Estimation of Missing DICOM Windowing Parameters in High-Dynamic-Range Radiographs Using Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Window Scaling
2.2. Dataset
2.3. Prior Work
- (i)
- Maximum bit scaling. The maxbit method linearly scales the entire pixel value range of the original image to 8-bit values. More precisely, 0 is mapped to 0, the maximum value in the pixel value histogram is scaled to 255, and the values in between are linearly interpolated to fit the range [0, 255]. This approach results in an image equivalent to the one obtained using uniform quantization. In this method, the lower window boundary is always 0, while the upper window boundary corresponds to the highest possible pixel value in the original image. Specifically, it is 1023 for a 10-bit image, 4095 for a 12-bit image, and 65,535 for a 16-bit image.
- (ii)
- Min-max scaling. As the name might suggest, the minmax approach utilizes the minimum and maximum values from the histogram of images’ pixel values. The lowest value in this histogram becomes the lower window boundary , while the highest value is used as the upper window boundary .
- (iii)
- Percentile scaling. The percentile scaling method sets the lower boundary to 10-th percentile of pixel value histogram, and upper boundary to 90-th percentile. These particular percentile values were chosen due to their performance in the baseline study [23].
- (iv)
- Maximum peak scaling. This approach, henceforth referred to as maxpeak, searches for peaks in the raw pixel value histogram. Maximum peak is defined as the maximum pixel value in a range. The process is defined as follows:
- (a)
- The histogram of pixel values H is first denoised by eliminating all pixel values that occur less than the 25-th percentile of all pixel value occurrences in the histogram. The obtained denoised histogram will be referred to as .
- (b)
- A candidate lower boundary is set as the first pixel value that appears after the occurrence of 10 consecutive non-zero pixel values in . In a similar manner, a candidate upper boundary is calculated as the last pixel value that appears before the occurrence of 10 consecutive non-zero pixel values in .
- (c)
- Once candidate window boundaries and have been identified as an area of interest, they are moved to the closest peak within range. This peak was searched for in a range of size , which was determined to be in the baseline study [23]. Finally, lower and upper window boundaries are calculated as
2.4. Objectives and Model Training
- (i)
- Edge Preservation. Edges are characterized by changes in pixel value, and in medical images, edges often correspond to boundaries of clinically relevant structures, such as bone contours or borders between healthy and diseased tissue. For example, fractures in X-ray images typically exhibit a visible change in pixel values compared with healthy bone tissue, meaning that an edge should be visible in the fracture area. Losing such information could result in the fracture becoming invisible in the exported 8-bit image, meaning that clinically relevant information was lost. It is also important to consider that windowing can also enhance certain edges, and such enhancement is not necessarily undesirable (it may even improve visual clarity).Approach: The Sobel operator [31], a standard technique for edge detection, can be used to measure a loss of edges after windowing. This involves applying the Sobel operator to both the original (raw) and the windowed images and then calculating the difference between the resulting edge maps via Mean Squared Error (MSE) or L1 distance; and a lower value would indicate better edge preservation. If desired, asymmetric L1 or MSE distances can be computed to penalise edge loss more heavily than edge enhancement.
- (ii)
- Structural Similarity Preservation. The exported image should retain the structural integrity of the original image. Excessive clipping of pixel values (such as restricting the value range too narrowly) can distort the underlying structure and clip important details.
- (iii)
- Perceptual Similarity. As the human eye can only perceive a limited range of colors and shades, if some of the colors (i.e., pixel values) are removed from the image through windowing, it should still remain perceptually similar to the original image.
