements across both datasets.
For the POC dataset, VGG16_SE yielded a notable improvement, achieving 86.65% accuracy and an F1-score of 0.8583. On the Brain Tumor dataset, EfficientNetB5_SE exhibited evident gains, with test accuracy rising from 80.50% to 84.37% and an F1-score of 0.8235. These results support the utility of channel recalibration in enhancing feature selectivity and advancing classification outcomes.
4.3. Selective SE Integration into Deeper Layers
Table 9 and
Table 10 show the results of employing SE blocks only in deeper layers, a method that lowers computational demands while maintaining advanced semantic feature refinement. The data reveal modest but consistent improvements over models with global SE integration.
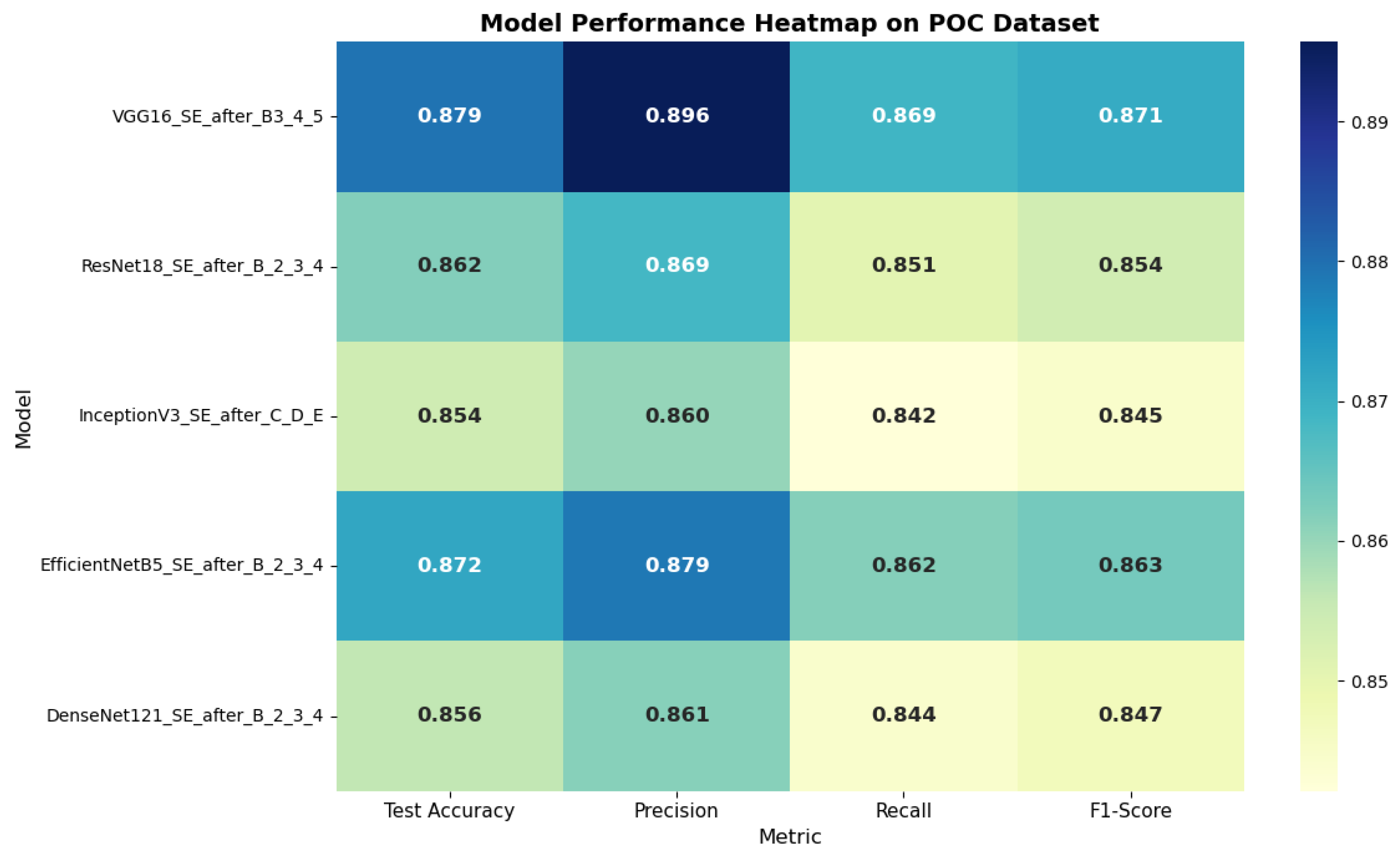
On the POC dataset, VGG16_SE_after_B3_4_5 achieved the best performance, with an F1-score of 87.08% and a test accuracy of 87.95%. For the Brain Tumor dataset, EfficientNetB5_SE_after_B2_3_4 delivered the highest results, reaching 86.53% accuracy and an F1-score of 85.11%, surpassing even the globally integrated SE variants. These outcomes indicate that applying attention to deeper layers plays a more critical role in decision-making than refining shallow features. The comparative performance of CNN architectures with selectively integrated SE modules on the POC dataset and brain tumor dataset is illustrated in
Figure 8 and
Figure 9. The heatmap clearly shows the variation in test accuracy, precision, recall, and F1-score across different model configurations.
4.4. Hybrid Attention: Combined SE and SA
