1. Introduction
Visual computing in medicine has been a topic of interest for researchers over the years. Visual computing in medicine involves handling the generation, processing, analysis, exploration, and interpretation of medical visual information, like MRI (magnetic resonance imaging) scans, X-rays, or even 3D (three-dimensional) models of organs [
1]. The ultimate purpose of visual computing in medicine is to monitor, explore, and interpret the target being visualized. Any process involving visual computing, including the acquisition, processing, analysis, interpretation, and visualization of medical visual information, can be formulated abstractly by an input–output system.
Traditionally, the related input–output relationship is achieved by constructing mathematical models on low-level visual representation space and iteratively solving optimization problems. These methods are supported by strong mathematical theories, but also have obvious limitations.
Machine learning, especially its subset “deep learning and neural networks”, is one of the largest diamonds ever discovered in the evolution of science, and has many facets, one of which is machine-learning-based visual computing—the central theme of this paper. It is the process of enabling computers to automatically learn from data and obtain certain knowledge, patterns, or input–output relationships. In visual computing in medicine, the tasks of acquisition, processing, analysis, and interpretation of visual information could be transformed into the tasks of modern machine learning. Nowadays, related methods of visual computing in medicine are based on knowledge, goals, and artificial intelligence, which are widely applicable and accomplished by solving a machine learning problem with the help of higher-level feature information. Reference [
2] highlighted the importance of information visualization in machine learning and its applications, setting the stage for further exploration in this area. Mathematically speaking, the foundation of learning-based visual computing is a rapidly growing field called machine learning theory. Therefore, the final visual exploration and interpretation is important for visual computing and is intrinsically connected to learning theory and vision-based artificial intelligence. A combination of acquisition, processing, analysis, visualization, and interpretation of medical visual information is often necessary for the task of artificial visual computing.
In recent years, the research community has witnessed an increased number of significant and novel methods and applications in mathematical modeling and machine learning for visual computing in medicine. However, there are few reviews that comprehensively introduce and survey the systematic implementation of machine-learning-based vision computing in medicine, and in relevant reviews, little attention has been paid to how to use machine learning methods to transform medical visual computing tasks (such as medical image reconstruction, analysis, and visualization) into data-driven learning problems with high-level feature representation, while exploring their effectiveness in key medical applications, such as image-guided surgery. This review paper addresses the above question and surveys fully and systematically the recent advancements, challenges, and future directions of machine-learning-based medical visual computing with high-level features. The remainder of this paper is organized as follows.
Section 2 introduces briefly the fundamentals and paradigm of visual computing in medicine. The functionality and interrelationships of visual computing in medicine are described in detail in this section, as well as its purpose.
Section 3 discusses the acquisition of medical visual information and medical image data.
Section 4 investigates the processing and analysis of medical visual information. Image segmentation, registration, and fusion of medical visual information are the focuses of this section.
Section 5 delves into methods for the exploration and interpretation of visual information.
Section 6 focuses on the exploration of visual information with visualization and augmented reality.
Section 7 is devoted to the overall application of visual computing, image-guided surgery.
Section 8 further summarizes the main challenges and future directions for visual computing in medicine. Finally,
Section 9 summarizes the conclusion with major research findings and future recommendations.
2. Paradigm for Visual Computing in Medicine
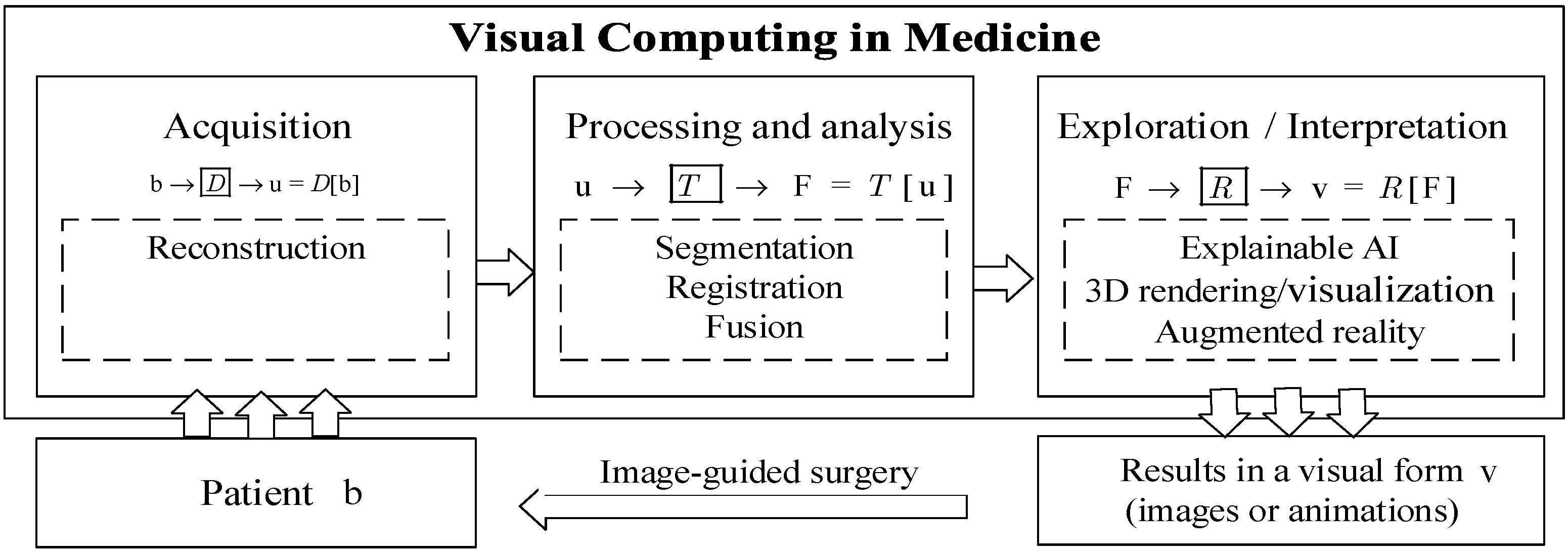
Figure 1 presents the data processing flowchart of visual computing in medicine.
Figure 1 also shows the systematic implementation process of overall medical visual computing based on machine learning. As shown in
Figure 1, it is often carried out as a pipeline of several steps: acquisition of visual data about patients, processing and analysis of visual information, and the exploration and interpretation of visual information [
1]. In the process of “acquisition of visual information”, medical image data
are acquired for tasks in medicine, such as diagnosis, therapy planning, intraoperative navigation, and post-operative monitoring. Tomographic image reconstruction methods are important components of it [
3]. The process of “acquisition of visual data” is followed by the process of “processing and analysis of visual information”. In the module of “processing and analysis of visual information” [
4], image segmentation, including the identification and delineation of relevant structures and regions, is the focus of the process, since visualization and many interaction techniques benefit from image segmentation. Registration and fusion are needed to integrate image data from different modalities or different points in time, e.g., pre- and intraoperative images. Image registration is the process of aligning image data from different sources or at different times so that their corresponding features are related. Image registration is an instance of an inverse problem where the transformation parameters are derived from the images. Finally, visual information can be explored and interpreted in the process of “exploration and interpretation of visual information”, by using representation learning, visualization [
5], virtual and augmented reality, visual ExAI (explainable artificial intelligence based on visualization) [
6], and image-guided application techniques [
1].
Let
denote the data acquired by the scan,
denote an image function,
denote the targeted features or patterns,
denote the visual results (images or animations),
the process of reconstruction,
the process of processing and analysis, and
the process of exploration and interpretation. Then, a typical process of solving medical visual computing problems can be expressed by the flowchart:
where these processes, either linear or nonlinear, can be formulated by an input–output system. The process of acquisition can be:
. The process of “processing and analysis” can be:
. The process of “visualization, exploration, and interpretation” can be:
.
Suppose the input is information
, output is feature
/class
/conditional probability
, and
is the model/input–output relationship (
), machine learning is the process and main tool to obtain the input–output relationship
by learning from data. The tasks involving visual computing in medicine often could be transformed into tasks of machine learning. That is, the input–output relationship and problem regarding
(reconstruction),
(segmentation, registration or fusion), or
(exploration and interpretation) can be learned and solved by using machine learning. So, as shown in
Figure 1, medical visual computing tasks can be transformed into data-driven learning problems with high-level feature representation by using machine learning methods.
3. Acquisition of Medical Visual Information
In the acquisition of medical visual information and tomographic reconstruction algorithms constitute the research hotspot.
3.1. Prior Information Guided Reconstruction
Tomography is nothing other than image reconstruction from measurement data indirectly related to hidden structures, so here, medical image reconstruction is a typical ill-posed inverse problem [
3]. As described in
Section 2, the process of image reconstruction can be:
;
In a simple case, data
acquired by the scan are related to an underlying image
through a mapping
. This mapping, as shown in Equation (1), is called the forward process:
Then, the reconstruction (inverse problem) is to calculate the image
from data
, given the imaging system matrix
. According to the Bayesian rule, the reconstruction is equivalent to an unconstrained optimization problem for a specific objective function (energy function). So, solving inverse problems in the Bayesian inference framework, the prior information-guided image reconstruction problem is equivalent to minimizing the objective function
via optimization, as shown in Equation (2).
where
can be presented as Equation (3),
is an image to be reconstructed, and
is the observed data of
.
For a prior information-guided image reconstruction problem, the image reconstruction task can be transformed into a machine learning task. The optimization objective function , as shown in Equation (3), can be taken as the objective function of machine-learning-based image reconstruction. In Equation (3), is the data fidelity term to measure the data fitting error corresponding to the likelihood function. According to different data models, the first term can be specialized into , , and , which are statistically well-suited to additive Gaussian noise, impulsive noise, and Poisson noise, respectively. The second term is the regularization term, which is the prior. Just like the first term, the second term can be also specialized according to the statistical model for . is the regularization parameter that balances these two terms. For tomographic imaging in particular, the first term is the logarithmic likelihood term, which mainly describes the statistical distribution of the original measured data.
A more general objective function
for regularization-based image reconstruction can be expressed as
where
is a forward operator.
is a suitable transform, which can be seen as the data fidelity. The term
is the regularization term, which is the prior information item, and is on some prior distribution of the images to be reconstructed. From the perspective of machine learning, the regularization consists of adding an extra term to the objective function. Regularization is any modification made to a learning algorithm that is intended to reduce its generalization error, but not its training error.
The composite optimization problem, shown as Equation (4), can be solved by using some classical optimization algorithms, such as ISTA (iterative soft-thresholding algorithm) and ADMM (alternating direction method of multipliers).
3.2. Traditional Reconstruction Methods
In the medical imaging process, image quality may be degraded by various factors, which may cause a reconstructed image to deteriorate with severe noise and artifacts. Over the past few decades, iterative reconstruction methods have been widely studied for noise and artifact reduction. With different priors, various regularization terms were developed. The sparse regularizer total variation (TV) [
7] in prior information-guided reconstruction is a powerful sparse regularizer [
8], as shown in Equation (5). Furthermore, the idea of nonlocal TV (NLTV) was adopted in several models for various imaging tasks [
9].