2.5. Evaluation
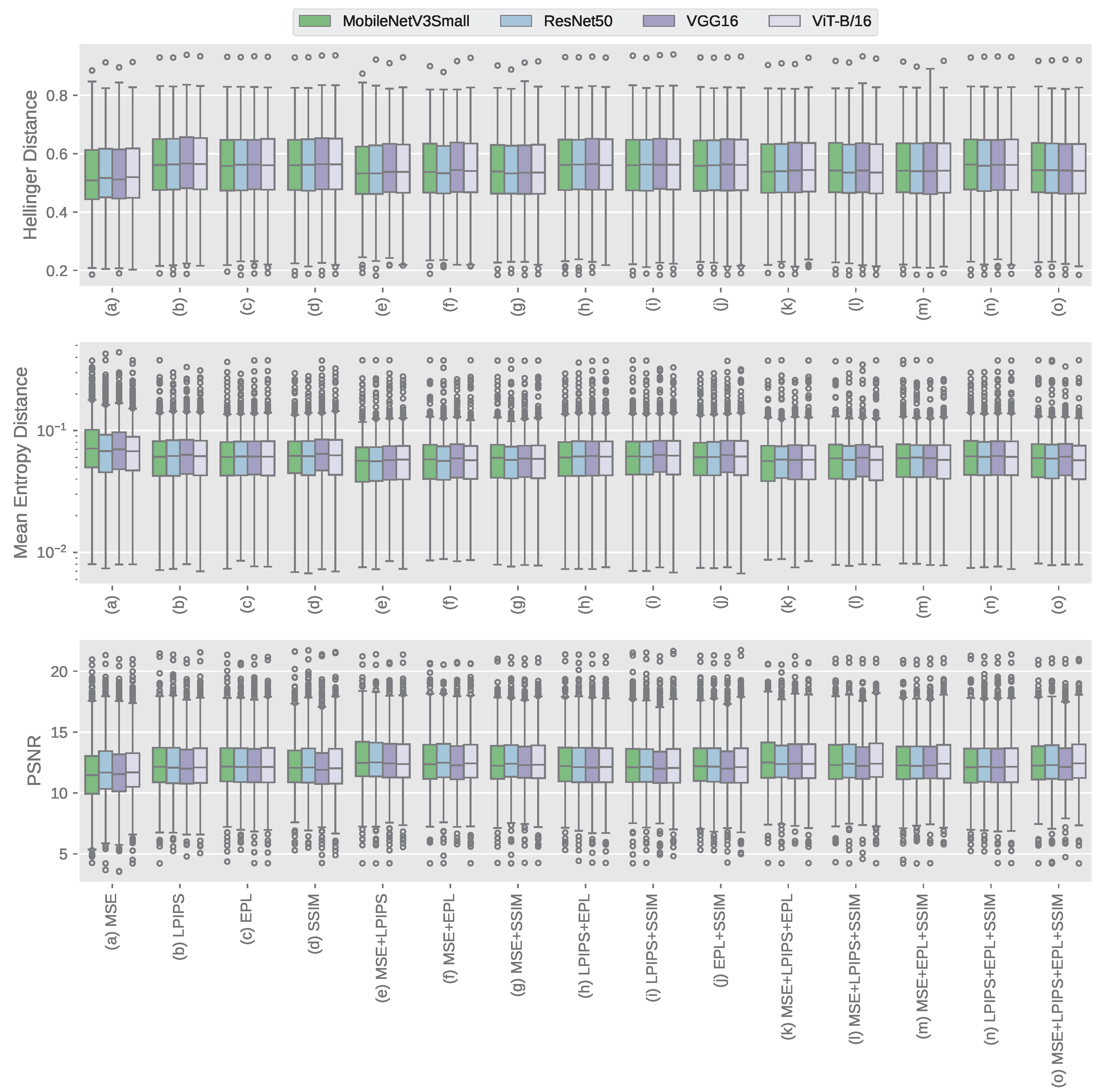
3. Results
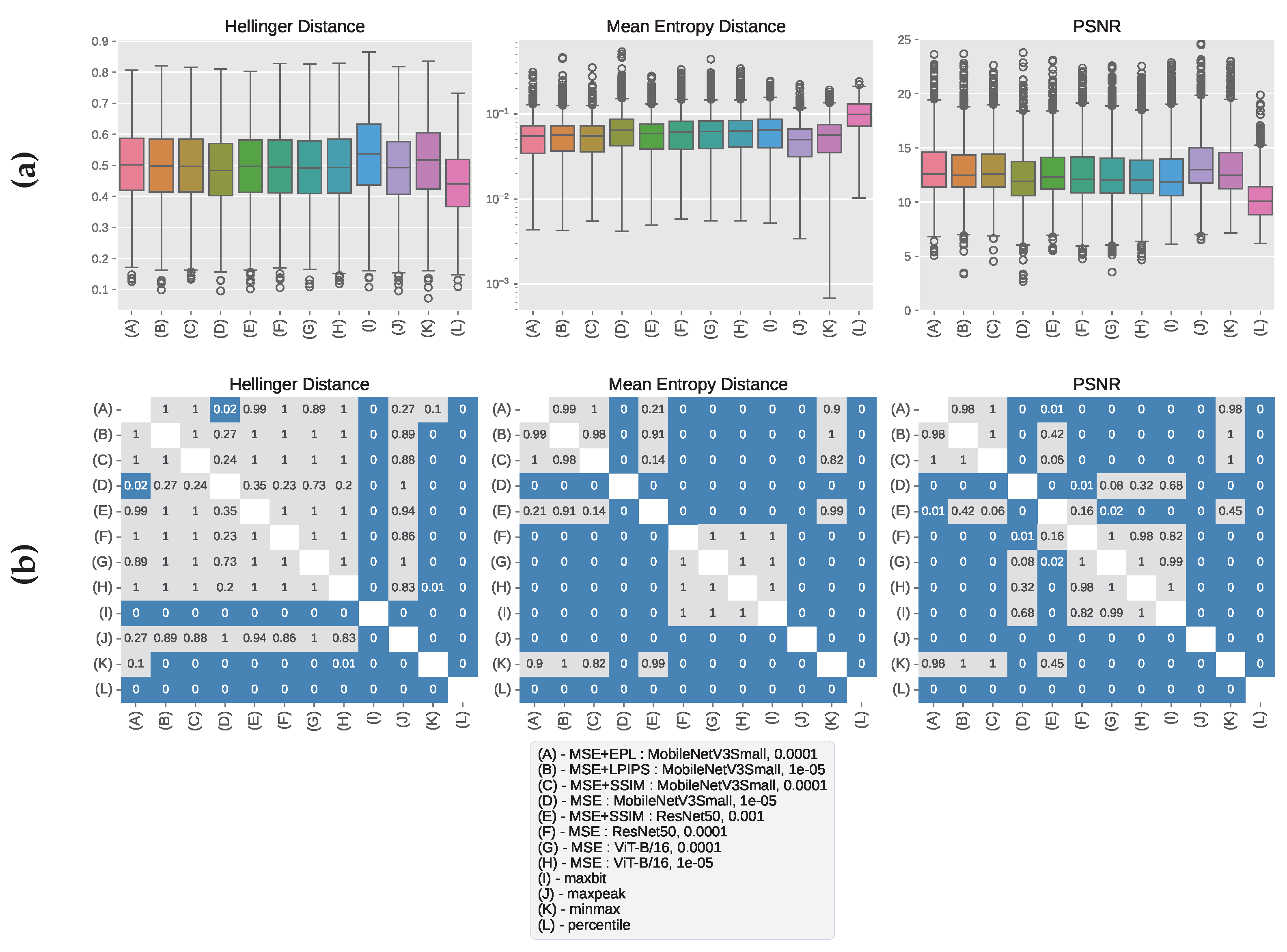
3.1. CHC Rijeka Results
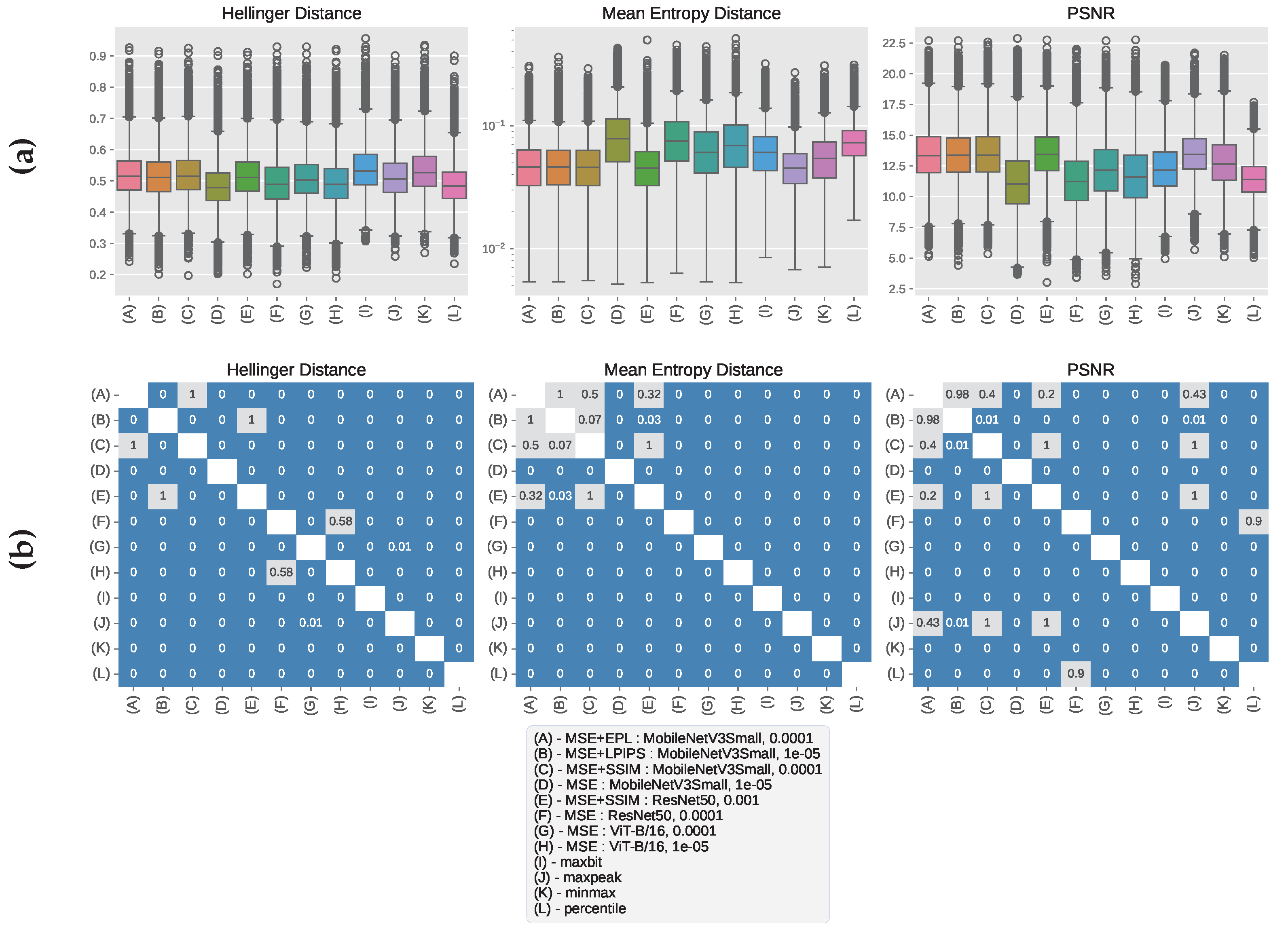
3.2. GRAZPEDWRI-DX Results
3.3. Qualitative Analysis of Windowed Image Outputs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mildenberger, P.; Eichelberg, M.; Martin, E. Introduction to the DICOM standard. Eur. Radiol. 2001, 12, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Mustra, M.; Delac, K.; Grgic, M. Overview of the DICOM standard. In Proceedings of the 2008 50th International Symposium ELMAR, Zadar, Croatia, 10–12 September 2008; Volume 1, pp. 39–44. [Google Scholar]
- Kimpe, T.; Tuytschaever, T. Increasing the Number of Gray Shades in Medical Display Systems—How Much is Enough? J. Digit. Imaging 2006, 20, 422–432. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef]
- Ali, S.; Li, J.; Pei, Y.; Khurram, R.; ur Rehman, K.; Mahmood, T. A Comprehensive Survey on Brain Tumor Diagnosis Using Deep Learning and Emerging Hybrid Techniques with Multi-modal MR Image. Arch. Comput. Methods Eng. 2022, 29, 4871–4896. [Google Scholar] [CrossRef]
