5.1. Test Instances, Parameters, and Testing Environment
We tested the approaches on randomly generated graphs. Instances were generated according to three parameters: , d, and l. For the number of nodes , the values considered were . The continuous parameter represents the graph density; that is, for a given value of d, the resulting graph has arcs. For , we would obtain a complete graph. Finally, the continuous parameter is a label density parameter, controlling the number of labels . In particular, for any choice of l, we obtain a graph with different labels. Clearly, for , each arc would have a different label, while fewer labels will be present in the graph for . The assignment of labels to arcs is random.
The values considered for both d and l were . Ten different instances were generated for each combination of parameters, resulting in 810 instances for each value of , and 7290 instances in total. For each instance, nodes s and t were selected randomly.
The tests were executed on a Windows 10 PC with an Intel i7-11700 processor running at 2.5 GHz, with 32 GB of RAM. Both VNS and BRKGA-MFML were coded in the C++ language. A library for BRKGAs [
27] was used in the implementation of BRKGA-MFML. Based on some preliminary tests, the following parameter values were chosen for BRKGA-MFML:
,
,
,
,
,
,
,
. The values chosen for
,
, and
are consistent with the recommendations given in [
19] for BRKGAs. The other parameter choices provided a favorable trade-off between solution quality and computational times. Indeed,
controls how many chromosomes belong to each population, while
,
, and
regulate how many iterations the method performs. Finally, parameters
and
affect the performances of the decoder operator, since lower
and higher
values would lead to additional executions of the Push Relabel algorithm in the solution completion and solution cleaning step, respectively.
VNS, as described in [
11], used a maximum number of iterations as a termination criterion. However, to obtain a fair comparison, we modified this criterion so that it iterates until a time limit is reached. In particular, for each of its 7290 tests, as a time limit for VNS, we used the computational time required by BRKGA-MFML on the same instance.
Formulations ILP1 and ILP2 were also coded in C++, embedding the solver IBM ILOG CPLEX 22.1.1 through its native library. A time limit of 3600 s was considered for each formulation and each of the 7290 instances. To compare with the results of the two heuristics, we use either the optimal solution or the best upper bound from either formulation for instances unsolved with respect to proven optimality.
5.2. Results and Discussion
Before focusing on the comparison between the two heuristics, we briefly discuss the performance of the ILP1 and ILP2 formulations. ILP1 solved all instances with , with times increasing up to about 600 s for instances with and , with a peak of 669.85 s. On larger instances, an increasing number of instances were not solved with regard to proven optimality, particularly for . Indeed, for these values of density, ILP1 did not solve 5, 24, 45, 67, 96, or 103 instances with 50, 60, 70, 80, 90, or 100 nodes, respectively. Due to its additional complexity, ILP2 had overall larger computational times and more unsolved instances. One instance was already not solved for , with the other computational times increasing up to 956.40 s. For , ILP2 did not solve optimality for 11, 34, 55, 86, 109, or 129 instances with 50, 60, 70, 80, 90, or 100 nodes, respectively. Overall, ILP1 found better solutions than ILP2 139 times, while the opposite happened 37 times. As mentioned, we compare our heuristics with either optimal solutions or the best upper bounds found.
We report average results in
Table 1,
Table 2 and
Table 3. We divided instances into three groups, based on the value of
: small instances (20 to 40 nodes), medium instances (50 to 70 nodes), and large instances (80 to 100 nodes). For each value of
d and
l, in
Table 1,
Table 2 and
Table 3, we report average values computed for the corresponding 30 instances. In particular, under
heuristic time (s), we report the average computational time required by BRKGA-MFML, which, as discussed, corresponds to the average computational time given as a time limit for VNS. The two columns under the
BRKGA-MFML %gaps heading contain the average gaps between BRKGA-MFML and either VNS or the best upper bound returned by any of the two models. In particular, for VNS and a given instance
x, each gap is computed as
, where
refers to the objective function value of method
on instance
x. Gaps computed with respect to solutions obtained by CPLEX are computed analogously.