Beyond TV, many other regularization methods have been proposed to improve iterative reconstruction results. Another popular sparse representation model is dictionary learning [
10], which has been demonstrated to be powerful in signal processing.
3.3. Deep-Learning-Based Reconstruction Methods
Recently, deep-learning-based image reconstruction methods have shown significant advancements.
Deep reconstruction methods denote the methods with data as an input and the reconstructed image as an output without an explicit pseudo-inversion operation. Deep learning was first introduced into iterative reconstruction in Ref. [
11] to obtain a mapping function
as a regularization term for MRI reconstruction, as shown in Equation (6).
where
indicates the Fourier encoding matrix normalized as
(where
denotes the identity matrix) and
denotes the Hermitian transpose operation.
is an image-to-image mapping function using the super-resolution convolutional neural network with the parameter vector . The network is trained in the image domain, whose input and label are the zero-filled and ground truth images, respectively.
Following this idea, as shown in Equation (6), based on some traditional iterative optimization methods (fast sampling and image reconstruction, ISTA, total variation regularization, ADMM, and primal-dual), there are many iterative reconstruction methods combined with deep learning, such as “learned primal–dual reconstruction, direct reconstruction via deep learning, ISTA-Net, and ADMM-Net”, variational reconstruction network, leveraging generic network structures, cascaded CNNs, and GAN (generative adversarial network)-based reconstruction networks”. Reference [
12] introduced ADMM-CSNet (alternating direction method of multipliers–compressive sensing network), combining traditional model-based compressive sensing methods and data-driven deep learning for image reconstruction.
Deep-learning-based medical image reconstruction is still an active research field. An overview of recent results in this field can be found in [
3].
3.4. Specific Practical Application Examples with Machine-Learning-Based Medical Image Reconstruction
Recently, the application of machine-learning-based medical image reconstruction has garnered significant attention, and various studies have demonstrated practical methods and frameworks to enhance imaging quality and diagnostic capabilities. Some specific practical application examples with machine-learning-based medical image reconstruction are as follows.
Reference [
13] provided an overview of machine learning approaches for parallel MRI reconstruction, including image-domain- and k-space-based techniques. Reference [
14] concluded that modularized deep neural network-based low-dose CT (computerized tomography) image reconstruction methods have strong competitive performance by comparing them with several traditional commercial methods. Reference [
15] explored the application of neural architecture search to MRI reconstruction. Their proposed network demonstrated an effective balance between computational efficiency and reconstruction quality in imaging applications. Reference [
16] focused on the ability of generative adversarial networks (GANs) to learn canonical medical image statistics, which can enhance medical imaging workflows and be leveraged for reconstruction tasks. Reference [
17] introduced a self-supervised deep learning network to enhance the quality of grating-based phase-contrast CT images. Their work illustrated how machine learning can effectively reduce noise in medical imaging, thereby improving diagnostic outcomes. Reference [
18] presented another avenue for applying machine learning in medical imaging. Their information theory analysis of multimodal images highlights the potential of machine-learning-based translation techniques to address various medical imaging challenges, further expanding the toolkit for image reconstruction.
In summary, the literature reveals a rich landscape of applications in machine-learning-based medical image reconstruction, ranging from prior statistical modeling and GANs to deep-learning-based multimodal translation. More and more advanced applications of machine-learning-based image reconstruction have entered the stage of routine clinical use.
4. Processing and Analysis of Visual Information
As described in
Section 2, the process of “processing and analysis of visual information” (e.g., segmentation, registration, or fusion) formulated by an input–output system [
4] is shown as Equation (7).
The input is an acquired image or visual information, which may degrade due to some problems. Even can contain more than one image, such as multimodal images, video processing, or follow-up monitoring of individual patients. Mathematically speaking, is the operator (any linear or nonlinear). The desired features/pattern/results can be produced by on the inputs.
Most problems in image processing and analysis are to recover from , thus are inverse problems. How to compute is usually an ill-posed inverse problem. In order to effectively solve the ill-posed problem, is often solved in a specific task-driven way. If the problem is transformed into a machine learning problem, regularization methods will be applied to reduce generalization errors.
4.1. Segmentation of Medical Visual Information
Medical image segmentation plays a crucial role in the analysis of visual information, enabling efficient disease diagnosis and treatment, and it is one of the most challenging problems in medical visual computing [
1]. The field of medical image segmentation continues to evolve with the development of novel segmentation methods, frameworks, and models aimed at improving segmentation accuracy and efficiency in various applications of medical visual computing.
For general image segmentation,
specifically denotes the process of “segmentation” here. It finds a visually meaningful partitioning of the domain from a domain [
4]. From the perspective of machine learning, image segmentation crucially bridges low-level and high-level computer vision. This means that image segmentation is processed in low-level representation space, and may further be processed in high-level representation space. In high-level computer vision, image segmentation can be taken as a classification task [
19,
20,
21,
22].
4.1.1. Traditional Methods and Machine-Learning-Based Methods
Traditional segmentation methods are often processed in low-level representation space. In the early stage, the threshold-based method is applied to medical segmentation, which divides pixels into foreground or background based on the intensity threshold. Such a method is ineffective when segmenting complex structures. Additionally, region-based methods were developed for more accurate segmentation results. Region-based methods iteratively combine pixels into one region under special rules. In addition, edge detection methods are used to solve the segmentation problem. However, the above methods cannot be directly used in application due to the weak segmentation ability for variable structures in medical images.
With the development of machine learning, feature-based and learning-based methods have become popular, showing better segmentation performance. As supervised segmentation methods, multi-atlas segmentation frameworks have been proposed for abdominal organ segmentation, utilizing spatial and appearance information from atlases to create benchmarks for large-scale automatic segmentation [
23]. Recent advancements in medical image segmentation include the proposal of structure boundary-preserving segmentation frameworks to address ambiguity and uncertainty in medical image segmentation [
24]. Uncertainty-aware multi-view co-training has been introduced for semi-supervised medical image segmentation [
25].
4.1.2. Deep-Learning-Based Segmentation Methods
With the great success of deep learning networks (convolutional neural networks and transformers), more and more deep-learning-based methods have been proposed to solve the problem of medical image segmentation in high-level representation space, which gained better accuracy than traditional methods. Based on different network structures, these methods can be grouped into three categories: CNN (convolutional neural network)-based [
20], transformer-based [
21], and SAM (segment anything model)-based [
22].
CNN-Based Segmentation Methods
Inspired by the powerful feature extraction ability of CNNs in image classification tasks, Ref. [
19] constructed a FCN (fully convolutional network) for image segmentation, which allows any size of input image. Based on FCN, Ref. [
20] added skip connections between the encoder and decoder to provide segmentation output with low-level details. Due to its simple, but efficient, network structure, U-shape design has gradually dominated the field of medical segmentation. Lots of works have been proposed to improve segmentation accuracy from the original U-net [
20]. There are four main ideas to improve U-net, encoder, decoder, bottleneck, and skip connection.
The encoder is used to extract high-level features and increase the receptive field. Some researchers tried to add new modules into the encoder to gain better feature representations. Inspired by the power of dense connections, Ref. [
26] constructed a hybrid dense connection U-Net for tumor segmentation.
The decoder stacks convolutional and up-sampling blocks to transform high-level features into low-level representation and restore the original image size. In order to gain better boundary segmentation performance, Ref. [
27] constructed an additional boundary prediction decoder. Additionally, Ref. [
28] proposed the multi-scale output consistency in the decoder to provide better semi-supervised signals.
The bottleneck contains the deepest features, which have a big impact on the final segmentation performance. Recently, the attention mechanism became popular due to its feature refinement ability. Inspired by this, Ref. [
29] added a spatial attention block to guide the bottleneck focus on valuable regions.
The skip connection is proposed to provide positional information for the decoder. Reference [
30] claimed that skip connections built at the same level may be not the best choice. They proposed U-net++, which constructed skip connections at different levels, and the experimental results demonstrated its better performance.
Segment Anything Model-Based Methods
Generalist vision foundation models, such as the SAM, have been applied to various medical image segmentation benchmarks, showing inconsistent zero-shot segmentation performance across different medical domains.
The differences between SAM-based and transformer-based segmentation in computer vision are mainly reflected in model architecture. SAM is a universal segmentation model based on Transformer, aimed at image segmentation through any hint (such as points, boxes, text descriptions, etc.). SAM learns from large-scale datasets through pre-training and can perform well in various segmentation tasks, especially in resource-limited environments, where it can still provide efficient and accurate segmentation results.
SAM, as a fundamental model, can perform well in various segmentation tasks by learning from large-scale datasets through pre-training. SAM [
22] has a strong zero-shot learning ability and performs well in natural images. However, medical images have different feature distributions across different modalities; thus, simply using SAM cannot gain satisfactory results in medical segmentation. Since many medical images are actually 3D data, one obvious idea is to make SAM learn inter-slice context in a 3D sequence [
32].
To better use SAM in different modalities, it is necessary to finetune SAM. Because full tuning consumes large amounts of computational resources, efficient tuning methods (projection tuning and adapter tuning) have become popular. Projection tuning has been proposed to use a new, task-specific projection head to replace the original decoder. Additionally, adapter tuning methods usually add an adapter module to the encoder, which allows for adjusting the encoder only by training a few parameters [
33].
Overall, the integration of machine learning, particularly deep learning methods, has significantly advanced medical image segmentation, offering more accurate and efficient solutions for various medical imaging tasks.
4.1.3. Specific Practical Application Examples with Machine-Learning-Based Medical Image Segmentation
The application of machine learning in medical image segmentation has seen significant advancements. Some specific practical application examples with machine-learning-based medical image segmentation are as follows.
Reference [
34] presented an active learning strategy for semi-supervised 3D image segmentation, which is specifically applied to COVID-19 pneumonia in CT scanning. Their method leveraged a query function to select informative samples from noisy teacher-generated data, demonstrating the adaptability of deep learning in addressing urgent medical challenges.
Reference [
35] reported the automatic classification of lung cancer subtypes using deep learning and radiomics analysis based on CT scans. Their study highlighted the reliability of automated diagnostic tools in radiomics, with a low failure rate in tumor lesion segmentation, further validating the effectiveness of deep learning in oncological imaging.
In summary, these studies collectively illustrate the diverse segmentation methods and practical implementations that are shaping the future of medical applications.
4.2. Registration of Medical Visual Information
As stated in
Section 2, image registration is the process of aligning image data from different sources or at different times so that their corresponding features are related. Medical image registration has become a pervasive tool in the diagnosis of clinical conditions and the guidance of therapeutic interventions (e.g., image-guided surgery). Data may come from different sensors, times, depths, or viewpoints. Medical image registration, including unimodal registration and multimodal registration, represents a fundamental and pivotal technology within the domain of medical image processing and analysis. Prior to the processing and analysis of medical images with the aim of obtaining comprehensive information, the initial challenge to be addressed is the precise registration of the images in question. The registration of images is a fundamental prerequisite for a number of subsequent processes, including image fusion, image segmentation, image comparison, three-dimensional reconstruction, and the identification and analysis of changes in targets. Consequently, in-depth research on the technology of medical image registration has significant theoretical implications and clinical application value.