- Radiya, K.; Joakimsen, H.L.; Mikalsen, K.Ø.; Aahlin, E.K.; Lindsetmo, R.O.; Mortensen, K.E. Performance and clinical applicability of machine learning in liver computed tomography imaging: A systematic review. Eur. Radiol. 2023, 33, 6689–6717. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Nahiduzzaman, M. Complex features extraction with deep learning model for the detection of COVID19 from CT scan images using ensemble based machine learning approach. Expert Syst. Appl. 2022, 195, 116554. [Google Scholar] [CrossRef]
- Wu, G.; Wang, Z.; Peng, J.; Gao, S. Coarse-to-Fine bone age regression by using multi-scale self-attention mechanism. Biomed. Signal Process. Control 2025, 100, 107029. [Google Scholar] [CrossRef]
- Singh, D.; Singh, B. Investigating the impact of data normalization on classification performance. Appl. Soft Comput. 2020, 97, 105524. [Google Scholar] [CrossRef]
- Morán-Fernández, L.; Sechidis, K.; Bolón-Canedo, V.; Alonso-Betanzos, A.; Brown, G. Feature selection with limited bit depth mutual information for portable embedded systems. Knowl.-Based Syst. 2020, 197, 105885. [Google Scholar] [CrossRef]
- Pawar, K.; Chen, Z.; Shah, N.J.; Egan, G.F. A Deep Learning Framework for Transforming Image Reconstruction Into Pixel Classification. IEEE Access 2019, 7, 177690–177702. [Google Scholar] [CrossRef]
- Yeganeh, H.; Wang, Z.; Vrscay, E.R. Adaptive Windowing for Optimal Visualization of Medical Images Based on a Structural Fidelity Measure. In Image Analysis and Recognition; Springer: Berlin/Heidelberg, Germany, 2012; pp. 321–330. [Google Scholar] [CrossRef]
- Gao, S.; Han, W.; Ren, Y.; Li, Y. High Dynamic Range Image Rendering with a Luminance-Chromaticity Independent Model. In Intelligence Science and Big Data Engineering. Image and Video Data Engineering; Springer International Publishing: Cham, Switzerland, 2015; pp. 220–230. [Google Scholar] [CrossRef]
