A Methodology to Extract Knowledge from Datasets Using ML
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Generation over a Probability Distribution: Dataset Feature Splitting (DFS)
2.2. An Objective Measure of Classification Performance: Ordered Lists of All Combinations of FS of a Given Size
2.3. Encoding Knowledge Relevance: Using Medical Expertise from LLMs
“Is there a synergic relationship between diabetes and anemia to suffer heart failure?”
2.4. How Well Do the Different ML Algorithms Classify the Most Relevant Binary FSs?
2.5. Validation Methodology
2.5.1. Validation of Method 1 (DFS)
2.5.2. Validation of Methods 2, 3, and 4
2.6. Application of the Methodology: Knowledge Extraction from the Original Datasets
3. Results
3.1. Results of Methods
3.2. Validation of Methods
3.2.1. Validation of Method 1 (DFS)
3.2.2. Validation of Methods 2, 3, and 4
4. Discussion
Limitations and Further Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. The Four Original Datasets Used
| HCV data. UCI machine learning repository. Hepatitis C prediction. Twelve features and 615 instances. List of numbered features. Hepatitis C. 1: AGE, 2: SEX, 3: ALB albumin, 4: ALP alkaline phosphatase, 5: ALT alanine amino-transferase, 6: AST aspartate amino-transferase, 7: BIL bilirubin, 8: CHE choline esterase, 9: CHO cholesterol, 10: CRE creatinine, 11: GGT gamma glutamyl transpeptidase, 12: PRO protein. Heart Failure. UCI machine learning repository. Heart failure clinical records. Twelve features and 299 instances. List of numbered features. Heart Failure. 1: AGE, 2: ANA anemia, 3: CPH creatinine phosphokinase, 4: DIA diabetes, 5: EJF ejection fraction, 6: HBP high blood pressure, 7: PLA platelets, 8: SCR serum creatinine, 9: SSO serum sodium, 10: SEX, 11: SMO smoking, 12: TIM time. Heart Disease. UCI machine learning repository. Heart-h. Twelve features and 299 instances. List of numbered features. Heart disease. 1: AGE, 2: SEX, 3: CP chest pain, 4: BPS blood pressure, 5: FBS fasting blood sugar, 6: ECG electrocardiographic results, 7: MHR maximum heart rate, 8: ANG exercise induced angina, 9: STD depression induced by exercise relative to rest, 10: SLO slope of the peak exercise ST segment, 11: CA number of major vessels colored using fluoroscopy, 12: CHO serum cholesterol. Chronic kidney disease. UCI machine learning repository. Chronic kidney disease. Thirteen features and 158 instances. List of numbered features. Chronic Kidney Disease. 1: AGE, 2: ALB albumin, 3: URE blood urea, 4: CRE serum creatinine, 5: SOD sodium, 6: POT potassium, 7: HEM hemoglobin, 8: WHI white blood cell count, 9: RED red blood cell count, 10: HTN hypertension, 11: DM diabetes mellitus, 12: CAD coronary artery disease, 13: ANE anemia. |
Appendix B. The Seven Families of Supervised ML Algorithms Used
| LR. Logistic Regression was implemented using the Scikit-learn 1.0.1 library written in Python. This algorithm belongs to the linear classification learning scientific family. It implements the linear regression algorithm, which is based on classical statistics, adapted to a categorical, i.e., non-numeric class targeted feature using a non-linear transformation. The maximum number of iterations was set to 1000. NB. Naïve Bayes was implemented using the Scikit-learn 1.0.1 library written in Python. This algorithm belongs to the Bayesian statistical learning scientific family. It is a non-linear algorithm that sometimes may behave linearly. SVM. Support Vector Machine was implemented using the Scikit-learn 1.0.1 library written in Python. The hyperparameters were adjusted to C from 0 to 100 at intervals of 10, and the 10 resulting scores were averaged. An ‘rbf’ kernel was used as the mapping transformation. This algorithm belongs at the same time to the linear classification and instance-based learning scientific families. It uses a