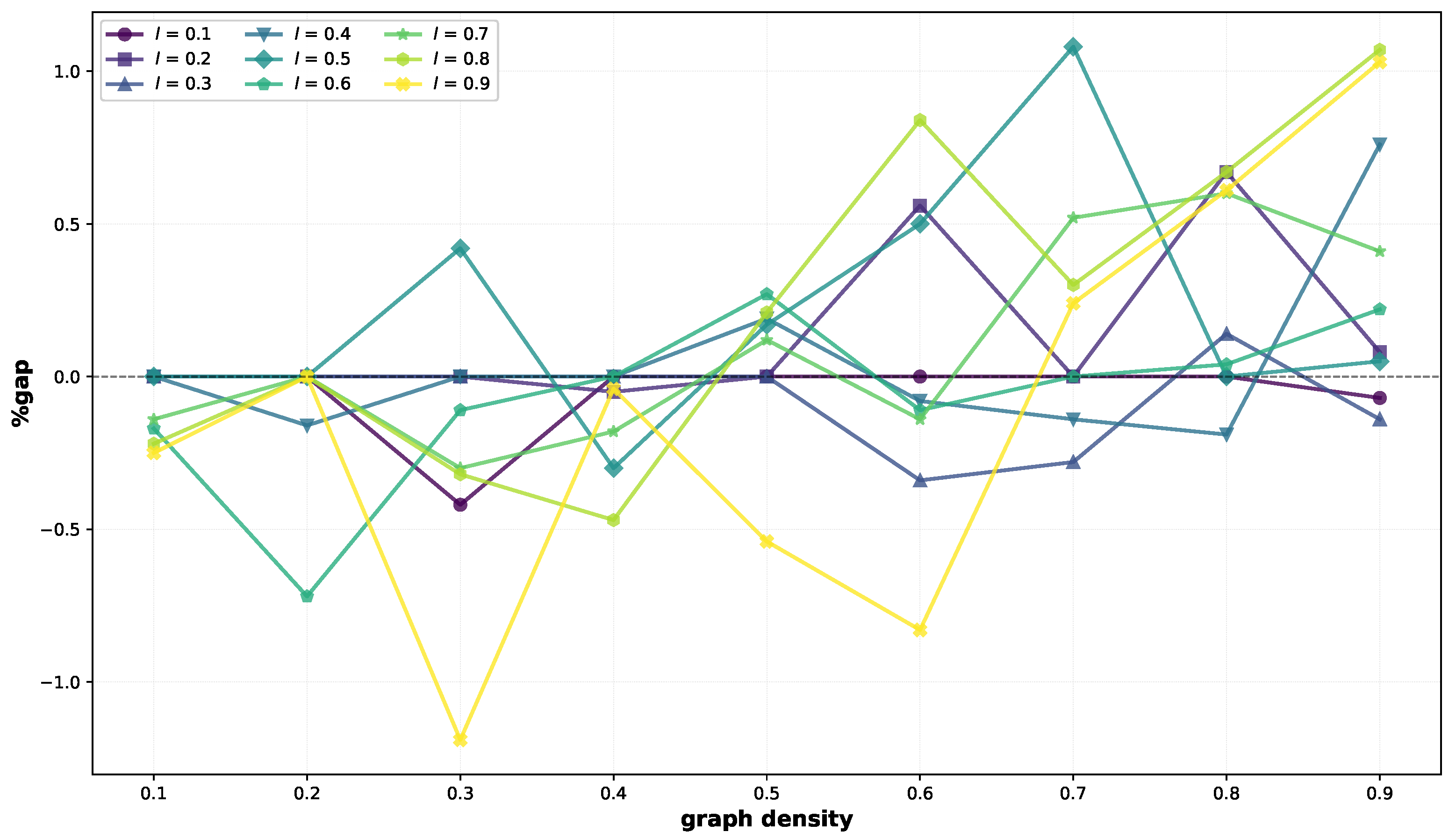
On small instances (
Table 1), the two heuristics perform very similarly. Indeed, they return the same solutions (VNS gap = 0) in 26 cases out of 81, BRKGA-MFML finds better solutions on average in 28 cases (negative value in the VNS gap column), and VNS finds better solutions in the remaining 27. Gaps are very small, varying between −1.19% and 1.08%. Average computational times remain within about
for all instances with a density up to
, reaching
on average for the instances with more edges and labels (
). The comparison between BRKGA-MFML and VNS on small graphs is illustrated visually in
Figure 1, where it can be seen that for most graph and label densities, gaps are close to 0, with peaks around 1% and −1%.