To enhance both the type of features emphasized and the spatial focus, SA modules were combined with SE blocks, as detailed in
Table 11 and
Table 12. This hybrid architecture consistently produced the most robust outcomes across both datasets.
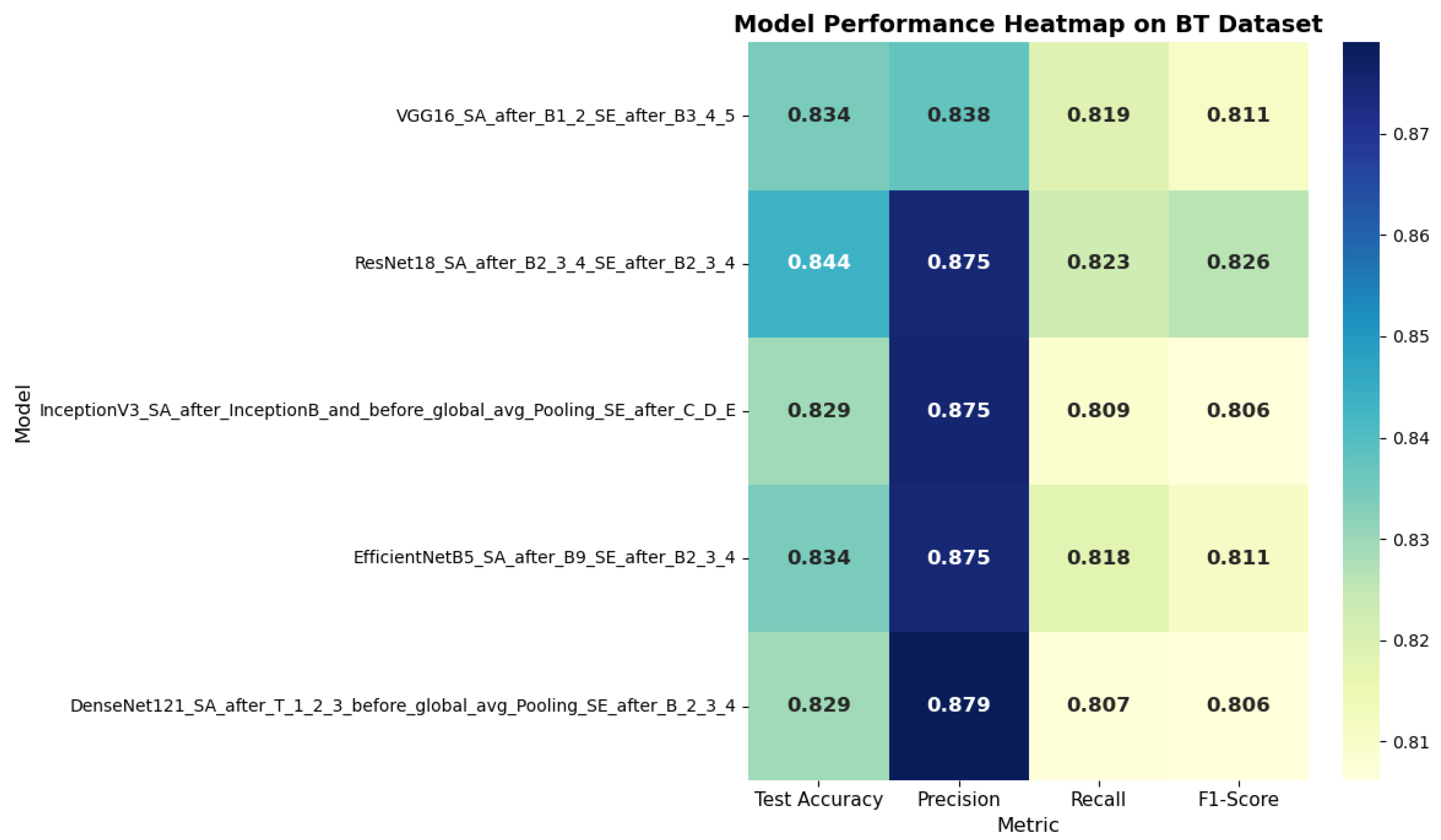
For the POC dataset, EfficientNetB5_SA_after_B9_SE_after_B2_3_4 acquired the best performance, reaching 89.97% accuracy with 89.72% F1-score. Similarly, on the Brain Tumor dataset, ResNet18 with hybrid attention attained 84.37% accuracy with 82.60% F1-score. These findings highlight the complementary roles of spatial and channel attention, where SA improves feature localization and SE strengthens channel relevance.
Figure 10 and
Figure 11 present the analysis of SA mechanisms on the POC dataset and brain tumor dataset by selectively integrating SE modules at different depths within CNN architectures. The heatmap highlights the variation in test accuracy, precision, recall, and F1-score across five configurations. EfficientNetB5 with selective SE placement achieved the highest overall performance, indicating the benefit of deeper SE integration for medical image classification.
4.5. Comparative Analysis and Key Observations
Our results provide several notable findings beyond the common assertion that “attention boosts accuracy.” The comparative performance of five CNNs across two medical imaging tasks demonstrates that the effect of attention mechanisms varies considerably based on both network depth and task characteristics. Specifically, attention introduced at earlier layers tends to improve feature discrimination for classification tasks, whereas mid or later layer attention enhances spatial coherence in segmentation. This suggests that the strategic placement of attention modules can support the development of more effective models for medical image analysis. By adhering to consistent preprocessing, training, and evaluation protocols, this study supplies a reproducible foundation for evaluating attention mechanisms in CNN-based models.
The experimental findings yield several key insights. First, EfficientNetB5 consistently outperformed other models, especially when enhanced with hybrid attention, owing to its balanced approach to scaling depth, width, and resolution. Second, hybrid attention yielded the highest overall metrics, emphasizing the advantage of integrating both channel-wise and SA within medical image classification. Third, selective integration demonstrated greater effectiveness than global application by providing similar performance gains with reduced computational requirements. Furthermore, attention mechanisms improved not only overall accuracy, but also class-specific metrics, as reflected in increased recall, precision, and F1-scores. These results collectively demonstrate that CNNs augmented with attention mechanisms offer significant improvements in both the reliability and accuracy of phenotypic pattern classification in spontaneous abortion and brain tumor diagnosis. Of all tested methodologies, selective hybrid attention applied to deeper layers was the most successful.