non-linear mapping transformation in order to treat non-linear problems. MLP. Multilayer Perceptron was implemented using the Scikit-learn 1.0.1 library written in Python. An architecture of [100 × 100] neurons was used. An activation function ‘tanh’ was performed for the hidden layer. An initial learning rate of 0.001 and a maximum number of iterations of 100,000 were used. A ‘random_state’ 0 argument was performed in order to have reproducible results across multiple function calls. This algorithm belongs to the linear classification learning scientific family. It implements the perceptron learning rule, which is the standard neural network architecture. Due to its multilayer structure, it is able to learn non-linear data concepts. DT. Decision Tree was implemented using the Scikit-learn 1.0.1 library written in Python. This algorithm belongs to the information theory learning scientific family. It creates step-function-like decision boundaries to learn from non-linear relationships in data. KN. K-nearest Neighbors was implemented using the Scikit-learn 1.0.1 library written in Python. This algorithm belongs to the instance-based learning scientific family. It is considered to create decision boundaries that are often non-linear. RF. Random Forest was implemented using the Scikit-learn 1.0.1 library written in Python. This algorithm belongs to the ensemble methods that combine the predictions of several base estimators built with a given learning algorithm in order to improve the generalizability and robustness over a single estimator. |
References
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Blum, A.L.; Langley, P. Selection of relevant features and examples in machine learning. Artif. Intell. 1997, 97, 245–271. [Google Scholar] [CrossRef]
- Kohavi, R.; John, G.H. Wrappers for feature selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Domingos, P. A few useful things to know about machine learning. Commun. ACM 2012, 55, 78–87. [Google Scholar] [CrossRef]
- Liu, H.; Yu, L. Toward integrating feature selection algorithms for classification and clustering. IEEE Trans. Knowl. Data Eng. 2005, 17, 491–502. [Google Scholar]
- Dash, M.; Liu, H. Feature Selection for Classification. Intell. Data Anal. 1997, 1, 131–156. [Google Scholar] [CrossRef]
- Kononenko, I. Estimating Attributes: Analysis and Extensions of RELIEF. In Proceedings of the European Conference on Machine Learning (ECML-94), Catania, Italy, 6–8 April 1994; Springer: Berlin/Heidelberg, Germany, 1994. [Google Scholar]
- Molnar, C. Interpretable Machine Learning, 2nd ed.; Lulu Enterprises, Inc.: Durham, NC, USA, 2022; Available online: https://christophm.github.io/interpretable-ml-book/ (accessed on 20 May 2025).
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006. [Google Scholar]
- Swallow, D.M. Genetic influences on lactase persistence and lactose intolerance. Annu. Rev. Genet. 2003, 37, 197–219. [Google Scholar] [CrossRef] [PubMed]
- UC Irvine Machine Learning Repository. Available online: https://archive.ics.uci.edu/ (accessed on 26 March 2025).
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann: Burlington, Massachusetts, MA, USA, 2016. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, Y.; Zhang, M.; Xu, C.; Guo, Y.; Zhan, Y.; Ding, S.; Wang, J.; Xu, K.; Fang, Y.; et al. Large Language Models for Disease Diagnosis: A Scoping Review. arXiv 2024, arXiv:2409.00097. [Google Scholar]
- Nazi, Z.A.; Peng, W. Large Language Models in Healthcare and Medical Domain: A review. arXiv 2024, arXiv:2401.06775. [Google Scholar] [CrossRef]
- OpenAI. ChatGPT (v2.0) [Large Language Model]. OpenAI. 2025. Available online: https://openai.com/chatgpt (accessed on 26 March 2025).
- Google AI. Gemini: A Tool for Scientific Writing Assistance. 2025. Available online: https://gemini.google.com/ (accessed on 26 March 2025).
