An Efficient Sparse Twin Parametric Insensitive Support Vector Regression Model
Abstract
1. Introduction
- An effective STPISVR model is introduced which encourages sparse solutions while retaining TPISVR’s ability for complex data structures, including heteroscedasticity.
- The optimization problems are reformulated as two sparse LPPs with constraints derived from the KKT conditions. One of these constraints corresponds to an L1-norm expression, which is shown to be a constant through further analysis of the KKT conditions. As a result, the explicit L1-norm regularization term is removed, and the resulting LPPs inherently produce sparse solutions due to their underlying geometric structure.
- This reformulation reduces computational complexity via two mechanisms: enhanced solution sparsity induced by the LPP structure and improved efficiency in solving LPPs instead of traditional QPPs.
- A two-stage hybrid parameter tuning strategy is adopted to improve both tuning accuracy and computational efficiency.
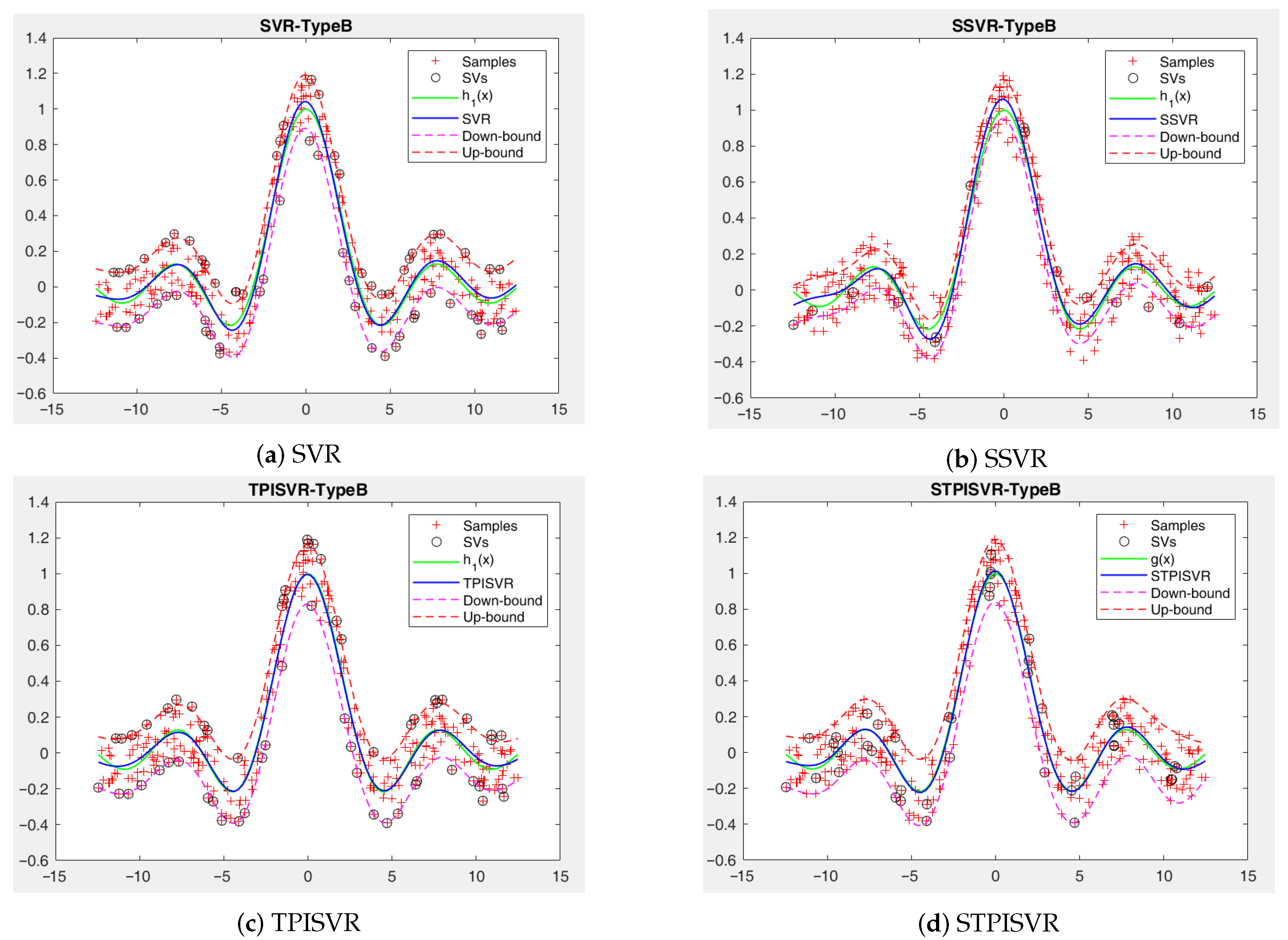
- Extensive experiments demonstrate that STPISVR significantly reduces SVs, enabling faster predictions without sacrificing accuracy, and achieves a superior trade-off among accuracy, sparsity, and computational efficiency, especially for complex data structures.