4.2.1. Mathematical Form of Medical Image Registration
Image registration can be formulated as shown in Equation (8), which is the process of learning the optimal spatial transformation parameters
that maximize the similarity measure under a given optimization strategy.
where
denotes the fixed image and
denotes the moving image. The spatial transformation
of the moving image
is performed continuously until the transformed moving image exhibits the greatest possible similarity to the fixed image
, and
is the optimal result of spatial transformation.
The core technology of the medical image registration algorithm encompasses four fundamental components: similarity assessment, spatial transformation, interpolation [
36], and optimization algorithms.
Similarity Assessment
Similarity measures can be classified into gray-scale-based measures or feature-based measures. In general, similarity measures used for non-rigid image registration comprise two components: voxel grayscale/structure-related and deformation field-related [
37]. Consequently, the cost function represents a compromise between the grayscale/structure and the constraints imposed on the deformation field. These constraints are frequently referred to as penalties or regular terms.
The most commonly utilized gray-scale-based similarity measures can be classified into three categories: gray-scale difference, inter-correlation, and information theory-based similarity measures. On the other hand, feature-based measurement methods [
38] necessitate the initial extraction of salient features from both images. Subsequently, the spatial topological relationship between these features and the grayscale of the feature region must be combined to establish a correspondence. In general, all of the aforementioned grayscale-based similarity measures can be utilized as feature-based similarity measures. The feature-based algorithm is immune to the effects of image grayscale attributes and noise, rendering it more robust.
Optimization Algorithms
Image registration can be regarded as a multi-parameter optimization problem, the selection of an appropriate optimization algorithm being of great consequence in view of the spatial transformation model.
In accordance with the parameters delineated within the domain of solution definition, multi-parameter optimization algorithms can be classified into local and global optimization algorithms.
Local optimization algorithms require specific information to ascertain a search direction. The most prevalent form of this information is the gradient value of the cost function. Examples of algorithms that utilize this information include steepest gradient descent [
39], conjugate gradient [
40], Powell’s algorithm [
41], and so on.
Global optimization algorithms are characterized by robust search capabilities and a robust theoretical foundation. They are particularly well-suited to registration models with a large number of transformation parameters to be solved. The most commonly used algorithms are the simulated annealing algorithm [
42], the particle swarm optimization algorithm [
43], and so on. However, these algorithms are computationally intensive, exhibit slow convergence, and are highly susceptible to misregistration due to factors such as the initial search point, search direction, and search strategy.
It is worth noting that optimization algorithms are merely a means of implementation for medical image registration algorithms. Consequently, the majority of medical image registration models utilize well-established optimization algorithms. To enhance search efficiency, some researchers have proposed a combination of local and global algorithms. The hybrid optimization algorithm is an effective means of accelerating convergence and improving alignment accuracy while maintaining reasonable computational efficiency.
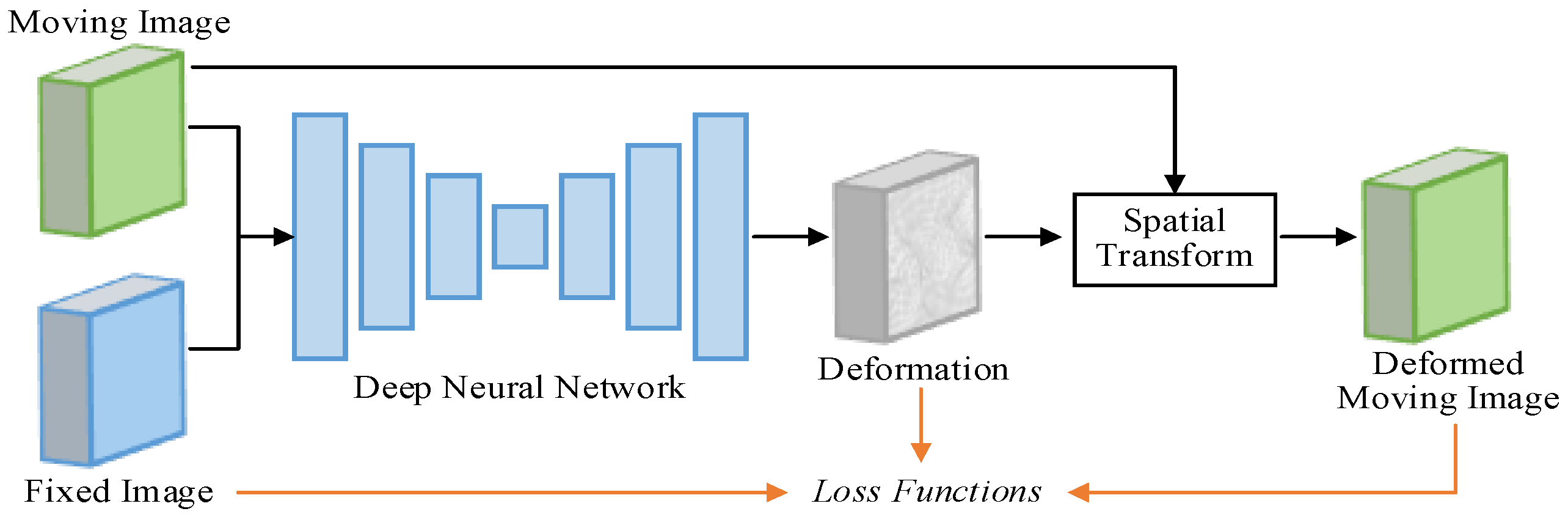
4.2.2. Deep-Learning-Based Registration
Loss Function
The loss function
is utilized as the objective function of the deep-learning-based registration, which can typically be expressed as Equation (9).
where
denotes the dissimilarity penalty term, used to penalize the dissimilarity between the fixed image
and warped moving image
; and
denotes the regularization term, used to encourage a realistic deformation field
.
The dissimilarity penalty loss function can guide the training of registration networks. It is worth noting here that several regions with ‘folds’ (here, the Jacobian determinant is negative) may occur in the estimated deformation fields. The deformation field is usually hoped to be diffeomorphic, that is, smooth and invertible. So, L2-norm, the dominant general-purpose regularization term, can be used in Equation (9).
Further, some extra anatomy loss, such as overlap of anatomical label maps, dice loss, cross-entropy, and focal loss, can be introduced in learning-based registration methods. And a more effective method is the registration network, introducing auxiliary anatomical information through multi-task learning [
54,
55].
Deep-learning-based medical image registration is still an active research field. An overview of recent results in the learning-based registration field can be found in [
51].
4.2.3. Specific Practical Application Examples with Machine Learn-Based Medical Image Registration
The application of machine learning in medical image registration has garnered significant attention in recent years, with various studies demonstrating its practical utility across different imaging modalities in different times. Some specific practical application examples with machine-learning-based medical image registration are as follows.
The development of tools such as “DeepInfer” by Ref. [
56] illustrated the practical deployment of deep learning models in established medical image analysis platforms. This toolkit aimed to bridge the gap between advanced machine learning techniques and their clinical applications, facilitating the integration of state-of-the-art models into routine medical workflows, including image registration tasks.
The exploration of self-similarity in medical images in Ref. [
57] had also contributed to this field, proposing a new image resolution enhancement method that could indirectly support the registration process by improving the quality of input images. This method emphasized the potential of utilizing machine learning to enhance image features, thereby promoting more accurate registration results.
Reference [
58] addressed the limitations of linear registration methods by training deep neural networks on virtual images generated from label maps. Their method allowed for anatomical structure-specific registration without the need for prior segmentation, thereby simplifying the registration process and improving accuracy.
In summary, recent studies reflect a dynamic and rapidly developing field, with significant contributions from both traditional and deep-learning-based approaches to medical image registration [
59]. These advancements not only improve the accuracy and efficiency of registration processes, but also expand the applicability of these techniques in various medical imaging modalities in different times.
4.3. Fusion of Medical Visual Information
Fusion is essential to analyze different image data in a common frame. In many medical scenarios, it is crucial to combine information related to the same patient extracted from different sources at different times to draw conclusions. This section explains what is technically feasible, along with the clinical motivation and use for these developments.
There are three main fusion strategies: pixel-level fusion, feature-level fusion, and decision-level fusion. As pixel-level fusion methods have been introduced in numerous studies on visual computing, this section will focus on feature-level fusion and decision-level fusion.
4.3.1. Feature-Level Fusion
In this paper, methods of medical visual computing are established by solving a machine learning problem with the help of high-level feature information. Feature-level fusion plays a crucial role in improving the performance of sensor-based systems by integrating relevant features from multiple sources, so it not only improves the accuracy of the obtained information, but also distinguishes itself from data and sensor fusion by focusing on extracting and combining meaningful high-level features, rather than just raw data. Therefore, the significance of feature-level fusion in medical visual computing lies in its ability to improve the accuracy and reliability of the information obtained from different sources. Feature-level fusion is particularly advantageous when dealing with heterogeneous medical information from various sensors, as it allows for the extraction of relevant important high-level features that can be effectively combined to form a unified representation. This is in contrast to data fusion, which may involve combining raw data or signals without necessarily extracting features first. Moreover, feature-level fusion can facilitate the integration of information from different sensing modalities, which is essential for medical applications requiring comprehensive situational awareness.
Feature-level fusion typically involves extracting features from multiple input modalities and fusing them into
(e.g., a single feature vector). In recent years, feature-level fusion strategies have received widespread attention, with methods based on the transform domain being particularly popular due to their multi-scale and multi-frequency information processing capabilities, often demonstrating good robustness. Consequently, they have been widely applied in feature-level fusion schemes for medical multimodal data [
60].
In feature-level fusion methods, an important challenge is to achieve the alignment of features extracted from heterogeneous multi-source information, and then fusion tasks in high-level feature space are implemented based on the completed alignment. Taking into account common issues (such as data imbalance, a small number of samples, and significant heterogeneity gaps) in medical multi-modal data fusion, researchers have introduced learning methods based on attention mechanisms to integrate complementary information from multiple sources. Reference [
61] designed a two-layer dynamic adaptive network for image fusion through a dynamic transfer mechanism to guide medical image fusion tasks. In this way, potential correlations between different modality data can be captured, reducing the impact of data distribution shifts and data feature redundancy on model performance. Reference [
62] proposed an end-to-end data fusion algorithm, which introduced self-normalizing neural network learning for data representation and feature selection, hence addressing the issue of data heterogeneity. Compared with traditional vector computation fusion methods, this model innovatively introduces a cross-attention mechanism, taking into account the gap between heterogeneous modalities. In addition, the way features are fused at intermediate layers also has a critical impact on model performance. Reference [
63] explored a method that uses intermediate fusion to integrate information from multiple modalities, embedding multiple modalities from extracted features and then using a multiplicative fusion approach to obtain combined embeddings.