- Elmore, J.G.; Longton, G.M.; Carney, P.A.; Geller, B.M.; Onega, T.; Tosteson, A.N.; Nelson, H.D.; Pepe, M.S.; Allison, K.H.; Schnitt, S.J.; et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 2015, 313, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Massey, F.J., Jr. The Kolmogorov-Smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951, 46, 68–78. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pearson, K. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Philos. Mag. 1900, 50, 157–175. [Google Scholar] [CrossRef]
- Friedman, M. A comparison of alternative tests of significance for the problem of m rankings. Ann. Math. Stat. 1940, 11, 86–92. [Google Scholar] [CrossRef]
- Demšar, J. Statistical comparisons of classifiers over multiple datasets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Bender, E.M.; Gebru, T.; McMillan-Major, A.; Shmitchell, S. On the Dangers of Stochastic Parrots: Can Language Models Be Too Big? In Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency, FAccT’21, Toronto, ON, Canada, 3–10 March 2021. [Google Scholar]
- Nori, H.; King, N.; McKinney, S.M.; Carignan, D.; Horvitz, E. Capabilities of GPT-4 on Medical Challenge Problems. arXiv 2023, arXiv:2303.13375. [Google Scholar]
- Liebrenz, M.; Schleifer, R.; Buadze, A.; Bhugra, D. Generative AI in Healthcare: Point of View of Clinicians; McKinsey & Company: New York, NY, USA, 2023. [Google Scholar]

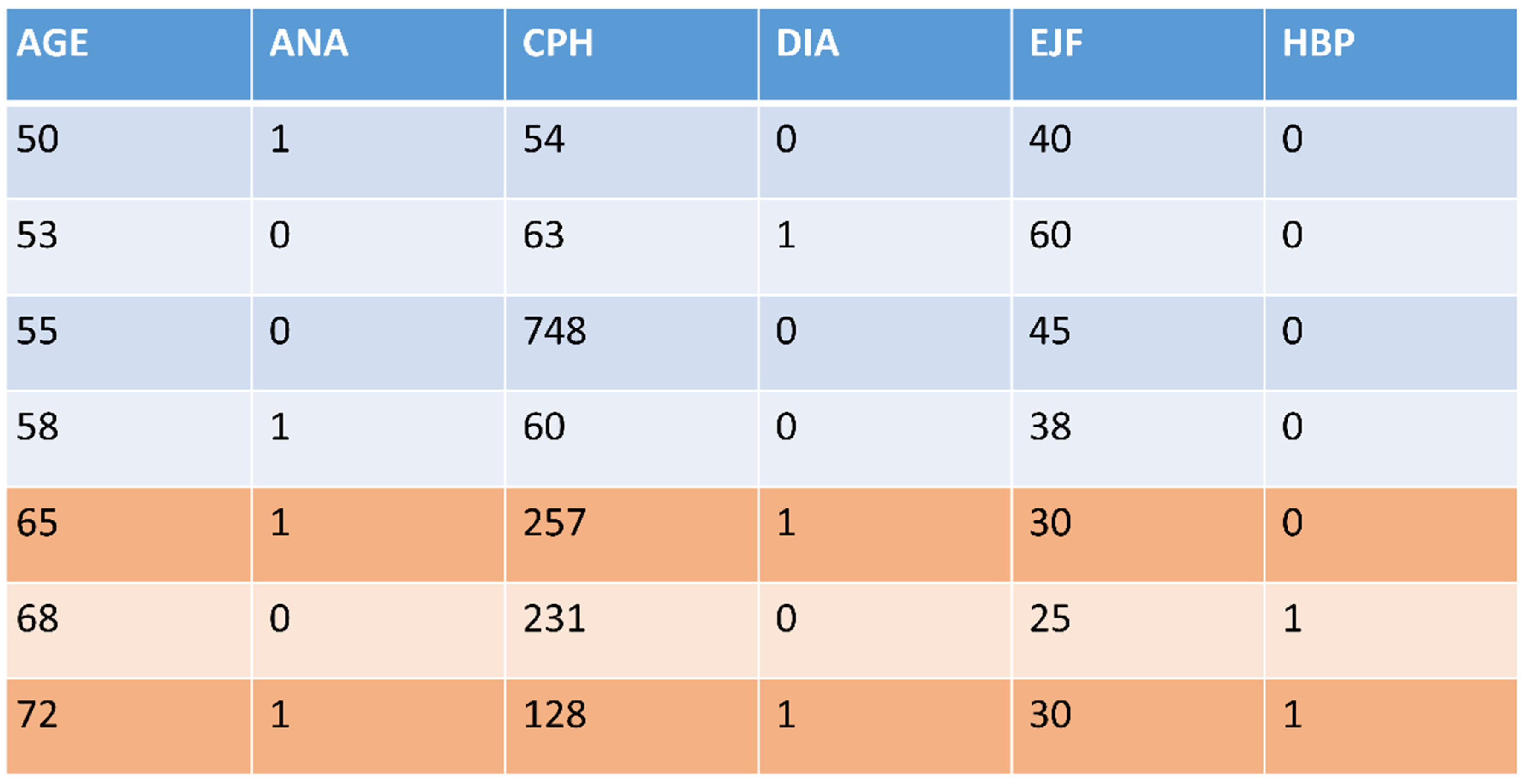

| Feature Types | Missing Data | Class Distribution | Instances | |
|---|---|---|---|---|
| HP-UCI | 1 CAT/9 NUM | NO | 75/540 | 615 |
| HF-UCI | 5 CAT/7 NUM | NO | 96/203 | 299 |
| HD-UCI | 4 CAT/8 NUM | NO | 138/161 | 299 |
| CKD-UCI | 4 CAT/9 NUM | NO | 43/115 | 158 |
| HP-UCI | HF-UCI | ||
|---|---|---|---|
| AGE H | (4, 5, 6, 7, 10, 11) | AGE H | (2, 3, 4, 6, 7, 9, 10, 11) |
| SEX H | (5) | ANA H | (1, 3, 4, 5, 6, 7, 8, 9, 10, 11) |
| SEX L | (3, 10) | CPH H | (1, 2, 4, 6, 9, 11) |
| ALB L | (4, 5, 6, 7, 11, 12) | DIA H | (1, 2, 3, 5, 6, 7, 8, 9, 10, 11) |
| ALP H | (1, 3, 5, 6, 7, 9, 10, 11, 12) | EJF L | (1, 2, 3, 4, 6, 7, 8, 10, 11) |