In accordance with the computational times, the number of generations needed to reach the best solution by BRKGA-MFML increases with d and l. In particular, it was always found in the initial population for and . The average number of generations to reach this solution remains always below 2 for and up to 1.20 for , and for , up to 1.87 for . The best solution is found after more than 10 generations on average only for , up to 11.90 generations.
Comparing with the CPLEX solutions, we can see that their values are very close. The CPLEX gap is 0 in 37 out of 81 cases and below 1% in 70 cases, increasing only twice over 2%, up to 2.63% ().
To conclude, both heuristics prove to be very suitable for solving small instances, providing accurate solutions within fast computational times.
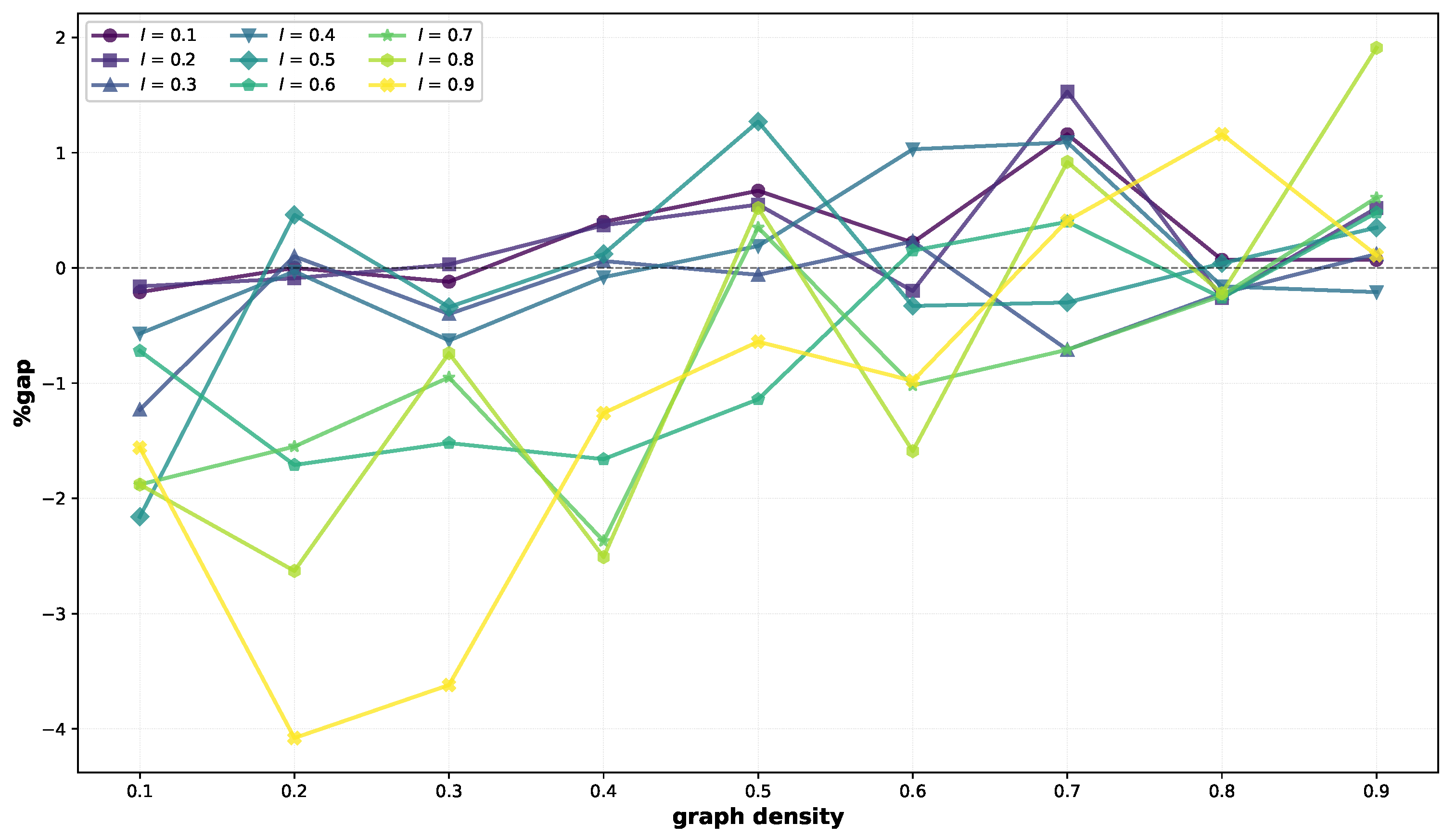
Let us now consider medium-sized instances (
Table 2). Compared to the small ones, there is a more pronounced difference in performance between BRKGA-MFML and VNS. Indeed, BRKGA-MFML finds better solutions in 46 out of 81 cases, with a maximum negative gap of −4.08% (
). VNS finds better solutions in 34 cases, with a maximum gap equal to 1.91%. The 46 negative gaps average −1%, while the positive gaps average 0.52%. We can see that BRKGA-MFML is particularly effective on the less dense scenarios, finding better solutions in 23 out of 27 scenarios for
, and in 36 out of 54 scenarios for
. While the performances of the two methods appeared comparable on small graphs, on the medium ones, we can see that the diverse solutions across the search space maintained in the population by BRKGA-MFML is often beneficial for escaping from local minima. These results are illustrated in
Figure 2, which shows that most gaps are negative, particularly for low graph densities and high graph densities. Similar trends will be observed for large graphs.
In terms of computational times, the heuristics run within 2 s on average for , within 10 s for , and within 30 s for . On the denser instances with more labels (), the average computational time reaches 40.70 s.
The average number of generations needed by BRKGA-MFML to find the best solution is below 2 in all cases except one for , where it grows to 2.63 (). It remains within 5 generations for , and within 15 generations for , increasing to over 20 generations only for (with a maximum of 25.47 for , ).
Comparing with CPLEX solutions, we observe that the gap is 0 in 8 cases, within 1% in 43 cases, and within 2% in 61 out of 81 cases, exceeding 4% only in 3 cases (up to 4.51% for ).
