1. Introduction
In data science and statistics, it is common to have data sets which mix cheap and easy-to-obtain information for a large sample of cases with more expensive and hard-to-acquire additional information for a small sample of cases. In an epidemiological study, for instance, a cohort may consist of a large number of individuals whose demographic characteristics and phenotypes have been recorded, together with a smaller number who additionally have biomarker data or genetic profiles. The purpose of this paper is to introduce a method which enhances analysis of the smaller group by making best use of data from the larger group.
One approach might be to consider the detailed information as being missing for the larger cohort, and then to adopt one of the many missing-data methodologies now available. Kang (2013) presents several techniques for handling missing data, including case deletion, mean substitution, and multiple imputation [
1]. Indeed, one of the most common and popular tools for handling missing data is the missing imputation tool MICE (Multiple Imputation by Chained Equations), which is an available library in a vast number of coding languages [
2]. However, when there is a large proportion of missing data, Multiple Imputation (MI) is not considered to be the most effective way of dealing with missing data issues [
3]. Many authors have attempted to provide ‘cutoff’ points for an acceptable amount of missing data that MI can handle [
4,
5]; however, these are found to be largely arbitrary and other factors need to be taken into account such as types of missingness and imputation mechanisms, although vast amounts of missing data are unsuited to MI. This paper explores how we can utilise the larger cohort of individuals to enhance what is learnt from the smaller cohort, without the need for imputation.
Let us assume that there are two data sets: Horizontal Data (denoted
) and Vertical Data (denoted
. For simplicity,
has a sample size of
with two scalar covariates
x and
z,
has a sample size of
with only one covariate
x, and in our case
≫
. Both data sets contain a response variable
y. In reality,
x and
z would represent a selection of covariates each as we will show in
Section 6. The validity of reducing multiple covariates into single
x and
z vectors is discussed later. Illustrated in
Figure 1 is how each data set may look in practice:
We see that the data set is tall and thin, while the data set is short and wide; hence the naming, Vertical and Horizontal data sets. Our overarching research question is therefore whether it is possible to utilise what we learn from to enhance the predictive performance and modelling upon observations within , thus improving our knowledge about response variable y.
To tackle this research question, we firstly display the knowledge and motivation for studying non-parametric modelling techniques in
Section 2, wherein we specifically focus upon smoothing methods and penalized regression modelling including B-Splines and P-Splines. We show that they display specific qualities that make them attractive for modelling our smaller cohort
and also show in
Section 3 that they can be adapted into a new model that is able to consider the larger cohort
through including a second penalty term which takes into account discrepancies in the marginal value of
x, i.e., covariates that exist in both
and
. We compare our twice penalized model structure against a linear B-Spline model and single penalty P-Spline estimation upon a series of controlled data simulations in
Section 4, before adapting our model further to take into account a binary response
y in
Section 5. We will finally apply our model upon a real healthcare data set in
Section 6, where we utilise a large cohort of individuals who have undergone baseline tests, alongside a smaller cohort who have extensive further testing in order to predict an individual’s risk of developing metabolic dysfunction associated with steatohepatitis (MASH). A discussion surrounding our work is presented in
Section 7, before concluding our work in
Section 8.
2. Background
2.1. Flexible Smoothing with Splines
Within this work, we focus solely upon non-parametric models specifically to take into account conditional models. Our motivation for going this route is shown in
Appendix A. Many non-parametric modelling techniques exist; one popular approach is smoothing, in particular spline methods. Eilers and Marx [
6] cite several reasons for their popularity including data sets being too complex to be modelled sufficiently through parametric models, and also an increasing demand for graphical representations and exploratory data analysis.
2.1.1. An Introduction to Smooth Functions
Let us assume that
x is a vector. A linear model therefore assumes:
A generalised linear model assumes that:
where
is some function.
When introducing generalised additive models (GAMs), first proposed by Hastie and Tibshirani (1986) [
7], it should first be noted that GAMs build upon familiar likelihood-based regression models in a way that provides more robustness and flexibility, such that more complex distributed data points can be modelled beyond linear or polynomial regression. If there is a single covariate, a GAM assumes a model that is of the form:
where
is a smooth function. With regards to how we select
x, one way is through a polynomial model that is of the form:
However, a more flexible alternative is provided by the use of basis functions:
evaluated at
,
,
and so on. Here,
, etc., are smooth basis functions which can be displayed with a basis matrix, with each row being evaluated at different values for
x.
2.1.2. An Introduction to Splines
Common basis functions are spline basis functions. Spline models split the
x-axis into separate intervals and assume a different model for each, as for example:
The joins between each interval are known as ‘knots’. In order for the function to be differentiable everywhere and therefore smooth at the knots, the following conditions must also hold:
In practice, it is usually sufficient that the derivatives up to
match at the knots; this is because the human eye struggles to detect higher order discontinuities [
8]. The number and placement of knots, the choice of smooth polynomial pieces that are fitted between two consecutive knots, and whether or not a penalty term is included, are what defines the type of spline—for now we focus upon non-penalized splines, specifically B-Splines.