4.3.2. Decision-Level Fusion
Decision-level fusion involves independent processing of each modality for the input, and ultimately fuses the output decisions (probability distributions, classification results, etc.) of each modality at the prediction result layer through voting or weighting mechanisms. Unlike feature-level fusion, which focuses on the extraction of intermediate features and the processing of raw data, the key to decision-level fusion lies in designing an appropriate fusion strategy to integrate the decision results of different modalities. Based on the type of output decisions of the modalities, decision-level fusion can be broadly divided into: fusion based on discrete outputs, fusion based on probabilistic outputs, and fusion based on scoring/confidence level outputs.
In the probabilistic output fusion strategies, the output of the modality is often the probability distribution obtained from a single modality prediction. Reference [
64] proposed a multimodal multi-level fusion model using Bayesian inference, where the performance indicators were superior to the classic radiomics-guided feature-level fusion model. In order to obtain the optimal weight vector for the weighted fusion of different modalities, Ref. [
65] used the strategy of optimal weighted average fusion to fuse the predictions of the modalities, achieving very good prediction results.
In addition, the fusion method based on scoring is also introduced to solve the problem of weight vector allocation in modality fusion; some researchers have incorporated decision thresholds into the model to determine the final decision [
66]. They fused the prediction outputs of multiple classifiers at the decision level based on likelihood ratios and set a decision threshold, with values greater than this indicating the presence of disease; values less than this indicate otherwise, thus obtaining the decision result.
4.3.3. Specific Practical Application Examples with Machine-Learning-Based Medical Visual Information Fusion
Machine-learning-based medical visual information fusion has attracted significant attention, leading to various practical applications that enhance decision-making, diagnostic, and treatment processes in healthcare. Some specific practical application examples with machine-learning-based medical visual information fusion are as follows.
Reference [
67] developed a universal framework that learns both specific and general feature representations. This method minimizes the need for manual selection in diverse applications, thereby simplifying the fusion process and enhancing its applicability in various medical environments.
Reference [
68] introduced a hybrid-scale contextual fusion network for medical image segmentation. This network captures richer spatial and semantic information, outperforming existing state-of-the-art methods on multiple public datasets, thus demonstrating its effectiveness in clinical diagnostics.
Reference [
69] introduced a hierarchical health decision fusion and support system that fused information from wearable medical sensors into clinical decision support systems. This innovative system comprised six disease diagnosis modules for four disease categories, demonstrating how machine learning can facilitate real-time health monitoring and diagnosis, thereby improving patient outcomes.
In summary, the reviewed studies illustrate a range of practical applications of machine learning in medical visual information fusion, from automating coding processes to enhancing decision support systems.
4.4. Specific Practical Application Examples with Machine-Learning-Based Medical Visual Analysis
The application of machine-learning-based medical visual analysis has seen significant advancements. Some specific practical application examples of machine-learning-based processing and analysis of visual information are as follows.
Reference [
56] developed “DeepInfer”, an open-source toolkit designed for the deployment of deep learning models within the 3D Slicer platform. This toolkit facilitated the application of deep learning in clinical medical image analysis.
Reference [
59] investigated the application of deep-learning-based breast cancer image analysis, covering various imaging modalities and databases. Their findings emphasized the role of deep learning in enhancing image interpretation and clinical results.
Reference [
70] provided a comprehensive overview of the application of deep-learning-based visual analysis in cardiovascular medicine. They emphasized the advantages of deep learning, such as automating image interpretation and enhancing clinical decision-making.
In summary, these studies collectively highlight the effectiveness and efficiency of machine-learning-based processing and analysis of visual information in various routine clinical applications.
5. Visualization, Augmented Reality, and Exploration of Visual Information
An essential trend is the combination of biomedical simulations with advanced visual exploration. As a consequence, processing and analysis results in visual computing can be explored with techniques and augmented reality. That is: .
Data analysis techniques and their integration are also relevant for volume classification, the basic process of assigning transfer functions to medical volume data. That is, the analysis and identification result regarding visual information, such as the classification result or class posterior probability, or even uncertainty, can be visualized and explored through transfer function assignment in visualization.
Visual exploration, visualization, and interpretation are partially the inverse problems of image acquisition. On the other hand, visualization, augmented reality, exploration, and interpretation attempt to reconstruct, explore, visualize, or interpret the 3D world in higher feature space from 2D visual information.
5.1. Visualization and Rendering
There are some basic medical visualization techniques, such as surface rendering, direct volume rendering, and hybrid volume and surface rendering.
Ray–surface intersection [
71], surface shading, and lighting [
72] are the issues of concern among surface rendering. Using the volume rendering integral based on the equations of radiative transfer, volume data are visualized and projected directly to the display. Due to stochastic sample collection at intervals, volume rendering is under-constrained, resulting in noise near the surfaces. Surface rendering only provides gradients on the surfaces of objects, and these gradients cannot propagate smoothly in the spatial domain. Further, the advantages of opaque surface-based renderers and volume renderers are combined by many differentiable renderers. An overview of recent results in the visualization field can be found in Ref. [
5].
The increasing size and complexity of medical image data motivated the development of visualization techniques that radically differ from the classical surface and volume rendering techniques. Conveying the complex information of medical flow data along with the relevant anatomy, for example, benefits from illustrative techniques that render the anatomy sparsely. Thus, illustrative rendering plays a more prominent role in discussing the extraction of various features from medical volume data and related meshes as a basis for rendering.
5.2. Visualization Techniques and Depth Estimation for Medical Augmented Reality
Virtual reality techniques are used to integrate reality and virtuality into an augmented scene. This integrated visualization is the goal of medical AR (augmented reality). In medicine, it is related to the registered and fused visualization of multi-modal data from the same patient, which requires exploring and presenting fusion information without excessive visual clutter.
There are different types of augmented reality systems and methods, such as projection-based augmented reality, augmented microscopes and binoculars, augmented endoscopes, and augmented reality windows. The systems based on GPU (graphics processing unit) are realized in real-time.
In AR, visual augmented representation requires not only appropriate display devices, but also advanced visualization and smart visibility techniques. Now, there are many advanced methods regarding visualization and smart visibility in medical augmented reality. An important problem therein that needs to be addressed is occlusion and proper depth perception.
Some widespread smart visibility techniques, such as focus-and-context visualizations, cutaways, and ghosted views, are used to show deep-seated tumors in the context of the enclosing organ.
Furthermore, vision depth estimation methods based on deep learning have been widely studied in recent years [
73,
74].
5.3. Specific Practical Application Examples with Machine-Learning-Based Visualization and Augmented Reality
Machine-learning-based medical visualization and augmented reality (AR) have made significant progress in visualization technology, enhancing both education and practical applications in the healthcare field. Some specific practical application examples of machine-learning-based medical visualization and AR are as follows.
A significant application of AR in medical education and surgical training is the “Complete Anatomy” application, which utilizes augmented reality to provide interactive and detailed visualization of human anatomy. AR has been applied in breast surgery education, where machine-learning-assisted processing and visualization techniques were expected to improve training results by providing real-time data and interactive learning environments [
75]. Reference [
76] discussed the broader meanings of VR and AR in spatial visualization, emphasizing their potential in medical fields. This foundational work laid the foundation for specific applications in medical environments, where immersive visualization can enhance understanding of anatomical structures and medical conditions.
Reference [
77] presented a mobile AR 3D model registration system designed for practical use in hospital environments. This system exemplified the shift toward patient-specific applications, allowing for real-time visualization of medical data in non-instrument environments. Such innovations were crucial for improving surgical accuracy and patient outcomes, as they enabled surgeons to directly visualize complex anatomical relationships in patients.
Additionally, a workflow for generating patient-specific 3D augmented reality models from medical imaging data was proposed, demonstrating practical applications in surgical planning and education [
78]. This method not only aided in preoperative preparation, but also served as a valuable educational tool for medical students and professionals, enhancing their spatial understanding of anatomy.
Despite these advancements, challenges remain in the effective implementation of AR and VR technologies in medicine. Issues such as depth perception in AR visualizations and the need for seamless integration with existing medical imaging systems are critical areas for further research [
78]. Addressing these challenges will be essential for maximizing the benefits of AR and VR in medical visualization.
In summary, recent studies emphasize the transformative potential of AR and VR in medical visualization, with specific examples demonstrating their applications in improving surgical accuracy, patient education, and comprehensive understanding of complex medical data. Continuous research and development in this field are crucial for overcoming existing challenges and fully realizing the capabilities of these technologies in clinical practice.
6. Interpretation of Medical Information with Visual Computing
6.1. Interpretation of Medical Information in High-Level Vision
As mentioned in the first section, image interpretation is considered as high-level visual computing. An ideal visual computing system in medicine shall be able to perform object differentiation and interpretation. A combination of image analysis and interpretation is often necessary for these tasks. Here, in should be the process of interpretation with visual computing.
Visual information is analyzed and classified by visual computing, followed by labeling, measuring, visualizing, and exploring with visualization and interaction techniques, as has been introduced in the last section. Further, those related to risk structures and targets (e.g., lesions and tumors), as well as their high-level features and representation, can be analyzed and interpreted by machine-learning-based visual computing.
6.2. Interpretation with Visual Computing
In fact, although visual computing based on machine learning/deep learning can achieve high accuracy, its clinical application in medicine is still limited. The most important challenges faced by existing methods are as follows: (1) the accuracy of existing interpretable methods is usually lower than that of black box models, resulting in limited applicability in the medical field; (2) existing methods may typically have insufficient interpretability; and (3) existing deep-learning-based models consist of millions of parameters and a ‘black box’ nature, which lead to complex decision-making mechanisms that lack interpretability. This is unacceptable for high-risk decision-making areas, especially in clinical medicine. Therefore, as deep-learning-based models have rapidly developed, the demand for explainable methods has become increasingly urgent. Such methods are not only required to provide accurate prediction results, but also to provide understandable prediction bases.
Researchers both domestically and internationally have conducted numerous studies on medical image analysis and visual computing based on interpretable artificial intelligence models. The explainable methods developed for medical image analysis and visual computing can be divided into model-based explainable methods and post hoc explanation methods [
79].
Model-based explainable methods are usually simple traditional machine learning models, such as linear regression and support vector machines, which have sparsity by nature. With the help of a limited number of features, these models can offer enough understanding of their inner working mechanisms to model users. In contrast, post hoc explanation methods are used to analyze trained models (such as deep learning models like neural networks) and provide visual explanations through mathematical computation to understand the model’s decision-making process. Such methods mainly include the representation and visual explanation of learned features and the importance of features. The most commonly used explainable methods in the field of medical image analysis include visual explanation and textual explanation [
6]. Visual explanation, also known as saliency mapping, is the most commonly used explanation method in medical image analysis, and it provides decision support by displaying the most significant parts of the image through a saliency map. Textual explanation provides textual descriptions of medical images, similar to a physician’s diagnostic report, such as the features of pulmonary nodules, spiculation, and lobulation. The model outputs the corresponding diagnostic text to achieve interpretability. For example, Ref. [
80] proposed a benign/malignant lung nodule classification method based on an improved SHapley additive exPlanations model. Through quantitative analysis of semantic features annotated by doctors and extracted radiomics features, the importance of each feature to the model’s decision-making is obtained; then, a tree-based ensemble classifier model is used for classification. Further, more researchers are adopting deep neural network-based methods for the study of the interpretable classification of visual information.