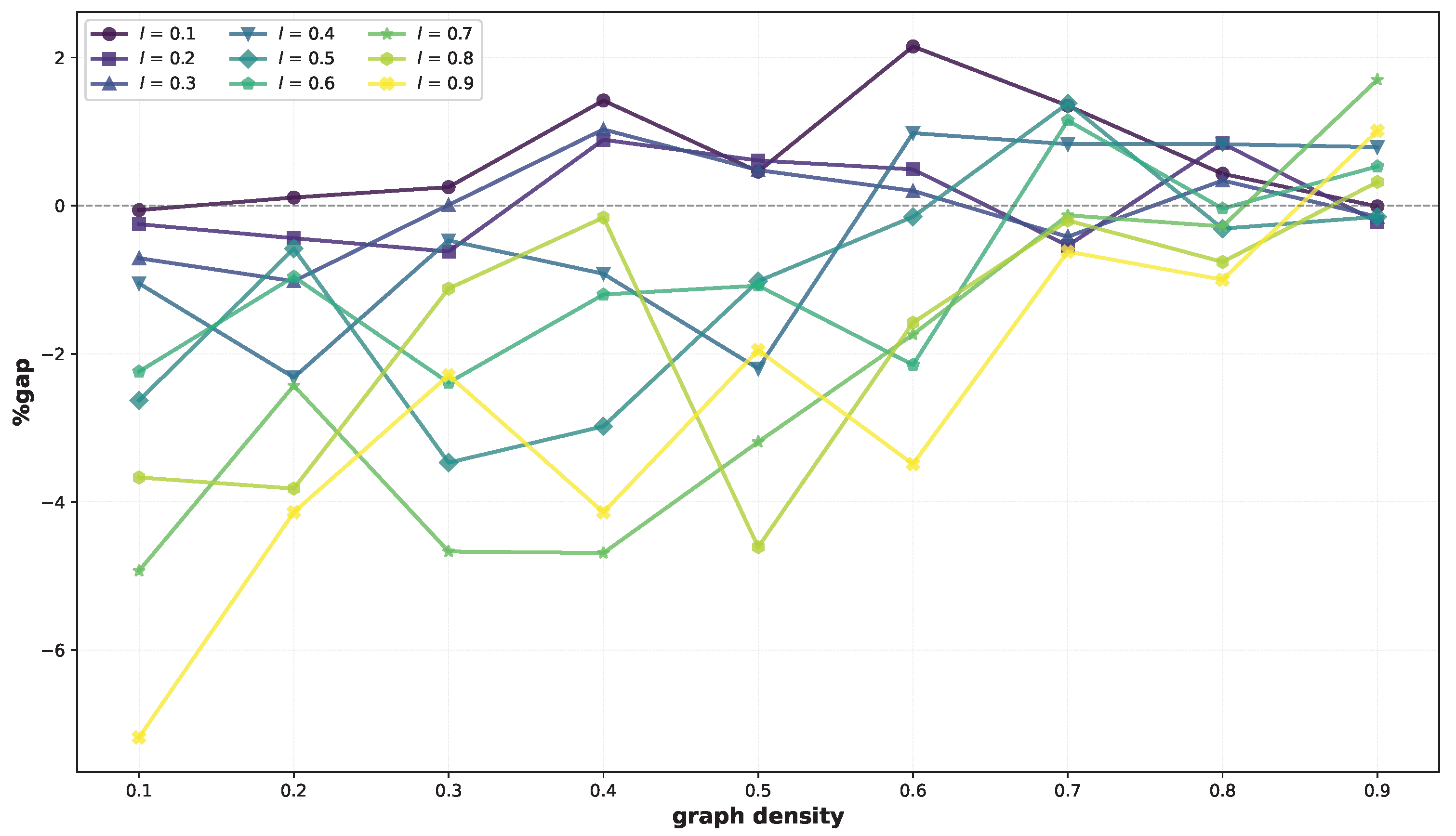
Finally, we consider the large instances. The results are reported in
Table 3. For these instances, BRKGA-MFML performs better on average in 55 out of 81 cases, while VNS performs better in 26 cases. Negative gaps reach −7.18% (
). Positive gaps reach 2.15% (
); however, they are higher than 2% only once. Again, we observe that BRKGA-MFML performs particularly well for low densities, finding better solutions in 24 out of 27 cases for
and 41 out of 54 cases for
. These trends, similar to those noted for medium graphs, are shown in
Figure 3.
While higher than those observed for the other instances, computational times remain reasonable. The heuristics run within about 6 s for , and within 60 s for . For , and , average computational times reach around 100 s, 170 s, and 250 s, respectively. As observed for the previous cases, computational times increase according to both the d and l parameters.
The best solution is found by BRKGA-MFML on average within 4.77 generations for , 5.20 generations for , and 8.40 generations for . The average number of generations needed exceeds 10 for and , 20 for and , and 30 for and , reaching 38.57 for and .
Finally, looking at CPLEX gaps, we can see that they reach 4.81% in the worst case. However, they remain below 3% in 64 out of 81 cases.