2.1.3. B-Splines
First proposed by Schoenberg (1946) [
9], B-Splines became an increasingly popular tool for mathematical smoothing in the 1970s following publications by De Boor [
10] and Cox [
11]. B-Splines are highly attractive in non-parametric modelling and indeed Cox writes that B-Splines are ‘eminently suitable for many numerical calculations’. Eilers and Marx [
6] and Perperoglou [
8] offer good summaries of the key properties and advantages of modelling with B-Splines, along with a review of the thousands of software packages that exist for spline procedures. A B-Spline of degree
q consists of
polynomial pieces each of degree
q, which join together at
q inner knots. At the inner knots, the derivatives up to
are continuous and therefore provide a smooth function. The B-Spline is positive upon the support that is spanned over
knots and is 0 everywhere else [
6]; this provides the advantage of high numerical stability and makes them relatively simple to compute.
Let us temporarily assume a model that is linear in selected spline functions of a single covariate,
x. Further, we assume
n independent replications, so now for
where
is a zero mean error, and exactly one of the spline terms corresponds to an intercept. This is a standard linear model, which can be written in vector form
where
y,
, and
are vectors of appropriate length, and
D is a design matrix with row
i corresponding to the spline vector of observation
i.
The standard least-squares estimator is
provided that
is invertible.
An advantage of expressing coefficient vector
in Equation (
9) as linear is that we can interpret the estimation of
y as an optimisation problem in
. This means that traditional estimation methods can be used for splines in generalized multivariable regression models [
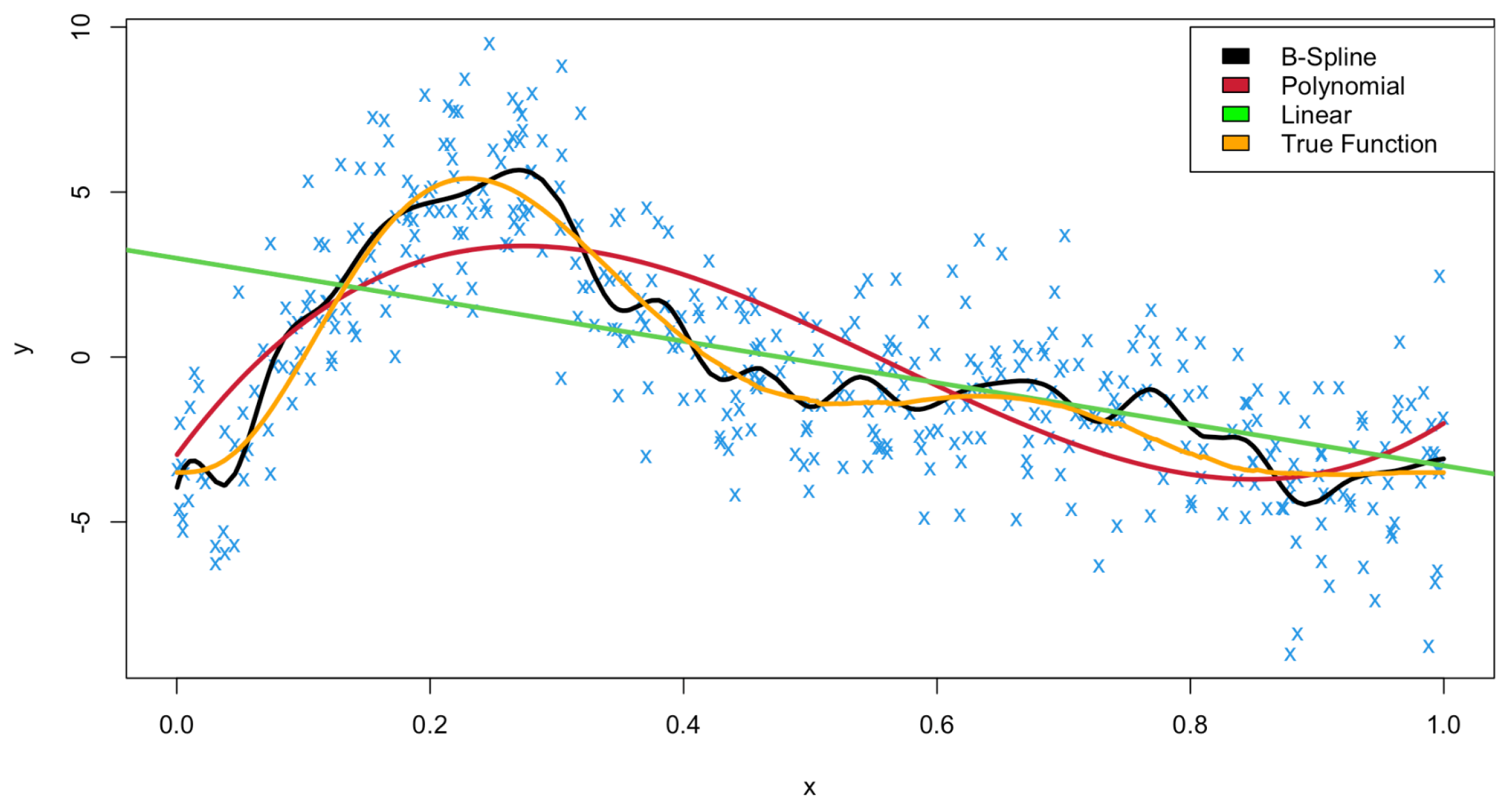
8]. Through fitting three kinds of model: linear regression (green), polynomial regression (red), and a B-Spline (black) along with the true relationship between
x and
y (orange) we illustrate in
Figure 2 how a B-Spline can flexibly and robustly fit data in which there is no obvious linear or polynomial relationship amongst bivariate data. All graphical outputs within this work were created using R. For details on how this data was generated, see
Appendix B.