Comparative Analysis Across Models
A thorough comparison of all CNN models, both with and without attention mechanisms, reveals notable trends regarding architectural design and the impact of attention strategies. EfficientNetB5 consistently emerged as the optimal backbone across both datasets, achieving the highest F1-scores and test accuracy when paired with hybrid attention that combined SE and spatial modules. Its compound scaling approach facilitates more efficient feature extraction with fewer parameters compared to traditional deep networks. For every architecture assessed, models enhanced with attention surpassed their corresponding baselines. The global application of SE blocks increased both accuracy and F1-scores; however, targeted SE deployment within deeper layers yielded even greater improvements, reinforcing that higher-level semantic features are especially crucial for classification. Hybrid attention demonstrated superior performance overall, for example, EfficientNetB5_SA_after_B9_SE_after_B2_3_4 produced the highest results, achieving 89.97% accuracy and an F1-score of 89.72% on the POC dataset. These results indicate that fusing channel and spatial attention modules offers complementary benefits, guiding networks to concentrate on both the most pertinent regions and the most informative feature channels.
Lighter models such as ResNet18 and VGG16 also experienced significant improvement following the integration of attention mechanisms. Despite having shallower architectures, incorporating SE and SA modules markedly enhanced their classification capability, suggesting that attention can compensate for the intrinsic constraints of simpler backbones. The improvements were especially prominent on the Brain Tumor dataset, where high intra-class variability and nuanced texture differences make attention-guided feature learning particularly impactful. Conversely, InceptionV3 demonstrated less uniform advances: although SE contributed to measurable performance gains, the addition of hybrid attention yielded only limited benefits. This may result from the multi-branch design of Inception modules, which inherently capture diverse receptive fields and spatial patterns. In summary, these findings demonstrate that attention mechanisms, especially when used selectively and in combination, enhance robustness and generalization across CNN frameworks. EfficientNetB5 augmented with hybrid attention stood out as the most effective model for both Brain Tumor and POC classification. More generally, this evidence highlights the importance of engineering lightweight yet attention-integrated architectures to ensure reliable results in real-world medical image analysis scenarios.
5. Limitations and Future Work
While integrating attention mechanisms into pretrained CNN architectures has demonstrated notable performance improvements for brain tumor and POC classification, several limitations should be considered. Firstly, this work evaluated only two datasets, specifically the Brain Tumor and POC datasets. The inclusion of more heterogeneous datasets is necessary to validate generalizability across diverse clinical settings and a wider range of imaging modalities. Secondly, although attention modules such as SE and hybrid spatial–channel mechanisms offer classification enhancements, their integration adds additional complexity and computational overhead, potentially restricting deployment in real-time or resource-constrained environments, including mobile health applications and embedded diagnostic devices. A further limitation arises from the predetermined, manually selected positions of attention modules within the network architectures. These choices were informed by prior literature and empirical observations, yet they might not represent the most effective design choices. Automated strategies for optimizing module placement could yield superior architectures. Lastly, while performance increased, there remains a risk of overfitting, particularly in deeper models trained on relatively limited datasets, highlighting the ongoing need for rigorous cross-validation protocols and the development of more effective regularization techniques. In future work, we aim to expand experimentation to encompass larger and more varied datasets that reflect a broader spectrum of imaging scenarios and patient cohorts. To address computational challenges, we intend to investigate model compression techniques including pruning, quantization, and knowledge distillation. We will also explore automated strategies for attention module integration, employing neural architecture search or reinforcement learning to identify optimal configurations. Moreover, integrating multimodal data, combining imaging with clinical information, may improve model robustness and support more contextually informed diagnostic systems. In conclusion, while the proposed attention-augmented CNN models exhibit considerable potential for tumor classification, systematically addressing the identified limitations remains essential for facilitating their adoption in clinical practice.