6.3. Post Hoc Explanation Methods Based on Deep Neural Network and Class Activation Mapping
The class activation map (CAM) [
81] is a method that visualizes which regions in an image contribute the most to the classification of a specific category through the feature maps of the last convolutional layer in a convolutional neural network. The CAM combines the feature maps of the last convolutional layer with the weights of the classification layer (SoftMax) using global average pooling (GAP), and outputs the class probabilities based on the weighted spatial average map values. Subsequently, the weight matrix of the fully connected layer is projected back onto the last convolutional layer to calculate the weighted feature maps and generate the class activation maps for identifying the importance of image regions.
For a given image, suppose a CNN model obtains
feature maps
, (width
and height
in the last convolutional layer). These feature maps undergo spatial pooling via GAP, and then a linear transformation is applied to them, yielding a score
for each class, as shown in Equation (10).
where
is the weight of the
-th feature map corresponding to class
c in the fully connected layer and
, which is a normalization factor. Define
as the class activation maps, and their spatial elements can be expressed as shown in Equation (11).
Gradient-weighted CAM (Grad-CAM) [
82] is an improved method based on CAM. It generates class activation maps by calculating the gradients of the target with respect to the feature maps. Different from the conventional CAM method, Grad-CAM can provide visual explanations for any model in the CNN family without the need for architectural modifications or retraining. Grad-CAM calculates the gradients of the target score with respect to the feature maps of the last convolutional layer.
Score-CAM [
83] is a gradient-free visual explanation method based on CAM. It generates class activation maps by calculating the contribution scores of each feature map to the target category. Different from Grad-CAM, Score-CAM does not need to calculate gradients. Instead, it assesses the importance of feature maps on the target category by perturbing them, and incorporates the importance as the increase in confidence, thus avoiding gradient saturation, gradient vanishing, and gradient noise that exist in gradient calculations. Specifically, Score-CAM first performs forward propagation on the input image through the CNN to obtain feature maps, upsamples them to the size of the input image, and then uses them as masks to perturb the input image.
LayerCAM [
84] is a class activation mapping method based on inter-layer correlations. Different from previous CAM methods, LayerCAM takes into account not only the feature maps of the last convolutional layer, but also those of the intermediate layers. LayerCAM generates separate weights for each spatial position in the feature maps by obtaining gradients through backpropagation. LayerCAM generates reliable class activation maps from the information of the final convolutional and shallow layers, enabling it to obtain both the rough spatial positions and fine-grained object details, so that more precise and complete class-specific object regions will be generated.
6.4. Specific Practical Application Examples of Machine-Learning-Based Interpretation of Medical Information with Visual Computing
The interpretation of complex medical information is crucial for accurate diagnosis and treatment planning. By enabling machines to understand and interpret medical information with machine-learning-based visual computing, healthcare providers can leverage these insights to enhance patient care and streamline workflows. This research area focuses on visual information integration and model interpretation, which are critical for developing robust systems that can assist in medical diagnostics and treatment strategies. Some specific practical application examples of machine-learning-based interpretation of medical information with visual computing are as follows.
Reference [
85] introduced a multi-modal transformer network designed to generate radiology reports by integrating chest X-ray images with patient demographic data. This method illustrated how visual computing enhanced the synthesis of patient-specific medical information, thereby improving the accuracy and relevance of radiological interpretations.
The role of machine-learning-based interpretation of medical information with visual computing extends beyond mere image analysis; further, it also involves the development of clinical decision support systems that leverage visual data to provide actionable insights to healthcare professionals [
86]. These systems can improve patient outcomes by facilitating timely and informed decisions based on comprehensive visual and contextual data.
The integration of explainable artificial intelligence (XAI) in medical applications has gained significant attention, particularly through the application of class activation mapping techniques, such as gradient-weighted class activation mapping (Grad-CAM). Reference [
87] investigated the role of Grad-CAM heatmaps in assisting medical professionals, especially radiologists, in making diagnoses. Their research findings indicated that radiologists found these heat maps to be beneficial, suggesting that visual interpretation could enhance the diagnostic process by providing insights into machine-learning-based classifications. This aligns with the broader goal of XAI to make AI (artificial intelligence) systems more interpretable and trustworthy in clinical settings.
Reference [
88] developed a deep learning model for classifying chest X-ray images into categories such as COVID-19, pneumonia, and tuberculosis. Their method emphasized the importance of maintaining classification accuracy while employing XAI techniques, thereby ensuring that the model’s predictions are not only accurate, but also interpretable.
In a comparative study, Ref. [
89] introduced a new XAI framework that provided feature interpretability for tumor decision-making in ultrasound data. They found that their framework, referred to as Explainer, outperformed Grad-CAM in identifying relevant features, highlighting the potential for advancements in XAI methodologies that can provide more precise explanations for medical imaging tasks.
Reference [
90] further explored the application of Grad CAM in the diagnosis of Alzheimer’s disease. Their study demonstrated how XAI techniques enhanced the interpretability of deep learning models, enabling clinical doctors to gain insights into the decision-making processes behind diagnoses. This is crucial for fostering trust in AI systems used in healthcare.
Reference [
91] combined Grad-CAM with the ResNet50 model to create a transparent framework for MRI image analysis and brain tumor detection. Their research emphasized the importance of interpretability in high-stakes medical diagnostics, where understanding the rationale behind AI predictions can significantly impact clinical outcomes.
Additionally, Ref. [
92] explored the application of feature injection in convolutional neural networks for melanoma diagnosis, using both qualitative and quantitative methods to explain model outputs. This study complemented the findings of other researchers by emphasizing the need for robust explanation methods that could elucidate the features driving AI predictions.
In summary, the practical applications of machine-learning-based interpretation of medical information through visual computing are extensive and influential, which can improve diagnostic accuracy and clinical decision-making. The advancements in computer vision not only facilitate a better understanding of medical images, but also contribute to improved healthcare outcomes by enabling more accurate and timely diagnoses. The ongoing research and development in this field are expected to further enhance the ability of medical professionals to interpret complex medical visual information.
7. Image-Guided Surgery as Overall Application of Visual Computing in Medicine
7.1. Overall Application of Visual Computing
Image-guided surgery, as one of the most essential and overall applications of visual computing in medicine, is becoming increasingly important, which is incorporating recent visual computing technology.
Image-guided surgery, providing navigation support in surgery, involves anatomical images obtained preoperatively and intraoperative images in the patient’s physical space. The planning results from the preoperative images will be applied intraoperation. Pathology is localized spatially in preoperative images; then, it is further aligned with features of interest in intraoperative images.
Visual computing applications, including visual information acquisition, multi-model data registration, as well as calibration and tracking, are prerequisites for intraoperative applications. These numerous recent advancements in visual computing have been introduced in previous sections “4. Processing and analysis of visual information” and “5. Visualization, augmented reality and exploration of visual information”. Via the augmented reality techniques introduced in
Section 5.2, relevant information from preoperative images is fused and augmented with live images (such as video images, intraoperative ultrasound, and so on) from surgery, and will be virtually represented in image-guided surgery.
There are still many potential challenges and risks of intraoperative guidance, such as the correct perception of virtual information, integrating AR into complex medical workflows, and so on [
93].
7.2. Specific Practical Application Examples with Machine-Learning-Based Image-Guided Surgery
The integration of machine learning and augmented reality (AR) in medicine has led to significant advancements in visualization techniques, enhancing both educational and practical applications in healthcare. Some specific practical application examples with machine-learning-based medical visualization and augmented reality are as follows.
Image-guided procedures have transformed procedural medicine with advanced imaging and technologies [
94]. An example of practical application was the combination of machine learning models with Raman spectroscopy for image guidance during prostate biopsy to detect prostate cancer [
95].
Integrating machine-learning-based visual computing and artificial intelligence (AI) into surgical practice is another growth area. Machine-learning-based visual computing technology is being used to enhance image-guided surgery, with applications ranging from virtual colonoscopy to real-time imaging analysis during the surgical process [
96]. With advances in many medical fields, the potential of machine-learning-based visual computing in improving surgical decision-making and outcomes is increasingly being recognized.
In addition, the American Society of Breast Surgeons has provided guidelines and resources that reflect the continuous development of image-guided surgery, particularly in the context of cancer treatment [
97].
In summary, recent advancements in image-guided surgery include various innovative technologies and methods, including machine-learning-based visual computing and robotic assistance. These developments have the potential to improve surgical precision, enhance patient outcomes, and shape the future of surgical oncology.
8. Challenges, Future Perspectives, and Discussion
In addition to those described in sections above, the main challenges and research gaps, as well as future directions for addressing these issues, are covered as follows.
8.1. Challenges and Future Perspectives Regarding Specific Medical Visual Computing Tasks
8.1.1. Medical Image Reconstruction
There are many challenges in medical image reconstruction. 1. From the perspective of processing objects, challenges include: (1) low dose and noise issues, and (2) motion artifact and dynamic imaging blur. Due to data quality and collection limitations in medical image reconstruction, the typical ill-posed inverse problem-solving is one of the main challenges in visual computing. 2. From the perspective of machine-learning-based reconstruction methods, challenges and research gaps include: (1) Algorithm complexity and efficiency: the bottleneck of traditional iterative methods is that iterative reconstruction algorithms based on physical models have high computational costs cannot easily meet real-time requirements (such as intraoperative imaging). (2) Insufficient generalization ability of deep learning: as described in
Section 1, since the medical image reconstruction task is transformed into a machine learning task, data-driven machine learning (especially deep learning) models are susceptible to device and protocol differences, and have weak cross-domain generalizability.
To address the above issues, challenges, and research gaps, future research directions include the following: (1) Physics-guided and task-driven deep learning: imaging physical models (such as CT projection operators, MRI k-space sampling) and task-driven models can be embedded into learning networks to improve reconstruction accuracy and robustness. (2) Unsupervised and limited learning: the machine-learning-based reconstruction methods being developed by using unsupervised and limited sample learning means that will reduce the reliance on large amounts of high-quality annotated data. (3) Real-time and lightweight reconstruction: model compression and distillation technology can be used in optimization of reconstruction methods, hence facilitating the real-time reconstruction of mobile terminal or edge computing equipment. (4) Collaborative and joint reconstruction: the medical image reconstruction task is achieved by using multi-task machine learning methods, which utilize complementary multimodal information to jointly optimize the reconstruction results, hence reducing generalization error.