2. Related Works
2.1. SVR Model
2.2. TPISVR Model
3. STPISVR
3.1. SSVR Model
3.2. STPISVR Model
| Algorithm 1 STPISVR training algorithm. |
Input: Training set ; parameter combination (Gaussian kernel parameter)) Output: Prediction function ; training time (Tr-time); number of SVs (Num-SVs) 1. Start timer. 2. Solve optimization problem (25) to obtain and . 3. Solve optimization problem (27) to obtain and . 4. Construct bound functions and using Equations (14) and (15), respectively. 5. Compute final regression function via Equation (16). 6. Stop timer, record Tr-time. 7. Count Num-SVs from and . 8. Return , Tr-time and Num-SVs. |
| Algorithm 2 STPISVR Testing Algorithm |
Input: Test set ; trained prediction function Output: Root mean squared error (RMSE); coefficient of determination (); test time (Te-time); Zone Width between insensitive boundaes (ZoneWidth, if needed) 1. Start timer. 2. For each test sample in , Compute prediction . 3. Calculate RMSE and . 4. Stop timer, record Te-time. 5. Return RMSE, , Te-time and ZoneWidth (if computed). |
3.3. Properties of the STPISVR Model
- (i)
- and are the lower bounds on the fractions of down-SVs and up-SVs, respectively.
- (ii)
- and are the upper bounds on the fractions of down-errors and up-errors, respectively.
- (i)
- for any data point if .
- (ii)
- for any data point if .
- (iii)
- for any data point if .
4. Discussion and Comparison
4.1. Sparsity
4.2. Prediction Speed
4.3. Computational Complexity
5. Numerical Experiments
| Algorithm 3 Two-stage parameter tuning strategy for STPISVR. |
Input: Full training set D; grid parameter space ; number of cross-validation folds K Output: Optimal parameter combination 1. Randomly select 50% of D as tuning subset . 2. Stage 1: Grid Search 3. For each in : 4. Perform K-fold cross-validation on : 5. For each fold: 6. Train STPISVR on training fold using Algorithm 1 with parameter . 7. Evaluate on validation fold using Algorithm 2 to obtain RMSE. 8. Compute average RMSE over all folds. 9. Select top 5 candidates with the lowest average RMSE. 10. Stage 2: Bayesian Optimization 11. Randomly split into training set (40%) and validation set (10%). 12. Define Bayesian parameter search space based on the top 5 candidates from grid search. 13. Initialize using the lowest RMSE of a top candidate from grid search, and set accordingly. 14. Set . 15. Repeat for up to 200 iterations: 16. Propose candidate parameter via Bayesian optimization. 17. Train STPISVR on using Algorithm 1 with . 18. Evaluate on using Algorithm 2 to obtain RMSE. 19. If RMSE < : 20. Update RMSE, , and reset . 21. Else: 22. Increment by 1. 23. Update Bayesian optimizer with . 24. If , break loop. 25. Return . |
5.1. Synthesis Datasets
5.2. Benchmark Datasets
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar]
- Drucker, H.; Burges, C.J.; Kaufman, L.; Smola, A.; Vapnik, V. Support vector regression machines. Adv. Neural Inf. Process. Syst. 1996, 9, 155–161. [Google Scholar]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Pontil, M.; Rifkin, R.; Evgeniou, T. From Regression to Classification in Support Vector Machines. 1998. Available online: https://dspace.mit.edu/bitstream/handle/1721.1/7258/AIM-1649.pdf?sequence=2&isAllowed=y (accessed on 30 May 2025).