5.1. Model Generalizability
Model generalizability plays a critical role in assessing the clinical utility of deep learning models. In this study, we assessed generalization by training and evaluating attention-augmented CNN architectures on two separate datasets: the Brain Tumor dataset and the POC dataset. The consistent improvements observed across both datasets demonstrate the robustness of models enhanced with attention mechanisms. These results suggest that attention mechanisms allow the networks to prioritize more discriminative and clinically relevant features, helping to minimize overfitting to dataset-specific characteristics. Pretrained architectures such as EfficientNetB5, ResNet18, and MobileNetV2 additionally supported improved generalization by leveraging knowledge acquired from large-scale natural image datasets and applying it in the medical context. Fine-tuning these models with medical data enabled the preservation of broad visual features while simultaneously adapting to domain-specific patterns. This approach to transfer learning is especially advantageous in situations where annotated medical datasets are scarce, as it facilitates superior performance on previously unseen instances. However, it is important to acknowledge that the generalizability of these models is most accurately established through evaluation on a wider array of datasets, encompassing images from various institutions, imaging equipment, and diverse patient demographics. Consequently, future research should focus on cross-dataset and cross-institutional validation to more thoroughly evaluate the models’ adaptability to real-world variability. In summary, the proposed attention-augmented CNNs demonstrate strong generalizability within the context of this research. With additional external validation and optimization, these models have significant promise for reliable use across a range of clinical environments.
5.2. Potential for Real-World Clinical Deployment
The promising outcomes from incorporating attention mechanisms into CNN architectures underscore their high potential for real-world clinical implementation in brain tumor and POC classification. Achievement of high classification accuracy, stable performance across both datasets, and improved generalizability via attention-based feature refinement establish these models as valuable assets for aiding radiologists and clinicians in diagnostic tasks. Notably, EfficientNetB5 with hybrid attention achieved the highest overall accuracy, while also demonstrating the capability to localize crucial imaging features, which is particularly important for detecting nuanced patterns in complex medical images. Utilizing pretrained backbones that are subsequently fine-tuned on medical datasets further enhances their clinical relevance, as this allows effective model training despite the limited availability of annotated samples common in healthcare contexts. This benefit of transfer learning, when integrated with modular attention mechanisms, enables models to adapt rapidly to novel tasks while requiring minimal annotation effort. Additionally, lightweight architectures such as ResNet18, when equipped with attention components, achieve efficient and precise predictions, rendering them ideal for use in portable or embedded diagnostic applications. However, several implementation challenges must be resolved before clinical deployment. Achieving real-time computation, ensuring interpretability, maintaining data security, and integrating seamlessly with hospital information systems are essential for successful adoption. Decision support tools should not only provide high levels of predictive accuracy but also generate interpretable explanations, such as visualizing attention-based feature maps, to build trust with healthcare practitioners. Moreover, regulatory certification and extensive clinical validation are imperative to establish reliability, safety, and conformance with medical guidelines.
In conclusion, CNN architectures augmented with attention mechanisms show considerable potential as the basis for clinical decision support systems. With ongoing enhancements aimed at improving interpretability, operational efficiency, and alignment with regulatory standards, these models may ultimately serve as highly effective tools in medical image-based diagnostics. Furthermore, in
Table 2 presents three variants of EfficientNet-B5 augmented with attention mechanisms. The first variant incorporates a custom Mobile Inverted Bottleneck Convolution (MBConv) block with an integrated SE module, placed after the pretrained EfficientNet-B5 feature extractor. This configuration replaces or complements the final processing layer with a block designed to adaptively recalibrate channel outputs, thereby increasing the model’s capacity to focus on salient features at the conclusion of the pipeline. The second variant preserves the native EfficientNet-B5 architecture while embedding SE modules after three critical convolutional blocks, specifically, blocks 4, 9, and 13. Through adaptive channel recalibration at multiple stages, this configuration progressively refines internal feature representations and maintains the integrity of the main convolutional pathway. The third variant expands upon this strategy by including SE modules at these positions alongside an additional SA block inserted following block 9. In this combined configuration, SE modules selectively enhance informative channels, and the SA module generates SA maps that delineate pertinent regions within the feature representations. This integrated attention strategy boosts the model’s discriminative power by addressing both the selection of key channels and the emphasis on significant spatial locations. Together, these variants exemplify distinct methodologies for embedding attention in EfficientNet-B5, ranging from modifying the final stage to implementing hierarchical channel recalibration and combining both channel and spatial-level attention to achieve more nuanced and robust feature enhancement.