| ALT H | (1, 2, 3, 4, 6, 7, 9, 10, 11, 12) | HBP H | (1, 2, 3, 4, 7, 9, 10, 11) |
| BIL H | (1, 3, 4, 5, 6, 11, 12) | PLA H | (1, 2, 4, 6, 10, 11) |
| CHO L | (4, 5, 6, 11) | SCR H | (1, 2, 3, 4, 5, 6, 7, 11) |
| CRE H | (1, 4, 5, 6, 12) | SSO H | (1, 2, 3, 4, 6, 10, 11) |
| PRO H | (3, 4, 5, 6, 7, 10, 11) | SEX H | (6, 7) |
| SEX L | (1, 2, 4, 9, 11) | ||
| HD-UCI | SMO H | (1, 2, 3, 4, 6, 7, 9, 10) | |
| AGE H | (2, 3, 4, 5, 7, 8, 9, 10, 11, 12) | ||
| SEX H | (5, 8, 10, 11, 12) | CKD-UCI | |
| SEX L | (1, 4, 6, 7, 9) | AGE H | (2, 3, 4, 6, 7, 8) |
| BPS H | (1, 2, 5, 6, 7, 8, 9, 10, 11, 12) | URE H | (1, 3, 4, 5, 6, 7) |
| FBS H | (1, 2, 4, 7, 9, 10, 11, 12) | CRE H | (1, 2, 4, 5, 6, 8) |
| ECG H | (2, 4, 7, 8, 9, 10) | SOD L | (1, 2, 3, 5, 6, 8) |
| MHR H | (1, 2, 4, 5, 6, 8, 9, 10, 11, 12) | POT H | (2, 3, 4, 6, 8) |
| ANG H | (1, 2, 4, 6, 7, 9, 10, 12) | HEM L | (1, 2, 3, 4, 5, 7, 8) |
| STD H | (1, 2, 4, 5, 6, 7, 8, 10, 11, 12) | WHI H | (1, 2, 6, 8) |
| SLO H | (1, 2, 4, 5, 6, 7, 8, 9, 11, 12) | RED L | (1, 3, 4, 5, 6, 7) |
| CA H | (1, 2, 4, 5, 7, 9, 10, 12) | ||
| CHO H | (1, 2, 4, 5, 7, 8, 9, 10, 11) |
| CRE H | |||||||
|---|---|---|---|---|---|---|---|
| size 3 | 156.3 (SVM) | 151.3 (RF) | 149.8 (DT) | 147.6 (NB) | 146.6 (KN) | 133.0 (LR) | 132.7 (MLP) |
| size 4 | 470.2 (SVM) | 446.8 (RF) | 445.0 (DT) | 443.2 (NB) | 441.9 (KN) | 424.4(MLP) | 408.9 (LR) |
| size 5 | 926.6 (SVM) | 892.5 (KN) | 879.2 (RF) | 878.4 (NB) | 874.6 (DT) | 841.0(MLP) | 825.3 (LR) |
| size 6 | 1273.0 (SVM) | 1245.4(KN) | 1218.3 (NB) | 1210.7 (RF) | 1206.0 (DT) | 1181.0(MLP) | 1157.0 (LR) |
| DFS Datasets | p-Values |
|---|---|
| ALP H | 1.1394834474 × 10−5 |
| CRE H | 4.2769255669 × 10−26 |
| HP-UCI | 4 out of 10 | HF-UCI | 10 out of 12 |
| AGE H | 1.0892849424 × 10−5 | AGE H | 0.00058360217226 |
| SEX H | 5.1118124321 × 10−6 | ANA H | 0.00352660750438 |
| SEX L | 1.9878513331 × 10−19 | CPH H | 0.59033279794372 |
| ALB L | 9.6008360198 × 10−26 | DIA H | 0.00765360026050 |
| ALP H | 1.1394834474 × 10−5 | EJF L | 4.7648532405 × 10−7 |
| ALT H | 4.3517379024 × 10−46 | HBP H | 0.00058360217226 |
| BIL H | 1.4937625445 × 10−5 | PLA H | 0.00018288992313 |