- Basak, D.; Pal, S.; Patranabis, D.C. Support vector regression. Neural Inf. Process.-Lett. Rev. 2007, 11, 203–224. [Google Scholar]
- Awad, M.; Khanna, R.; Awad, M.; Khanna, R. Support vector regression. In Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers; Apress: Berkeley, CA, USA, 2015; pp. 67–80. [Google Scholar]
- Zhang, F.; O’Donnell, L.J. Support vector regression. In Machine Learning; Academic Press: Cambridge, MA, USA, 2020; pp. 123–140. [Google Scholar]
- Huang, H.; Wei, X.; Zhou, Y. An overview on twin support vector regression. Neurocomputing 2022, 490, 80–92. [Google Scholar] [CrossRef]
- Peng, X. TSVR: An efficient twin support vector machine for regression. Neural Netw. 2010, 23, 365–372. [Google Scholar] [CrossRef]
- Hao, P.Y. Pair-v-svr: A novel and efficient pairing nu-support vector regression algorithm. IEEE Trans. Neural Netw. Learn. Syst. 2016, 28, 2503–2515. [Google Scholar] [CrossRef]
- Peng, X. Efficient twin parametric insensitive support vector regression model. Neurocomputing 2012, 79, 26–38. [Google Scholar] [CrossRef]
- Hao, P.Y. New support vector algorithms with parametric insensitive/margin model. Neural Netw. 2010, 23, 60–73. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.J.; Williamson, R.C.; Bartlett, P.L. New support vector algorithms. Neural Comput. 2000, 12, 1207–1245. [Google Scholar] [CrossRef]
- Khemchandani, R.; Chandra, S. Twin support vector machines for pattern classification. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 905–910. [Google Scholar]
- Peng, X. TPMSVM: A novel twin parametric-margin support vector machine for pattern recognition. Pattern Recognit. 2011, 44, 2678–2692. [Google Scholar] [CrossRef]
- Zhu, J.; Rosset, S.; Tibshirani, R.; Hastie, T. 1-norm support vector machines. Adv. Neural Inf. Process. Syst. 2003, 16, 49–56. [Google Scholar]
- Tang, Q.; Li, G. Sparse L0-norm least squares support vector machine with feature selection. Inf. Sci. 2024, 670, 120591. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Huang, K.; Zheng, D.; Sun, J.; Hotta, Y.; Fujimoto, K.; Naoi, S. Sparse learning for support vector classification. Pattern Recognit. Lett. 2010, 31, 1944–1951. [Google Scholar] [CrossRef]
- Bi, J.; Bennett, K.; Embrechts, M.; Breneman, C.; Song, M. Dimensionality reduction via sparse support vector machines. J. Mach. Learn. Res. 2003, 3, 1229–1243. [Google Scholar]