8.1.2. Segmentation
There are many challenges in medical image segmentation. 1. From the perspective of processing objects, challenges include: (1) low contrast and ambiguity in low-level feature space, (2) anatomical and appearance variability, (3) noise and artifacts, and (4) dynamic and moving structures. 2. From the perspective of machine-learning-based segmentation methods, challenges and research gaps include the following: (1) Limited training data: Medical datasets are often small due to privacy concerns, high costs, and the need for expert annotations. Limited training data can hinder the performance of machine-learning-based segmentation models. (2) Data limitations (lack of diversity, high annotation cost): Medical image data are limited by rare cases, equipment differences (such as different MRI/CT manufacturers), imaging parameters, etc., resulting in limited model generalizability. Relying on expert annotation (such as tumor boundary delineation) is time-consuming and labor-intensive, and is easily influenced by subjectivity. (3) Class imbalance: Some regions of interest may occupy only a small portion of the image, leading to class imbalance during training. This can reduce the model’s ability to detect rare or small structures. (4) Lack of standardization: There is no universal standard for defining or annotating medical image segments. This lack of standardization can lead to inconsistencies in segmentation results. (5) Model robustness and generalization across domains: Segmentation models trained on one dataset may not generalize well to other datasets or patient populations. Domain adaptation techniques are often required to address this issue. (6) Small sample learning: How to achieve high-precision segmentation with limited annotations for rare diseases or new modal data (such as photoacoustic imaging). (7) Interpretability and explainability: Automated segmentation models, especially deep-learning-based ones, often lack interpretability. (8) Continuous model updates: As new imaging techniques and pathologies emerge, segmentation models need to be continuously updated and retrained. (9) Scalability, computational cost, and real-time processing: In clinical settings, segmentation often needs to be performed in real-time to aid decision-making. Scalable and efficient algorithms are needed to handle large datasets in real-time.
To address the above issues, challenges, and research gaps, future research directions include the following. (1) Representation learning, heterogeneous data fusion, and segmentation in high-level feature space: how to effectively extract good high-level features, by integrating multi-modal information, or by combining with other tasks (such as representation learning, classification, and prognosis prediction), can improve segmentation performance. (2) Self-supervised and weakly supervised learning: Pre-train with unlabeled data (such as contrastive learning), or reduce dependence on fully annotated data through weak supervision. (3) Cross-modal generation and data argument: Generative adversarial networks or diffusion models are used to achieve data argument and enhance the model’s generalizability. (4) Interpretability and credibility: Develop visualization tools (such as attention maps) to help doctors understand the decision-making logic of models. (5) Dynamic and real-time segmentation: Combining temporal information or reinforcement learning to achieve real-time segmentation and adaptive adjustment during surgery.
8.1.3. Medical Image Registration
There are many challenges in medical image registration. 1. From the perspective of processing objects, challenges include the following. (1) Difficulty in multimodal alignment: Images from different imaging devices (such as MRI, CT, or ultrasound) or at different time points have differences in contrast, resolution, and anatomical structure, leading to a decrease in registration accuracy. (2) Pathological interference: Abnormal anatomical structures in the lesion area (such as tumors and edema) may mislead the registration model, and targeted optimization of deformation constraints is needed. (3) Non-rigid deformation modeling: Organ movements (such as respiration and heartbeat) or pathological deformations (such as tumor growth) require complex nonlinear registration algorithms, which are computationally expensive and prone to overfitting. 2. From the perspective of machine-learning-based registration methods, challenges and research gaps include the following. (1) Lack of gold standard, annotation dependency, and weak supervision: True labels require manual annotation by experts (such as anatomical landmarks or deformation fields), but the annotation process is time-consuming and subjective, especially for dynamic or pathological data. It is difficult for existing methods to achieve high-precision registration solely through image similarity measures (such as mutual information), and more robust weakly supervised strategies need to be explored. Computational efficiency and real-time performance: The registration of 3D/4D medical images (such as dynamic MRI) requires the processing of massive amounts of data, and it is difficult for traditional methods to meet real-time requirements (such as intraoperative navigation). (2) Domain generalization and robustness: The generalization and robustness of the registration algorithm in domain shift issues need to be addressed.
To address the above issues, challenges, and research gaps, future research directions include the following. (1) Self-supervised and unsupervised learning: Develop a registration framework based on the intrinsic similarity of images, such as contrastive learning and generative adversarial networks, to reduce reliance on annotated data. (2) Optimization and regularization: Develop spatiotemporal joint modeling methods (such as optical flow methods) for dynamic imaging. Optimize the parameterization methods of deformation fields (such as sparse representation and low-rank decomposition) to reduce computational complexity. By combining clinical prior knowledge (such as organ movement patterns) to constrain the deformation field, ensure that the results conform to anatomical logic. (3) Multi-task joint learning: Design a unified framework to simultaneously handle tasks such as registration, segmentation, and reconstruction, and utilize collaborative effects between tasks to improve performance (such as segmentation mask-guided registration). (4) Lightweight and real-time algorithms: Develop efficient models based on neural architecture search or knowledge distillation to adapt to edge computing devices (such as surgical robots). Explore the application of real-time dynamic registration in interventional therapy, such as radiotherapy target tracking.
8.2. Challenges, Future Perspectives, and Discussion of Visual Computing in Medicine
8.2.1. Challenges and Future Perspectives of Overall Medical Visual Computing
There are many challenges and research gaps in overall medical visual computing below. (1) Data complexity and heterogeneity: Most machine learning models are data-hungry, but medical data are scarce. Fusing diverse medical imaging data (e.g., CT, MRI, ultrasound) with clinical text or genomic data while preserving critical diagnostic features. There are limited labeled data, high annotation costs for medical datasets (e.g., pixel-level segmentation in pathology slides), and class imbalances in rare diseases. (2) Model generalizability, interpretability, and trust: Learning-based visual computing models are often black box models, yet clinicians require explainable visual computing models to validate diagnostic decisions. The interaction relationship between different modalities cannot easily be clearly revealed by existing explanatory methods. Models trained on narrow datasets may fail in diverse populations or atypical cases. (3) Real-time clinical integration: There are delays in rendering 3D/4D reconstructions or augmented reality overlays during surgery or emergency care. (4) Ethical and regulatory barriers: There are barriers in data-sharing for model training, privacy protection, and regulatory approval.
To address the above issues, challenges, and research gaps, in addition to the key points mentioned in the previous sections, other future research directions include the following. (1) Cross-modal representation learning and deep learning-driven data fusion: Leverage transformers or contrastive learning to extract shared features from heterogeneous modalities. Apply attention mechanisms to prioritize useful features and clinically significant regions during fusion. Combine visual, auditory, and haptic feedback for immersive experiences. (2) Integration of model design and interpretability: Consider interpretability during the model design phase, such as by introducing interpretable structures or rule embeddings. Develop a new model architecture that enhances the interpretability of the model itself without relying on external interpretation tools. Integrate domain knowledge (such as computer vision) with deep learning models to enhance the credibility and accuracy of explanations. Using knowledge graphs or symbolic reasoning methods, link the decision-making process of the model with a human-understandable knowledge system. (3) Federated learning and collaborative AI (artificial intelligence): Build a distributed training framework that allows hospitals to share model parameters instead of raw data. (4) Edge computing: Deploy lightweight visual computing models on portable devices. (5) Augmented reality (AR) innovations: Holographic guidance and context-aware interfaces are used in the surgical phase. (6) Robotic integration: Autonomous subsystems, haptic feedback, and reinforcement learning are used.
8.2.2. Discussion of Machine-Learning-Based Visual Computing in Medicine
Over the past decade, machine-learning-based methods for visual computing in medicine have been attracting increasing research interest. In this paper, the systematic implementation of machine-learning-based vision computing in medicine is comprehensively introduced and surveyed. This paper focuses on how to use machine learning methods to transform medical visual computing tasks (such as medical image reconstruction, analysis, and visualization) into data-driven learning problems with high-level feature representation, while exploring their effectiveness in key medical applications, such as image-guided surgery. Meanwhile, this review paper investigates fully challenges and explores future directions regarding machine-learning-based medical visual computing with high-level features.
However, there are still some limitations in this study as follows. (1) Due to the limitations of journal topics, research areas, and length requirements, this study does not provide a detailed discussion of imaging physics systems, despite the importance of imaging devices in medical visual computing. (2) Due to length requirements, the focus of this paper is on overall medical visual computing, high-level feature learning, and machine-learning-based medical visual computing with high-level features. However, tasks in machine learning, such as its detailed implementation, optimization computing, and classification recognition in feature space, have not been discussed in detail.
9. Conclusions
This review paper fully captures and analyzes systematically the recent advancements in visual computing in medicine. Beginning with a review of the fundamentals of learning-based visual computing, this investigation incorporated widely used and novel machine learning methods for visual computing in medicine.
To summarize, the major research findings of this review paper are as follows. (1) Machine-learning-based overall vision computing in medicine, as well as its systematic implementation, is comprehensively introduced and surveyed in this survey. (2) Meanwhile, this paper reviews how to use machine learning methods to transform medical visual computing tasks into data-driven learning problems with high-level feature representation, while exploring their effectiveness in key medical applications, such as image-guided surgery. (3) Furthermore, insights are provided into challenges and future directions regarding machine-learning-based medical visual computing with high-level features, aiming to guide future research in this rapidly evolving field.
Future recommendations for visual computing in medicine include: (1) developing a more intuitive and dynamic visual computing platform, AR innovations, and robotic integration to effectively and efficiently handle complex, high-dimensional data and high-level feature information; (2) enhancing interoperability and fusion between different visualization modalities and extracting generalized and discriminative features integrating regularized machine learning methods can reduce generalization error and further improve clinical decision-making; (3) ensuring transparency in visual feature representations and developing interpretability in machine learning based visual computing are crucial for medical applications and clinical adoption.