| CHO L | 1.8795587161 × 10−19 | SCR H | 0.59033279794372 |
| CRE H | 4.2769255669 × 10−26 | SSO H | 4.7648532405 × 10−7 |
| PRO H | 2.2151976407 × 10−22 | SEX H | 0.00018288992313 |
| SEX L | 0.00178955624413 | ||
| HD-UCI | 9 out of 12 | SMO H | 0.12566168302727 |
| AGE H | 0.66297421392268 | ||
| SEX H | 0.01899680622953 | CKD-UCI | 0 out of 8 |
| SEX L | 1.4733907068 × 10−8 | AGE H | 4.6913601105 × 10−31 |
| BPS H | 0.00468751862537 | URE H | 1.6302989587 × 10−33 |
| FBS H | 0.01116685752824 | CRE H | 2.0841286358 × 10−30 |
| ECG H | 0.00204213738762 | SOD L | 1.6087231294 × 10−42 |
| MHR H | 8.6014095603 × 10−11 | POT H | 2.0841286358 × 10−30 |
| ANG H | 0.01899680622953 | HEM L | 3.0897650452 × 10−47 |
| STD H | 1.1199214677 × 10−6 | WHI H | 2.1831400120 × 10−21 |
| SLO H | 0.22253198035961 | RED L | 3.4672730092 × 10−40 |
| CA H | 6.8454149810 × 10−9 | ||
| CHO H | 0.07620693805287 |
| HP-3 | AGE H | SEX H | SEX L | ALB L | ALP L | ALT H | BIL H | CHO H | CRE H | PROTH |
|---|---|---|---|---|---|---|---|---|---|---|
| LR | 172.6 | 24.36 | 50.31 | 185.37 | 256.16 | 280.24 | 199.22 | 133.61 | 132.97 | 209.00 |
| NB | 173.12 | 29.19 | 44.66 | 182.4 | 254.48 | 282.13 | 210.93 | 133.49 | 147.63 | 201.87 |
| SVM | 191.0 | 31.05 | 50.31 | 192.34 | 264.6 | 291.9 | 216.22 | 137.22 | 156.27 | 218.21 |
| MLP | 182.65 | 25.48 | 43.1 | 185.91 | 258.53 | 280.4 | 199.61 | 133.59 | 132.66 | 212.15 |
| DT | 193.09 | 29.07 | 51.15 | 193.26 | 268.13 | 293.11 | 217.32 | 135.70 | 149.81 | 220.55 |
| KN | 192.90 | 29.92 | 52.43 | 191.4 | 265.88 | 289.31 | 211.28 | 137.35 | 146.64 | 219.80 |
| RF | 193.36 | 29.42 | 52.47 | 192.82 | 270.33 | 294.63 | 219.22 | 133.73 | 151.34 | 221.07 |
| Size 3 | Size 4 | Size 5 | Size 6 | |
|---|---|---|---|---|
| HP | 6.981233 × 10−8 | 9.286800 × 10−8 | 1.877473 × 10−6 | 0.0001678653 |
| HF | 9.261686 × 10−7 | 2.554832 × 10−8 | 2.544815 × 10−7 | 3.296192 × 10−7 |
| HD | 0.0769675283 | 0.0325569482 | 0.0325569482 | 0.1000616722 |
| CKD | 0.3713981664 | 0.4664793246 | 0.2683203192 | 0.1442754291 |
| HP-UCI | HF-UCI | HD-UCI |
|---|---|---|
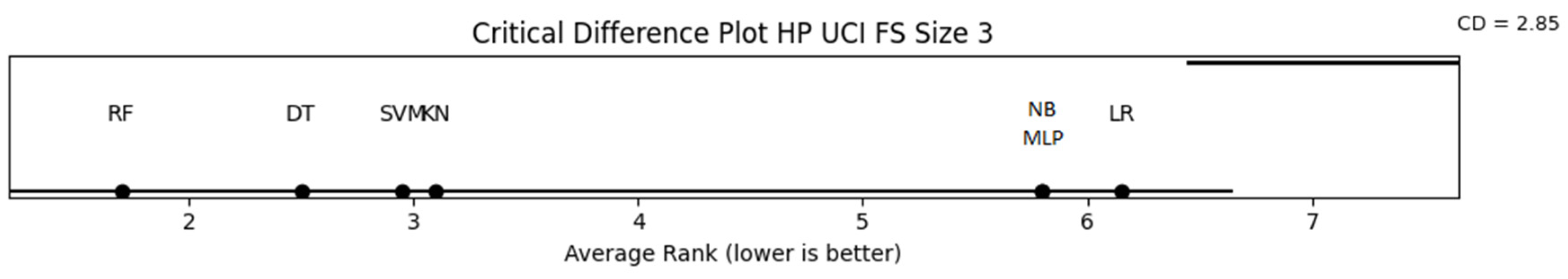