- Santos, J.D.A.; Barreto, G.A. Novel sparse LSSVR models in primal weight space for robust system identification with outliers. J. Process Control 2018, 67, 129–140. [Google Scholar] [CrossRef]
- Cui, L.; Shen, J.; Yao, S. The Sparse Learning of The Support Vector Machine. J. Phys. Conf. Ser. 2021, 2078, 012006. [Google Scholar] [CrossRef]
- Moosaei, H.; Mousavi, A.; Hladík, M.; Gao, Z. Sparse l1-norm quadratic surface support vector machine with universum data. Soft Comput. 2023, 27, 5567–5586. [Google Scholar] [CrossRef]
- Qu, S.; De Leone, R.; Huang, M. Sparse Learning for Linear Twin Parameter-margin Support Vector Machine. In Proceedings of the 2024 3rd Asia Conference on Algorithms, Computing and Machine Learning, Shanghai, China, 22–24 March 2024; pp. 50–55. [Google Scholar]
- Zhang, Z.; Zhen, L.; Deng, N.; Tan, J. Sparse least square twin support vector machine with adaptive norm. Appl. Intell. 2014, 41, 1097–1107. [Google Scholar] [CrossRef]
- Smola, A.; Scholkopf, B.; Ratsch, G. Linear programs for automatic accuracy control in regression. In Proceedings of the 1999 Ninth International Conference on Artificial Neural Networks ICANN 99. (Conf. Publ. No. 470), Edinburgh, UK, 7–10 September 1999; Volume 2, pp. 575–580. [Google Scholar]
- Breiman, L. Prediction games and arcing algorithms. Neural Comput. 1999, 11, 1493–1517. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, P. A duality theorem for non-linear programming. Q. Appl. Math. 1961, 19, 239–244. [Google Scholar] [CrossRef]
- Nesterov, Y.; Nemirovskii, A. Interior-Point Polynomial Algorithms in Convex Programming; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 1994. [Google Scholar]
- Shahriari, B.; Swersky, K.; Wang, Z.; Adams, R.P.; de Freitas, N. Taking the human out of the loop: A review of Bayesian optimization. Proc. IEEE 2015, 104, 148–175. [Google Scholar] [CrossRef]










| Notation | Description |
|---|---|
| The real space | |
| The feature space | |
| The cardinality of a set | |
| The L1 norm: sum of absolute values of a vector’s components | |
| The L2 norm: Euclidean norm of a vector | |
| I | The index set of samples: |
| n | The dimension of samples |
| the pth sample, | |
| X | The samples matrix: † |
| The response value of | |
| Y | The response vector, |
| e | The vector of opportune dimensions with all components equal to 1 |
| The normal vectors | |
| The mapping function from a original input space to a feature space | |
| The mapping matrix: | |
| The kernel function: are all random n-dimensional vector | |