- Echabbi, K.; Zemmouri, E.; Douimi, M.; Hamdi, S. A General Preprocessing Pipeline for Deep Learning on Radiology Images: A COVID-19 Case Study. In Progress in Artificial Intelligence; Springer International Publishing: Cham, Switzerland, 2022; pp. 232–241. [Google Scholar] [CrossRef]
- Rudolph, J.; Schachtner, B.; Fink, N.; Koliogiannis, V.; Schwarze, V.; Goller, S.; Trappmann, L.; Hoppe, B.F.; Mansour, N.; Fischer, M.; et al. Clinically focused multi-cohort benchmarking as a tool for external validation of artificial intelligence algorithm performance in basic chest radiography analysis. Sci. Rep. 2022, 12, 12764. [Google Scholar] [CrossRef] [PubMed]
- Skurowski, P.; Wicher, K. High Dynamic Range in X-ray Imaging. In Information Technology in Biomedicine; Springer International Publishing: Cham, Switzerland, 2018; pp. 39–51. [Google Scholar] [CrossRef]
- Lederer, A.; Kunzelmann, K.; Hickel, R.; Litzenburger, F. Transillumination and HDR Imaging for Proximal Caries Detection. J. Dent. Res. 2018, 97, 844–849. [Google Scholar] [CrossRef]
- Murphy, A.; Feger, J.; Ismail, M.A. Windowing (CT). 2017. Available online: https://radiopaedia.org/articles/52108 (accessed on 1 May 2025).