Author Contributions
Conceptualization, B.L.; formal analysis, B.L.; investigation, B.L., S.F., J.Z., G.C., S.H., S.L. and Y.Z.; resources, B.L.; writing—original draft preparation, B.L., S.F., J.Z., G.C., S.H., S.L. and Y.Z.; writing—review and editing, B.L. and S.L.; visualization, B.L. and S.F.; project administration, B.L.; funding acquisition, B.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research is funded by the National Natural Science Foundation of China, grant number 62273155, the Key Laboratory of Autonomous Systems and Network Control of Ministry of Education (SCUT of China).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Preim, B.; Botha, C. Visual Computing for Medicine: Theory, Algorithms, and Applications, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 626–663. [Google Scholar]
- Zhang, D.; Orgun, M.A.; Zhang, K. Special issue on Information Visualization in Machine Learning and Applications. J. Vis. Lang. Comput. 2016, 33, 1–2. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, Y.; Ye, X.; Mou, X. Machine Learning for Tomographic Imaging; IOP Publishing: Bristol, UK, 2020; pp. 5.1–5.4. [Google Scholar]
- Chan, T.F.; Shen, J. Image Processing and Analysis; SIAM: Philadelphia, PA, USA, 2005; pp. 4–6. [Google Scholar]
- Xie, Y.; Takikawa, T.; Saito, S.; Litany, O.; Yan, S.; Khan, N.; Tombari, F.; Tompkin, J.; Sitzmann, V.; Sridhar, S. Neural fields in visual computing and beyond. Comput. Graph. Forum 2022, 41, 641–676. [Google Scholar] [CrossRef]
- Van der Velden, B.H.M.; Kuijf, H.J.; Gilhuijs, K.G.A.; Viergever, M.A. Explainable Artificial Intelligence (XAI) in Deep Learning-Based Medical Image Analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef] [PubMed]
- Rudin, L.I.; Osher, S.; Fatemi, E. Nonlinear total variation based noise removal algorithms. Phys. D 1992, 60, 259–268. [Google Scholar] [CrossRef]
- Niu, S.; Gao, Y.; Bian, Z.; Huang, J.; Chen, W.; Yu, G.; Liang, Z.; Ma, J. Sparse-view x-ray CT reconstruction via total generalized variation regularization. Phys. Med. Biol. 2014, 59, 2997–3017. [Google Scholar] [CrossRef]
- Liu, J.; Ding, H.; Molloi, S.; Zhang, X.; Gao, H. TICMR: Total image constrained material reconstruction via nonlocal total variation regularization for spectral CT. IEEE Trans. Med. Imaging 2016, 35, 2578–2586. [Google Scholar] [CrossRef]
- Zhang, Y.; Mou, X.; Wang, G.; Yu, H. Tensor-based dictionary learning for spectral CT reconstruction. IEEE Trans. Med. Imaging 2016, 36, 142–154. [Google Scholar] [CrossRef]
- Wang, S.; Su, Z.; Ying, L.; Peng, X.; Zhu, S.; Liang, F.; Feng, D.; Liang, D. Accelerating magnetic resonance imaging via deep learning. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 514–517. [Google Scholar]
- Yang, Y.; Sun, J.; Li, H.; Xu, Z. ADMM-CSNet: A deep learning approach for image compressive sensing. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 42, 521–538. [Google Scholar] [CrossRef]
- Knoll, F.; Hammernik, K.; Zhang, C.; Moeller, S.; Pock, T.; Sodickson, D.K.; Akçakaya, M. Deep-learning methods for parallel magnetic resonance imaging reconstruction: A survey of the current approaches, trends, and issues. IEEE Signal Process. Mag. 2020, 37, 128–140. [Google Scholar] [CrossRef]
- Shan, H.; Padole, A.; Homayounieh, F.; Kruger, U.; Khera, R.D.; Nitiwarangkul, C.; Kalra, M.K.; Wang, G. Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction. Nat. Mach. Intell. 2019, 1, 269–276. [Google Scholar] [CrossRef]
- Yan, J.; Chen, S.; Zhang, Y.; Li, X. Neural architecture search for compressed sensing magnetic resonance image reconstruction. Comput. Med. Imaging Graph. 2020, 85, 101784. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, V.; Gotsis, D.; Brooks, F.; Prabhat, K.; Myers, K.; Zeng, R.; Anastasio, M. Assessing the ability of generative adversarial networks to learn canonical medical image statistics. IEEE Trans. Med. Imaging 2023, 42, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Wirtensohn, S.; Schmid, C.; Berthe, D.; John, D.; Heck, L.; Taphorn, K.; Flenner, S.; Herzen, J. Self-supervised denoising of grating-based phase-contrast computed tomography. Sci. Rep. 2024, 14, 32169. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, Y.; Li, Y.; Du, Y.; Liang, Z. Information-theoretic analysis of multimodal image translation. IEEE Trans. Med. Imaging 2025. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the 16th European Conference on Computer Vision (ECCV), Tel Aviv, Israel, 23–27 October 2022. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 2–6 October 2023. [Google Scholar]
- Burke, R.P.; Xu, Z.; Lee, C.P.; Baucom, R.B.; Poulose, B.K.; Abramson, R.G.; Landman, B.A. Multi-atlas segmentation for abdominal organs with Gaussian mixture models. In Proceedings of the SPIE Medical Imaging 2015: Biomedical Applications in Molecular, Structural, and Functional Imaging, San Diego, CA, USA, 17 March 2015. [Google Scholar]
- Lee, H.; Kim, J.U.; Lee, S.; Kim, H.G.; Ro, Y.M. Structure boundary preserving segmentation for medical image with ambiguous boundary. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- Xia, Y.; Yang, D.; Yu, Z.; Liu, F.; Cai, J.; Yu, L.; Zhu, Z.; Xu, D.; Yuille, A.; Roth, H. Uncertainty-aware multi-view co-training for semi-supervised medical image segmentation and domain adaptation. Med. Image Anal. 2020, 65, 101766. [Google Scholar] [CrossRef]
- Li, I.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.-W.; Heng, P.-A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Qiu, J.; Li, B.; Liao, R.; Mo, H.; Tian, L. A dual-task region-boundary aware neural network for accurate pulmonary nodule segmentation. J. Vis. Commun. Image Represent. 2023, 96, 103909. [Google Scholar] [CrossRef]
- Luo, K.; Wang, G.; Liao, W.; Chen, J.; Song, T.; Chen, Y.; Zhang, S.; Metaxas, D.N.; Zhang, S. Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency. Med. Image Anal. 2022, 80, 102517. [Google Scholar] [CrossRef]
- Guo, B.; Szemenyei, M.; Yi, Y.; Wang, W.; Chen, B.; Fan, C. Sa-unet: Spatial attention u-net for retinal vessel segmentation. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NeurIPS), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Gong, S.; Zhong, Y.; Ma, W.; Li, J.; Wang, Z.; Zhang, J.; Heng, P.-A.; Dou, Q. 3DSAM-Adapter: Holistic adaptation of SAM from 2D to 3D for promptable medical image segmentation. Med. Image Anal. 2024, 98, 103324. [Google Scholar] [CrossRef]
- Chen, C.; Miao, J.; Wu, D.; Zhong, A.; Yan, Z.; Kim, S.; Hu, J.; Liu, Z.; Sun, L.; Li, X.J. MA-SAM: Modality-agnostic SAM adaptation for 3D medical image degmentation. Med. Image Anal. 2024, 98, 103310. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Mirikharaji, Z.; Momeny, M.; Marhamati, M.; Neshat, A.; Garbi, R.; Hamarneh, G. Active deep learning from a noisy teacher for semi-supervised 3D image segmentation: Application to COVID-19 pneumonia infection in CT. Comput. Med. Imaging Graph. 2022, 102, 102127. [Google Scholar] [CrossRef] [PubMed]
- Dunn, B.; Pierobon, M.; Wei, Q. Automated classification of lung cancer subtypes using deep learning and CT-Scan based radiomic analysis. Bioengineering 2023, 10, 690. [Google Scholar] [CrossRef] [PubMed]
- Allasia, G.; Cavoretto, R.; De Rossi, A. Local interpolation schemes for landmark-based image registration: A comparison. Math. Comput. Simul. 2014, 106, 1–25. [Google Scholar] [CrossRef]
- Li, J.; Shi, Y.; Tran, G.; Dinov, I.; Wang, D.J.; Toga, A. Fast local trust region technique for diffusion tensor registration using exact reorientation and regularization. IEEE Trans. Med. Imaging 2014, 33, 1005–1022. [Google Scholar] [CrossRef]
- Savva, A.D.; Economopoulos, T.L.; Matsopoulos, G.K. Geometry-based vs. intensity-based medical image registration: A comparative study on 3D CT data. Comput. Biol. Med. 2016, 69, 120–133. [Google Scholar] [CrossRef]
- Sun, W.; Poot, D.H.J.; Smal, I.; Yang, X.; Niessen, W.J.; Klein, S. Stochastic optimization with randomized smoothing for image registration. Med. Image Anal. 2017, 35, 146–158. [Google Scholar] [CrossRef]
- Flach, B.; Brehm, M.; Sawall, S.; Kachelrieß, M. Deformable 3D-2D registration for CT and its application to low dose tomographic fluoroscopy. Phys. Med. Biol. 2014, 59, 7865–7887. [Google Scholar] [CrossRef]
- Du, X.G.; Dang, J.W.; Wang, Y.P.; Wang, S.; Lei, T. A parallel nonrigid registration algorithm based on B-spline for medical images. Comput. Math. Methods Med. 2016, 2016, 7419307. [Google Scholar] [CrossRef]
- Becker, K.; Stauber, M.; Schwarz, F.; Beißbarth, T. Automated 3D-2D registration of X-Ray microcomputed tomography with histological sections for dental implants in bone using chamfer matching and simulated annealing. Comput. Med. Imaging Graph. 2015, 44, 62–68. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Fakhry, A.E.; El-Henawy, I.; Qiu, T.; Sangaiah, A.K. Feature and intensity based medical image registration using particle swarm optimization. J. Med. Syst. 2017, 41, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wolterink, J.M.; Zwienenberg, J.C.; Brune, C. Implicit neural representations for deformable image registration. In Proceedings of the International Conference on Medical Imaging with Deep Learning (MIDL 2022), Zurich, Switzerland, 6–8 July 2022. [Google Scholar]
- Byra, M.; Poon, C.; Shimogori, T.; Skibbe, H. Implicit neural representations for joint decomposition and registration of gene expression images in the marmoset brain. In Proceedings of the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Vancouver, BC, Canada, 8–12 October 2023. [Google Scholar]
- Yang, X.; Kwitt, R.; Styner, M.; Niethammer, M. Quicksilver: Fast predictive image registration—A deep learning approach. NeuroImage 2017, 158, 378–396. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, G.; Zhao, A.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. VoxelMorph: A learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 2019, 38, 1788–1800. [Google Scholar] [CrossRef] [PubMed]
- Dalca, A.V.; Balakrishnan, G.; Guttag, J.; Sabuncu, M.R. Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces. Med. Image Anal. 2019, 57, 226–236. [Google Scholar] [CrossRef]
- Kim, B.; Kim, D.H.; Park, S.H.; Kim, J.; Lee, J.-G.; Ye, J.C. CycleMorph: Cycle consistent unsupervised deformable image registration. Med. Image Anal. 2021, 71, 102036. [Google Scholar] [CrossRef]