| K | The kernel matrix: with |
| The column vector: | |
| The slack variables | |
| The bias terms | |
| The regularization parameters | |
| Lagrange multiplier vectors, the dual variables in SVR or SSVR | |
| The pth element of | |
| The qth element of | |
| Lagrange multiplier vectors, the dual variables in TPISVR or STPISVR | |
| The pth element of | |
| The qth element of | |
| The insensitive tube parameter |
| Type of Noise | Evaluation Index | SVR | SSVR | TPISVR | STPISVR |
|---|---|---|---|---|---|
| Type A | RMSE | 0.1085 ± 0.0053 | 0.1063 ± 0.0026 | 0.1110 ± 0.0043 | 0.1028 ± 0.0041 |
| Noisy test | 0.9010 ± 0.0146 | 0.9102 ± 0.0102 | 0.8965 ± 0.0143 | 0.9111 ± 0.0133 | |
| Num-SVs | 107.60 ± 7.8060 | 18.80 ± 1.0328 | 154.10 ± 3.6347 | 46.60 ± 1.5776 | |
| Tr-time (s) | 0.4377 ± 0.0435 | 0.2509 ± 0.0220 | 0.8406 ± 0.0253 | 0.3270 ± 0.0213 | |
| Te-time (s) | 0.0116 ± 0.0013 | 0.0028 ± 0.0008 | 0.0186 ± 0.0029 | 0.0060 ± 0.0012 | |
| Noiseless test | RMSE | 0.0447 ± 0.0050 | 0.0377 ± 0.0035 | 0.0487 ± 0.0049 | 0.0366 ± 0.0049 |
| 0.9815 ± 0.0038 | 0.9844 ± 0.0012 | 0.9781 ± 0.0034 | 0.9873 ± 0.0042 | ||
| Te-time (s) | 0.0117 ± 0.0014 | 0.0021 ± 0.0004 | 0.0172 ± 0.0006 | 0.0051 ± 0.0005 | |
| Type B | RMSE | 0.1281 ± 0.0040 | 0.1194 ± 0.0048 | 0.1261 ± 0.0045 | 0.1171 ± 0.0038 |
| Noisy test | 0.8645 ± 0.0143 | 0.8820 ± 0.0153 | 0.8648 ± 0.0169 | 0.8866 ± 0.0151 | |
| Num-SVs | 214.50 ± 11.3652 | 15.30 ± 0.9487 | 164.30 ± 4.3474 | 45.50 ± 1.7159 | |
| Tr-time (s) | 0.4605 ± 0.0423 | 0.2753 ± 0.0349 | 0.7615 ± 0.0324 | 0.3232 ± 0.0113 | |
| Te-time (s) | 0.0268 ± 0.0038 | 0.0026 ± 0.0005 | 0.0198 ± 0.0032 | 0.0059 ± 0.0009 | |
| Noiseless test | RMSE | 0.0548 ± 0.0069 | 0.0277 ± 0.0055 | 0.0476 ± 0.0051 | 0.0140 ± 0.0036 |
| 0.9719 ± 0.0073 | 0.9927 ± 0.0029 | 0.9787 ± 0.0056 | 0.9981 ± 0.0010 | ||
| Te-time (s) | 0.0244 ± 0.0025 | 0.0022 ± 0.0006 | 0.0207 ± 0.0025 | 0.0048 ± 0.0004 | |
| Type A † | RMSE | 0.1089 ± 0.0049 | 0.1076 ± 0.0030 | 0.1115 ± 0.0045 | 0.1070 ± 0.0040 |
| Noisy test | 0.9003 ± 0.0119 | 0.9033 ± 0.0111 | 0.8957 ± 0.0145 | 0.9065 ± 0.0143 | |
| Num-SVs | 173.00 ± 8.3267 | 17.00 ± 0.4714 | 138.50 ± 3.0277 | 101.50 ± 1.9579 | |
| Tr-time (s) | 0.4255 ± 0.0443 | 0.9490 ± 0.0558 | 0.8805 ± 0.0484 | 0.8482 ± 0.0442 | |
| Te-time (s) | 0.0198 ± 0.0032 | 0.0016 ± 0.0005 | 0.0170 ± 0.0025 | 0.0125 ± 0.0016 | |
| Noiseless test | RMSE | 0.0448 ± 0.0065 | 0.0385 ± 0.0033 | 0.0503 ± 0.0047 | 0.0379 ± 0.0058 |