- Masoudi, S.; Harmon, S.A.A.; Mehralivand, S.; Walker, S.M.; Raviprakash, H.; Bagci, U.; Choyke, P.L.; Turkbey, B. Quick guide on radiology image pre-processing for deep learning applications in prostate cancer research. J. Med. Imaging 2021, 8, 010901. [Google Scholar] [CrossRef]
- Mangone, M.; Diko, A.; Giuliani, L.; Agostini, F.; Paoloni, M.; Bernetti, A.; Santilli, G.; Conti, M.; Savina, A.; Iudicelli, G.; et al. A Machine Learning Approach for Knee Injury Detection from Magnetic Resonance Imaging. Int. J. Environ. Res. Public Health 2023, 20, 6059. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, T.; Liu, H.; Zhu, G.; Zou, X. Automatic Windowing for MRI With Convolutional Neural Network. IEEE Access 2019, 7, 68594–68606. [Google Scholar] [CrossRef]
- Hržić, F.; Napravnik, M.; Baždarić, R.; Štajduhar, I.; Mamula, M.; Miletić, D.; Tschauner, S. Estimation of Missing Parameters for DICOM to 8-bit X-ray Image Export. In Proceedings of the 2022 International Conference on Electrical, Computer, Communications and Mechatronics Engineering (ICECCME), Maldives, Maldives, 16–18 November 2022; pp. 1–6. [Google Scholar]
- Suganyadevi, S.; Seethalakshmi, V.; Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retr. 2021, 11, 19–38. [Google Scholar] [CrossRef]
- Mikulić, M.; Vičević, D.; Nagy, E.; Napravnik, M.; Štajduhar, I.; Tschauner, S.; Hržić, F. Balancing Performance and Interpretability in Medical Image Analysis: Case study of Osteopenia. J. Imaging Inform. Med. 2024, 38, 177–190. [Google Scholar] [CrossRef]
- Chowdhury, M.E.H.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Emadi, N.A.; et al. Can AI Help in Screening Viral and COVID-19 Pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 2021, 128, 104115. [Google Scholar] [CrossRef]
- Abbaoui, W.; Retal, S.; Ziti, S.; El Bhiri, B. Automated Ischemic Stroke Classification from MRI Scans: Using a Vision Transformer Approach. J. Clin. Med. 2024, 13, 2323. [Google Scholar] [CrossRef]
- Napravnik, M.; Hržić, F.; Tschauner, S.; Štajduhar, I. Building RadiologyNET: An unsupervised approach to annotating a large-scale multimodal medical database. BioData Min. 2024, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Choplin, R.H.; J M Boehme, N.; Maynard, C.D. Picture archiving and communication systems: An overview. RadioGraphics 1992, 12, 127–129. [Google Scholar] [CrossRef]
- Sobel, I.; Feldman, G. A 3×3 isotropic gradient operator for image processing. Pattern Classif. Scene Anal. 1973, 271–272. [Google Scholar]
- Brunet, D.; Vrscay, E.R.; Wang, Z. On the Mathematical Properties of the Structural Similarity Index. IEEE Trans. Image Process. 2012, 21, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Gallo, O.; Frosio, I.; Kautz, J. Loss Functions for Image Restoration With Neural Networks. IEEE Trans. Comput. Imaging 2017, 3, 47–57. [Google Scholar] [CrossRef]