- Jaderberg, M.; Simonyan, K.; Zisserman, A.; Kavukcuoglu, K. Spatial transformer networks. In Proceedings of the 28th International Conference on Neural Information Processing Systems (NeurIPS), Montreal, QC, Canada, 7–12 December 2015. [Google Scholar]
- Chen, J.; Liu, Y.; Wei, S.; Bian, Z.; Subramanian, S.; Carass, A.; Prince, J.L.; Du, Y. A survey on deep learning in medical image registration: New technologies, uncertainty, evaluation metrics, and beyond. Med. Image Anal. 2024, 100, 103385. [Google Scholar] [CrossRef]
- Avants, B.B.; Epstein, C.L.; Grossman, M.; Gee, J.C. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008, 12, 26–41. [Google Scholar] [CrossRef]
- Hernandez, M.; Bossa, M.N.; Olmos, S. Registration of anatomical images using paths of diffeomorphisms parameterized with stationary vector field flows. Int. J. Comput. Vis. 2009, 85, 291–306. [Google Scholar] [CrossRef]
- Xu, Z.; Niethammer, M. DeepAtlas: Joint semi-supervised learning of image registration and segmentation. In Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Khor, H.G.; Ning, G.; Sun, Y.; Lu, X.; Zhang, X.; Liao, H. Anatomically constrained and attention-guided deep feature fusion for joint segmentation and deformable medical image registration. Med. Image Anal. 2023, 88, 102811. [Google Scholar] [CrossRef]
- Mehrtash, A.; Pesteie, M.; Hetherington, J.; Behringer, P.; Kapur, T.; Wells, W.; Rohling, R.; Fedorov, R.; Abolmaesumi, P. DeepInfer: Open-source deep learning deployment toolkit for image-guided therapy. In Proceedings of the SPIE Medical Imaging, Orlando, FL, USA, 11–16 February 2017; Volume 10135. [Google Scholar]
- Xu, Z.; Chen, Z.; Hou, W. A self-similarity based method for medical image resolution enhancement. In Proceedings of the Tenth International Conference on Graphics and Image Processing (ICGIP 2018), Chengdu, China, 12–14 December 2018. [Google Scholar]
- Hoffmann, M.; Hoopes, A.; Fischl, B.; Dalca, A. Anatomy-specific acquisition-agnostic affine registration learned from fictitious images. In Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 19–24 February 2023. [Google Scholar]
- Debelee, T.; Schwenker, F.; Ibenthal, A.; Yohannes, D. Survey of deep learning in breast cancer image analysis. Evol. Syst. 2020, 11, 143–163. [Google Scholar] [CrossRef]
- Singh, S.; Anand, R.S. Multimodal medical image fusion using hybrid layer decomposition with CNN-based feature mapping and structural clustering. IEEE Trans. Instrum. Meas. 2020, 69, 3855–3865. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, H.; Quan, X.; Wang, J. A two-level dynamic adaptive network for medical image fusion. IEEE Trans. Instrum. Meas. 2022, 71, 1–17. [Google Scholar] [CrossRef]
- Li, M.; Wu, C.; Zuo, X.; Cheng, L.; Zhang, J. Multi-modal data fusion algorithm for cancer survival analysis. In Proceedings of the 2024 International Conference on Electrical, Computer and Energy Technologies (ICECET), Sydney, Australia, 10–12 July 2024. [Google Scholar]
- Singh, A.; Shi, W.; Wang, M.D. Multi-modal deep feature integration for Alzheimer’s disease staging. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Turkey, 5–8 December 2023. [Google Scholar]
- Liu, Q.; Yao, J.; Yao, L.; Chen, X.; Zhou, J.; Lu, L.; Zhang, L.; Liu, Z.; Huo, Y. M2Fusion: Bayesian-based multimodal multi-level fusion on colorectal cancer microsatellite instability prediction. In Proceedings of the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Vancouver, BC, Canada, 8–12 October 2023. [Google Scholar]
- Sahu, S.K.; Chowdhury, A.S. Multi-modal multi-class parkinson disease classification using CNN and decision level fusion. In Proceedings of the Pattern Recognition and Machine Intelligence, Kolkata, India, 12–15 December 2023. [Google Scholar]
- Gokcay, D.; Eken, A.; Baltaci, S. Binary classification using neural and clinical features: An application in Fibromyalgia with likelihood-based decision level fusion. IEEE J. Biomed. Health Inform. 2019, 23, 1490–1498. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhao, W. Learning specific and general realm feature representations for image fusion. IEEE Trans. Multimed. 2020, 23, 2745–2756. [Google Scholar] [CrossRef]
- Bao, H.; Zhu, Y.; Li, Q. Hybrid-scale contextual fusion network for medical image segmentation. Comput. Biol. Med. 2023, 152, 106439. [Google Scholar] [CrossRef]
- Yin, H.; Jha, N. A health decision support system for disease diagnosis based on wearable medical sensors and machine learning ensembles. IEEE Trans. Multi-Scale Comput. Syst. 2017, 3, 228–241. [Google Scholar] [CrossRef]
- Krittanawong, C.; Johnson, K.W.; Rosenson, R.S.; Wang, Z.; Aydar, M.; Baber, U.; Min, J.K.; Tang, W.H.W.; Halperin, J.L.; Narayan, S.M. Deep learning for cardiovascular medicine: A practical primer. Eur. Heart J. 2019, 40, 2058–2073. [Google Scholar] [CrossRef]
- Galin, E.; Guérin, E.; Paris, A.; Peytavie, A. segment tracing using local Lipschitz bounds. Comput. Graph. Forum 2020, 39, 545–554. [Google Scholar] [CrossRef]
- Wizadwongsa, S.; Phongthawee, P.; Yenphraphai, J.; Suwajanakorn, S. Nex: Real-Time View Synthesis with Neural Basis Expansion. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Virtual Conference, 19–25 June 2021. [Google Scholar]
- Guizilini, V.; Ambrus, R.; Pillai, S.; Raventos, A.; Gaidon, A. 3D Packing for Self-Supervised Monocular Depth Estimation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Virtual Conference, 14–19 June 2020. [Google Scholar]
- Guizilini, V.; Vasiljevic, I.; Chen, D.; Ambruș, R.; Gaidon, A. Towards Zero-Shot Scale-Aware Monocular Depth Estimation. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Paris, France, 2–6 October 2023. [Google Scholar]
- Gouveia, P.; Luna, R.; Fontes, F.; Pinto, D.; Mavioso, C.; Anacleto, J.; Timóteo, R.; Santinha, J.; Marques, T.; Cardoso, F.; et al. Augmented reality in breast surgery education. Breast Care 2023, 18, 182–186. [Google Scholar] [CrossRef]
- Edler, P.; Kersten, T. Virtual and augmented reality in spatial visualization. J. Cartogr. Geogr. Inf. 2021, 71, 221–222. [Google Scholar] [CrossRef]
- Figueira, I.; Ibrahim, M.; Majumder, A.; Gopi, M. Augmented Reality Patient-Specific Registration for Medical Visualization. In Proceedings of the 28th ACM Symposium on Virtual Reality Software and Technology (VRST 2022), Tsukuba, Japan, 29 November–1 December 2022. [Google Scholar]
- Wake, N.; Rosenkrantz, A.B.; Huang, W.C.; Wysock, J.S.; Taneja, S.S.; Sodickson, D.K.; Chandarana, H. A Workflow to Generate Patient-Specific Three-Dimensional Augmented Reality Models from Medical Imaging Data and Example Applications in Urologic Oncology. 3D Print. Med. 2021, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Abuhmed, T.; El-Sappagh, S.; Muhammad, K.; Alonso-Moral, J.M.; Confalonieri, R.; Guidotti, R.; Ser, J.D.; Díaz-Rodríguez, N.; Herrera, F. Explainable Artificial Intelligence (XAI): What We Know and What Is Left to Attain Trustworthy Artificial Intelligence. Inf. Fusion 2023, 99, 101805. [Google Scholar] [CrossRef]
- He, W.; Li, B.; Liao, R.; Mo, H.; Tian, L. An ISHAP-Based Interpretation-Model-Guided Classification Method for Malignant Pulmonary Nodule. Knowl.-Based Syst. 2022, 237, 107778. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. ScoreCAM: Score-Weighted Visual Explanations for Convolutional Neural Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Virtual Conference, 14–19 June 2020. [Google Scholar]
- Jiang, P.-T.; Zhang, C.-B.; Hou, Q.; Cheng, M.-M.; Wei, Y. LayerCAM: Exploring Hierarchical Class Activation Maps for Localization. IEEE Trans. Image Process. 2021, 30, 5875–5888. [Google Scholar] [CrossRef]
- Aksoy, N.; Ravikumar, N.; Frangi, A.F. Radiology Report Generation Using Transformers Conditioned with Non-Imaging Data. In Proceedings of the SPIE Medical Imaging 2023: Physics of Medical Imaging, San Diego, CA, USA, 19–23 February 2023. [Google Scholar]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An Overview of Clinical Decision Support Systems: Benefits, Risks, and Strategies for Success. NPJ Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef]
- Chien, J.-C.; Lee, J.-S.; Hu, C.-H.; Wu, C.-H. The Usefulness of Gradient-Weighted CAM in Assisting Medical Diagnoses. Appl. Sci. 2022, 12, 7748. [Google Scholar] [CrossRef]
- Bhandari, M.; Shahi, T.; Siku, B.; Neupane, A. Explanatory Classification of CXR Images into COVID-19, Pneumonia and Tuberculosis Using Deep Learning and XAI. Comput. Biol. Med. 2022, 150, 106156. [Google Scholar] [CrossRef]
- Song, D.; Yao, J.; Jiang, Y.; Shi, S.; Cui, C.; Wang, L.; Wang, L.; Wu, H.; Tian, H.; Ye, X.; et al. A New XAI Framework with Feature Explainability for Tumors Decision-Making in Ultrasound Data: Comparing with Grad-CAM. Comput. Methods Programs Biomed. 2023, 235, 107527. [Google Scholar] [CrossRef]
- Mahmud, T.; Barua, K.; Habiba, S.; Sharmen, N.; Hossain, M.S.; Andersson, K. An Explainable AI Paradigm for Alzheimer’s Diagnosis Using Deep Transfer Learning. Diagnostics 2024, 14, 345. [Google Scholar] [CrossRef]
- Mohamed, M.M.; Mahesh, T.M.; Vinoth, K.K.; Guluwadi, S.B. Enhancing Brain Tumor Detection in MRI Images through Explainable AI Using Grad-CAM with ResNet-50. BMC Med. Imaging 2024, 24, 107. [Google Scholar]
- Grignaffini, F.; De Santis, E.; Frezza, F.; Rizzi, A. An XAI Approach to Melanoma Diagnosis: Explaining the Output of Convolutional Neural Networks with Feature Injection. Information 2024, 15, 783. [Google Scholar] [CrossRef]
- Navab, N.; Blum, T.; Wang, L.; Okur, A.; Wendler, T. First Deployments of Augmented Reality in Operating Rooms. IEEE Comput. 2012, 45, 48–55. [Google Scholar] [CrossRef]
- Brian, J.; Stephen, J.; Charles, M.; Gregory, J.; Bradford, J.; Terence, P. Augmented and Mixed Reality: Technologies for Enhancing the Future of IR. J. Vasc. Interv. Radiol. 2022, 31, 1074–1082. [Google Scholar]
- Picot, F.; Shams, R.; Dallaire, F.; Sheehy, G.; Trang, T.; Grajales, D.; Birlea, M.; Trudel, D.; Ménard, C.; Kadoury, S.; et al. Image-Guided Raman Spectroscopy Navigation System to Improve Transperineal Prostate Cancer Detection. Part 1: Raman Spectroscopy Fiber-Optics System and In Situ Tissue Characterization. J. Biomed. Opt. 2022, 27, 095003. [Google Scholar] [CrossRef]
- Hashimoto, D.; Rosman, G.; Rus, D.; Meireles, O. Artificial intelligence in surgery: Promises and perils. Ann. Surg. 2018, 268, 70–76. [Google Scholar] [CrossRef]
- Official Statements|ASBrS. Available online: https://www.breastsurgeons.org/resources/statements/ (accessed on 1 May 2025).
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).