| 0.9809 ± 0.0039 | 0.9838 ± 0.0013 | 0.9766 ± 0.0037 | 0.9864 ± 0.0034 | ||
| Te-time (s) | 0.0191 ± 0.0023 | 0.0019 ± 0.0002 | 0.0167 ± 0.0011 | 0.0059 ± 0.0006 |
| Type of Noise | Evaluation Index | SVR | SSVR | TPISVR | STPISVR |
|---|---|---|---|---|---|
| Type C | RMSE | 0.0518 ± 0.0027 | 0.0520 ± 0.0022 | 0.0508 ± 0.0025 | 0.0505 ± 0.0023 |
| Noisy test | 0.7950 ± 0.0163 | 0.7950 ± 0.0170 | 0.8040 ± 0.0114 | 0.8070 ± 0.0104 | |
| Num-SVs | 133.10 ± 5.8395 | 4.30 ± 0.4830 | 43.0 ± 0.6667 | 26.0 ± 0.6667 | |
| Tr-time (s) | 0.2779 ± 0.0243 | 0.1143 ± 0.0202 | 0.5414 ± 0.0420 | 0.1721 ± 0.0946 | |
| Te-time (s) | 0.0096 ± 0.0012 | 0.0008 ± 0.0005 | 0.0036 ± 0.0007 | 0.0020 ± 0.0004 | |
| Noiseless test | RMSE | 0.0146 ± 0.0029 | 0.0135 ± 0.0020 | 0.0097 ± 0.0021 | 0.0061 ± 0.0020 |
| 0.9795 ± 0.0077 | 0.9827 ± 0.0056 | 0.9910 ± 0.0036 | 0.9962 ± 0.0021 | ||
| Te-time (s) | 0.0093 ± 0.0010 | 0.0007 ± 0.0004 | 0.0035 ± 0.0011 | 0.0020 ± 0.0002 | |
| Type D | RMSE | 0.0455 ± 0.0025 | 0.0447 ± 0.0024 | 0.0447 ± 0.0024 | 0.0445 ± 0.0025 |
| Noisy test | 0.8350 ± 0.0222 | 0.8410 ± 0.0213 | 0.8410 ± 0.0203 | 0.8419 ± 0.0222 | |
| Num-SVs | 78.90 ± 4.4083 | 6.20 ± 0.4216 | 117.50 ± 3.2745 | 78.70 ± 0.6749 | |
| Tr-time (s) | 0.2892 ± 0.0154 | 0.5111 ± 0.0322 | 0.5069 ± 0.0183 | 0.1587 ± 0.0174 | |
| Te-time (s) | 0.0052 ± 0.0005 | 0.0007 ± 0.0003 | 0.0081 ± 0.0011 | 0.0059 ± 0.0005 | |
| Noiseless test | RMSE | 0.0116 ± 0.0020 | 0.0088 ± 0.0028 | 0.0087 ± 0.0031 | 0.0085 ± 0.0025 |
| 0.9872 ± 0.0043 | 0.9923 ± 0.0055 | 0.9924 ± 0.0044 | 0.9928 ± 0.0037 | ||
| Te-time (s) | 0.0052 ± 0.0005 | 0.0006 ± 0.0001 | 0.0073 ± 0.0008 | 0.0060 ± 0.0005 |
| Dataset | Evaluation Index | SVR | SSVR | TPISVR | STPISVR |
|---|---|---|---|---|---|
| D1 | RMSE | 0.7107 ± 0.0868 | 0.7121 ± 0.1000 | 0.7073 ± 0.0825 | 0.7034 ± 0.0548 |
| (398 × 8) | 0.4721 ± 0.1705 | 0.4519 ± 0.1373 | 0.4765 ± 0.0846 | 0.4897 ± 0.606 | |
| Num-SVs | 196.20 ± 5.1651 | 32.60 ± 3.0258 | 266.70 ± 5.5187 | 188.80 ± 4.4920 | |
| Tr-time (s) | 0.0215 ± 0.0074 | 0.3734 ± 0.0596 | 0.9445 ± 0.0260 | 0.5137 ± 0.1133 | |
| Te-time (s) | 0.0017 ± 0.0014 | 0.0003 ± 0.0002 | 0.0019 ± 0.0003 | 0.0015 ± 0.0002 | |
| D2 | RMSE | 0.3801 ± 0.2137 | 0.3741 ± 0.1654 | 0.3940 ± 0.1720 | 0.3712 ± 0.1163 |
| (159 × 16) | 0.7813 ± 0.1543 | 0.7958 ± 0.0944 | 0.7761 ± 0.1426 | 0.7983 ± 0.1179 | |
| Num-SVs | 79.60 ± 3.0258 | 52.80 ± 2.8206 | 106.50 ± 4.1433 | 37.10 ± 2.5582 | |
| Tr-time (s) | 0.0269 ± 0.0079 | 0.0868 ± 0.0125 | 0.5812 ± 0.0244 | 0.1352 ± 0.0806 | |