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The Unreasonable Effectiveness of Deep Features as a Perceptual Metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Ding, K.; Ma, K.; Wang, S.; Simoncelli, E.P. Comparison of Full-Reference Image Quality Models for Optimization of Image Processing Systems. Int. J. Comput. Vis. 2021, 129, 1258–1281. [Google Scholar] [CrossRef]
- Chang, Q.; Li, X.; Li, Y.; Miyazaki, J. Multi-directional Sobel operator kernel on GPUs. J. Parallel Distrib. Comput. 2023, 177, 160–170. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Fixing Weight Decay Regularization in Adam. arXiv 2017, arXiv:1711.05101. [Google Scholar] [CrossRef]
- Yu, T.; Kumar, S.; Gupta, A.; Levine, S.; Hausman, K.; Finn, C. Gradient Surgery for Multi-Task Learning. In Advances in Neural Information Processing Systems; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M., Lin, H., Eds.; Curran Associates, Inc.: Sydney, NSW, Australia, 2020; Volume 33, pp. 5824–5836. [Google Scholar]
- Wu, Y.; Zhou, Y.; Saveriades, G.; Agaian, S.; Noonan, J.P.; Natarajan, P. Local Shannon entropy measure with statistical tests for image randomness. Inf. Sci. 2013, 222, 323–342. [Google Scholar] [CrossRef]
- González-Castro, V.; Alaiz-Rodríguez, R.; Alegre, E. Class distribution estimation based on the Hellinger distance. Inf. Sci. 2013, 218, 146–164. [Google Scholar] [CrossRef]
- Al-Shaykh, O.; Mersereau, R. Lossy compression of noisy images. IEEE Trans. Image Process. 1998, 7, 1641–1652. [Google Scholar] [CrossRef]
- Nagy, E.; Janisch, M.; Hržić, F.; Sorantin, E.; Tschauner, S. A pediatric wrist trauma X-ray dataset (GRAZPEDWRI-DX) for machine learning. Sci. Data 2022, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Fekri-Ershad, S. Cell phenotype classification using multi threshold uniform local ternary patterns in fluorescence microscope images. Multimed. Tools Appl. 2021, 80, 12103–12116. [Google Scholar] [CrossRef]
- Fekri-Ershad, S.; Ramakrishnan, S. Cervical cancer diagnosis based on modified uniform local ternary patterns and feed forward multilayer network optimized by genetic algorithm. Comput. Biol. Med. 2022, 144, 105392. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Napravnik, M.; Bakotić, N.; Hržić, F.; Miletić, D.; Štajduhar, I. Estimation of Missing DICOM Windowing Parameters in High-Dynamic-Range Radiographs Using Deep Learning. Mathematics 2025, 13, 1596. https://doi.org/10.3390/math13101596
Napravnik M, Bakotić N, Hržić F, Miletić D, Štajduhar I. Estimation of Missing DICOM Windowing Parameters in High-Dynamic-Range Radiographs Using Deep Learning. Mathematics. 2025; 13(10):1596. https://doi.org/10.3390/math13101596
Chicago/Turabian StyleNapravnik, Mateja, Natali Bakotić, Franko Hržić, Damir Miletić, and Ivan Štajduhar. 2025. "Estimation of Missing DICOM Windowing Parameters in High-Dynamic-Range Radiographs Using Deep Learning" Mathematics 13, no. 10: 1596. https://doi.org/10.3390/math13101596
APA StyleNapravnik, M., Bakotić, N., Hržić, F., Miletić, D., & Štajduhar, I. (2025). Estimation of Missing DICOM Windowing Parameters in High-Dynamic-Range Radiographs Using Deep Learning. Mathematics, 13(10), 1596. https://doi.org/10.3390/math13101596