| RF, DT, SVM, KN | DT, NB, LR, RF | RF, KN, SVM, NB, DT |
| HP | |||||||
| size 3 | 170.5 (KN) | 170.0 (RF) | 169.5 (DT) | 167.7 (SVM) | 157.6 (LR) | 155.7 (NB) | 155.3 (MLP) |
| size 4 | 759.0 (KN) | 757.3 (DT) | 755.9 (SVM) | 755.1 (RF) | 719.6 (MLP) | 713.6 (LR) | 700.0 (NB) |
| size 5 | 2011.6 (SVM) | 2003.8 (KN) | 1997.4 (DT) | 1995.8 (RF) | 1943.9 (MLP) | 1912.5 (LR) | 1864.4 (NB) |
| size 6 | 3511.9 (SVM) | 3477.1 (RF) | 3477.1 (KN) | 3474.2 (DT) | 3402.4 (MLP) | 3365.4 (LR) | 3268.4 (NB) |
| HF | |||||||
| size 3 | 268.9 (DT) | 264.2 (RF) | 244.5 (KN) | 227.0 (NB) | 218.8 (SVM) | 211.9 (MLP) | 203.5 (LR) |
| size 4 | 1208.5 (DT) | 1190.6 (RF) | 1072.8 (KN) | 1022.9 (NB) | 1008.2 (SVM) | 968.1 (MLP) | 937.9 (LR) |
| size 5 | 3179.7 (DT) | 3115.8 (RF) | 2776.2 (KN) | 2744.1 (SVM) | 2706.2 (NB) | 2600.5 (MLP) | 2553.0 (LR) |
| size 6 | 5481.8 (DT) | 5391.9 (RF) | 4872.2 (SVM) | 4826.8 (KN) | 4712.2 (NB) | 4587.9 (MLP) | 4547.1 (LR) |
| HD | |||||||
| size 3 | 229.3 (KN) | 226.1 (RF) | 225.3 (MLP) | 224.3 (LR) | 223.9 (DT) | 223.6 (NB) | 223.2 (SVM) |
| size 4 | 1051.3 (KN) | 1048.2 (RF) | 1043.9 (MLP) | 1042.0 (SVM) | 1041.5 (LR) | 1035.2 (NB) | 1031.5 (DT) |
| size 5 | 2849.9 (MLP) | 2847.7 (RF) | 2840.4 (LR) | 2839.7 (KN) | 2837.7 (SVM) | 2830.7 (NB) | 2792.5 (DT) |
| size 6 | 5069.7 (MLP) | 5054.8 (RF) | 5051.1 (NB) | 5043.6 (KN) | 5041.6 (SVM) | 5036.4 (LR) | 4958.1 (DT) |
| HP-UCI | ||||
|---|---|---|---|---|
| FS size 3 | (5, 6, 7) KN | (4, 5, 6) RF | (4, 5, 6) DT | (5, 6, 9) SVM |
| FS size 4 | (4, 5, 6, 12) KN | (4, 5, 6, 10) DT | (5, 6, 9, 12) SVM | (4, 5, 6, 10) RF |
| FS size 5 | (4, 6, 9, 11, 12) SVM | (4, 5, 6, 10, 12) KN | (4, 5, 6, 7, 11) DT | (5, 6, 7, 10, 11) RF |
| FS size 6 | (3, 4, 5, 6, 11, 12) SVM | (4, 5, 6, 7, 10, 11) RF | (4, 5, 6, 9, 11, 12) KN | (3, 4, 5, 6, 10, 11) DT |
| HF-UCI | ||||
| FS size 3 | (5, 8, 11) DT | (2, 3, 8) RF | (1, 5, 12) NB | |
| FS size 4 | (4, 5, 8, 11) DT | (3, 8, 9, 10) RF | (2, 3, 5, 12) NB | |
| FS size 5 | (3, 4, 8, 9, 10) DT | (3, 4, 8, 9, 10) RF | ||
| FS size 6 | (1, 3, 6, 8, 9, 11) DT | (3, 5, 6, 7, 8, 10) RF | ||
| HD-UCI | ||||