| Te-time (s) | 0.0010 ± 0.0006 | 0.0006 ± 0.0010 | 0.0006 ± 0.0005 | 0.0003 ± 0.0002 | |
| D3 | RMSE | 0.3911 ± 0.0928 | 0.3812 ± 0.1029 | 0.3556 ± 0.1025 | 0.3426 ± 0.0896 |
| (506 × 14) | 0.8412 ± 0.0516 | 0.8422 ± 0.0631 | 0.8516 ± 0.0853 | 0.8554 ± 0.0583 | |
| Num-SVs | 273.20 ± 5.3707 | 61.50 ± 2.9533 | 307.10 ± 5.3635 | 226.20 ± 3.3928 | |
| Tr-time (s) | 0.1777 ± 0.0215 | 0.8818 ± 0.0502 | 1.1177 ± 0.0544 | 0.8554 ± 0.0583 | |
| Te-time (s) | 0.0020 ± 0.0005 | 0.0009 ± 0.0005 | 0.0029 ± 0.0004 | 0.0018 ± 0.0004 | |
| D4 | RMSE | 0.3384 ± 0.0610 | 0.3372 ± 0.0533 | 0.4223 ± 0.0504 | 0.3902 ± 0.0465 |
| (1030 × 9) | 0.8783 ± 0.0491 | 0.8795 ± 0.0459 | 0.8173 ± 0.0489 | 0.8424 ± 0.0413 | |
| Num-SVs | 603.70 ± 7.3944 | 156.90 ± 4.0947 | 634.60 ± 12.1546 | 485.6 ± 4,8808 | |
| Tr-time (s) | 0.8718 ± 0.0904 | 9.4158 ± 0.3500 | 4.7170 ± 0.1532 | 3.1539 ± 0.2175 | |
| Te-time (s) | 0.0110 ± 0.0009 | 0.0036 ± 0.0009 | 0.0122 ± 0.0018 | 0.0103 ± 0.0009 | |
| D5 | RMSE | 0.1058 ± 0.0875 | 0.1108 ± 0.1047 | 0.1012 ± 0.1313 | 0.1013 ± 0.1299 |
| (60 × 10) | 0.9566 ± 0.0574 | 0.9536 ± 0.0863 | 0.9659 ± 0.0387 | 0.9657 ± 0.0361 | |
| Num-SVs | 36.90 ± 1.8529 | 11.60 ± 0.8433 | 39.00 ± 2.3570 | 23.80 ± 0.8756 | |
| Tr-time (s) | 0.0070 ± 0.0013 | 0.0102 ± 0.0019 | 0.5196 ± 0.0463 | 0.0805 ± 0.0021 | |
| Te-time (s) | 0.0007 ± 0.0004 | 0.0004 ± 0.0007 | 0.0003 ± 0.0004 | 0.0002 ± 0.0002 | |
| D6 | RMSE | 0.0878 ± 0.0686 | 0.0906 ± 0.0859 | 0.1816 ± 0.2815 | 0.1824 ± 0.1871 |
| (209 × 7) | 0.9880 ± 0.0046 | 0.9845 ± 0.0141 | 0.9549 ± 0.0536 | 0.9546 ± 0.0234 | |
| Num-SVs | 30.90 ± 1.5239 | 18.90 ± 1.2649 | 122.30 ± 19.3861 | 103.40 ± 1.2293 | |
| Tr-time (s) | 0.0664 ± 0.0330 | 0.1188 ± 0.0167 | 0.6926 ± 0.1372 | 0.1327 ± 0.0395 | |
| Te-time (s) | 0.0012 ± 0.0015 | 0.0004 ± 0.0005 | 0.0006 ± 0.0004 | 0.0004 ± 0.0002 | |
| D7 | RMSE | 0.5786 ± 0.1970 | 0.6029 ± 0.2414 | 0.5576 ± 0.2119 | 0.5362 ± 0.1878 |
| (107 × 4) | 0.5454 ± 0.1982 | 0.5429 ± 0.1541 | 0.5555 ± 0.2720 | 0.5569 ± 0.2134 | |
| Num-SVs | 73.30 ± 2.6331 | 10.40 ± 1.1738 | 49.10 ± 2.9364 | 33.70 ± 2.0248 | |
| Tr-time (s) | 0.0180 ± 0.0061 | 0.0215 ± 0.0061 | 0.5677 ± 0.0446 | 0.0861 ± 0.0026 | |
| Te-time (s) | 0.0009 ± 0.0007 | 0.0003 ± 0.0005 | 0.0004 ± 0.0005 | 0.0003 ± 0.0002 | |
| D8 | RMSE | 0.4619 ± 0.1120 | 0.4616 ± 0.1238 | 0.4531 ± 0.1012 | 0.4398 ± 0.0918 |
| (133 × 2) | 0.6967 ± 0.3489 | 0.6969 ± 0.2139 | 0.7055 ± 0.1560 | 0.8256 ± 0.2867 | |
| Num-SVs | 74.80 ± 2.4060 | 13.40 ± 0.6992 | 66.80 ± 1.6364 | 35.40 ± 0.8756 | |
| Tr-time (s) | 0.0138 ± 0.0267 | 0.0555 ± 0.0159 | 0.5382 ± 0.0209 | 0.1007 ± 0.0094 | |
| Te-time (s) | 0.0008 ± 0.0006 | 0.0004 ± 0.0004 | 0.0007 ± 0.0004 | 0.0004 ± 0.0002 | |
| D9 | RMSE | 0.1008 ± 0.0211 | 0.1391 ± 0.0223 | 0.1764 ± 0.0737 | 0.1692 ± 0.0663 |
| (308 × 6) | 0.9846 ± 0.0033 | 0.9705 ± 0.0054 | 0.9450 ± 0.0396 | 0.9613 ± 0.0405 | |
| Num-SVs | 73.20 ± 3.7947 | 61.10 ± 1.7029 | 156.00 ± 1.8257 | 85.00 ± 2.9814 | |
| Tr-time (s) | 0.0445 ± 0.0425 | 0.3344 ± 0.0564 | 0.8192 ± 0.0430 | 0.2916 ± 0.0124 | |
| Te-time (s) | 0.0008 ± 0.0005 | 0.0006 ± 0.0005 | 0.0010 ± 0.0004 | 0.0007 ± 0.0004 | |
| D10 | RMSE | 0.6999 ± 0.0780 | 0.6848 ± 0.0742 | 0.6681 ± 0.1020 | 0.6429 ± 0.0711 |
| (536 × 8) | 0.4870 ± 0.0834 | 0.5089 ± 0.0761 | 0.5343 ± 0.0808 | 0.5428 ± 0.0547 | |
| Num-SVs | 475.20 ± 2.5298 | 16.90 ± 1.1005 | 288.40 ± 2.7568 | 238.50 ± 1.3540 | |
| Tr-time (s) | 0.0378 ± 0.0162 | 0.5449 ± 0.0505 | 1.3134 ± 0.1316 | 0.5132 ± 0.0426 | |
| Te-time (s) | 0.0019 ± 0.0010 | 0.0005 ± 0.0004 | 0.0023 ± 0.0005 | 0.0020 ± 0.0002 | |
| D8 † | RMSE | 0.4911 ± 0.1115 | 0.4759 ± 0.1057 | 0.4673 ± 0.1027 | 0.4464 ± 0.1179 |
| (133 × 2) | 0.5718 ± 0.4510 | 0.6161 ± 0.3490 | 0.6189 ± 0.5790 | 0.6572 ± 0.5867 | |
| Num-SVs | 102.20 ± 1.3160 | 11.00 ± 0.4714 | 43.20 ± 0.7888 | 32.50 ± 0.8498 | |
| Tr-time (s) | 0.3333 ± 0.0431 | 0.3548 ± 0.0323 | 0.5215 ± 0.0283 | 0.5054 ± 0.0165 | |
| Te-time (s) | 0.0017 ± 0.0015 | 0.0004 ± 0.0005 | 0.0006 ± 0.0004 | 0.0005 ± 0.0003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, S.; Guo, Y.; De Leone, R.; Huang, M.; Li, P. An Efficient Sparse Twin Parametric Insensitive Support Vector Regression Model. Mathematics 2025, 13, 2206. https://doi.org/10.3390/math13132206
Qu S, Guo Y, De Leone R, Huang M, Li P. An Efficient Sparse Twin Parametric Insensitive Support Vector Regression Model. Mathematics. 2025; 13(13):2206. https://doi.org/10.3390/math13132206
Chicago/Turabian StyleQu, Shuanghong, Yushan Guo, Renato De Leone, Min Huang, and Pu Li. 2025. "An Efficient Sparse Twin Parametric Insensitive Support Vector Regression Model" Mathematics 13, no. 13: 2206. https://doi.org/10.3390/math13132206
APA StyleQu, S., Guo, Y., De Leone, R., Huang, M., & Li, P. (2025). An Efficient Sparse Twin Parametric Insensitive Support Vector Regression Model. Mathematics, 13(13), 2206. https://doi.org/10.3390/math13132206