| FS size 3 | (3, 10, 11) KN | (3, 10, 11) RF | ||
| FS size 4 | (3, 4, 10, 11) KN | (3, 8, 10, 11) RF | (3, 8, 10, 11) SVM | |
| FS size 5 | (2, 3, 9, 11, 12) MLP | (2, 5, 8, 10, 11) RF | (3, 4, 5, 10, 11) KN | |
| FS size 6 | (1, 3, 8, 9, 10, 11) MLP | (1, 2, 3, 8, 10, 11) RF | (2, 3, 7, 8, 9, 11) NB | (3, 4, 8, 9, 10, 11) KN |
| HP-UCI | HF | HD | |||
|---|---|---|---|---|---|
| 4, 5, 6 | ALP, ALT, AST | 5, 8, 11 | EJF, SCR | 3, 10, 11 | CP, SLO, CA |
| 4, 5, 6, 10 | CRE | 3, 8, 9, 10 | CPH, SSO, SEX | 3, 8, 10, 11 | ANG |
| 4, 5, 6, 10, 12 | PRO | 3, 4, 8, 9, 10 | DIA | 3, 4, 5, 10, 11 | BPS, FBS |
| 4, 5, 6, 10, 11, 12 | GGT | 3, 4, 5, 8, 9, 10 |
| HP-UCI | HF-UCI | ||
|---|---|---|---|
| Size 3 | RF, DT, SVM, KN, NB, MLP, LR | Size 3 | DT, NB, LR, RF, MLP, KN, SVM |
| Size 4 | RF, DT, SVM, KN, MLP, NB, LR | Size 4 | NB, DT, LR, RF, KN, MLP, SVM |
| Size 5 | RF, SVM, DT, KN, MLP, NB, LR | Size 5 | NB, DT, LR, RF, MLP, KN, SVM |
| Size 6 | RF, SVM, KN, DT, MLP, LR, NB | Size 6 | NB, DT, LR, RF, MLP, KN, SVM |
| HD-UCI | CKD-UCI | ||
| Size 3 | RF, KN, SVM, NB, DT, MLP, LR | Size 3 | N/A |
| Size 4 | RF, DT, KN, NB, SVM, MLP, LR | Size 4 | N/A |
| Size 5 | RF, DT, NB, SVM, KN, MLP, LR | Size 5 | N/A |
| Size 6 | RF, SVM, NB, DT, KN, MLP, LR | Size 6 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-de-Madariaga, R.; Pascual Carrasco, M.; Muñoz Carrero, A. A Methodology to Extract Knowledge from Datasets Using ML. Mathematics 2025, 13, 1807. https://doi.org/10.3390/math13111807
Sánchez-de-Madariaga R, Pascual Carrasco M, Muñoz Carrero A. A Methodology to Extract Knowledge from Datasets Using ML. Mathematics. 2025; 13(11):1807. https://doi.org/10.3390/math13111807
Chicago/Turabian StyleSánchez-de-Madariaga, Ricardo, Mario Pascual Carrasco, and Adolfo Muñoz Carrero. 2025. "A Methodology to Extract Knowledge from Datasets Using ML" Mathematics 13, no. 11: 1807. https://doi.org/10.3390/math13111807
APA StyleSánchez-de-Madariaga, R., Pascual Carrasco, M., & Muñoz Carrero, A. (2025). A Methodology to Extract Knowledge from Datasets Using ML. Mathematics, 13(11), 1807. https://doi.org/10.3390/math13111807