The B-Spline fit is comprised of polynomials of degree 3; hence, they are cubic polynomials, and the number of knots is set to 50—this splits the domain [0,1] into 51 equidistant parts where a cubic spline is fitted within each subinterval and fused together at each knot by the conditions outlined above. Interestingly, the polynomial fit is fitted also by using a B-Spline basis; however, the number of knots is set to zero, and the result is ultimately a standard cubic polynomial fit. The linear fit is simply a straight-line relationship between
x and
y. We see from
Figure 2 that the B-Spline provides a far more flexible, robust, and accurate modelling interpretation of the data, fitting more closely to the true function in orange than the linear or polynomial fits. Indeed, when comparing the sum of squares for the fitted values, we find that the B-Spline has a value of 139.0, compared to values of 726.4 for the polynomial fit and 2095.3 for the linear fit. Our B-Spline fit in this case provides a 93% improvement over the linear model and an 80% improvement upon the polynomial model.
Selecting the number of knots is important: too high a number of knots can result in overfitting with high variance, whereas if the number of knots is too low, this can result in an underfit with high bias where the relationship is not properly observed [
8].
Figure 3 demonstrates four cubic splines with varying numbers of knots. We see from here that when the number of knots is equal to 1, there is an underfit, and there is a gross overfit when the number of knots is equal to 100. The spline fits where the number of knots is equal to 10 and 25, providing a more appropriate fit of the real data.
2.1.4. P-Splines (Penalized B-Splines)
Whilst unpenalized splines (also known as ‘regresssion splines’) have their flexibility controlled by the number of knots, penalized splines also known as ‘smoothing splines’ have theirs controlled by a penalty term, meaning that less emphasis is required on the choice of the number and position of the knots in order to avoid a potential under/over-fit of the data. One example of which is a P-Spline, short for penalized B-Spline. B-Splines are formed sequentially and ordered; therefore, for a smooth function, we expect neighbouring coefficients to be similar. Eilers and Marx in their work
“Flexible Smoothing with B-Splines and Penalties” (1996) [
6] proposed a penalty term based upon the higher order finite differences of these coefficient terms of adjacent B-Splines. This approach is a generalisation of O’Sullivan’s work in 1986 who created a penalty based upon the second derivative of the fitted curve [
12]. The formed objective function, i.e., the sum of squares (
SS), is therefore represented as follows:
in which the first term before the additive is the sum of squares between the observed data and the fitted B-Splines, and the second term after the additive is the penalty term which is controlled by smoothing parameter
. The
penalty term determines the level of smoothness that occurs, with smaller values resulting in a more jagged rougher spline, and larger values leading to smoother straighter curves.
is a difference operator with
and
is the second order difference
The penalties are therefore squared linear combinations of the coefficients. We can collect the coefficients into a matrix
C to give
a quadratic in
, just as for the first term. It therefore follows that the sum of squares (
SS) for a B-Spline with the Eilers and Marx higher order difference penalty is
The estimated coefficients
can be found through minimising the
SS. We therefore are able to find an equation for a fitted curve using a B-Spline with a high order difference penalty:
Using the same data created within
Figure 2 and
Figure 3, we fit four P-Splines with varying magnitudes of penalty terms in
Figure 4. Note that the number of knots is set at 50 for each fit.
As we can see from
Figure 4, where
and therefore no difference penalty term is applied, the resulting spline overfits the data and is particularly rough. As the penalty parameter
increases, the splines become smoother and move closer to the true function. Selection of an optimal penalty parameter is something discussed in later sections of this work.
As their creators, Eilers and Marx provide several properties of P-Splines that make them particularly advantageous to use over the standard B-Spline. The key advantage is naturally the reduced need to focus on the number and position of knots that are necessary to create an appropriate fit to data; the implementation of P-Splines is encouraged through selecting a large number of knots and then simply using
to control the level of smoothness within the fitted curve [
13]. P-Splines also display no boundary effects, i.e., an erratic behaviour of the spline when modelled beyond the data’s support; this is because the penalty term implements linearity constraints at the outer knots [
8]. They are also able to conserve moments of the data, meaning that the estimated density’s mean and variance will be equal to that of the data itself—often other types of smoothing such as a kernel smoother struggle to preserve variance to the data’s level [
6]. This property allows for valuable insights into the data’s shape, distribution, and central tendency.
The level of research that has been undertaken with P-Splines is extensive. This research covers a broad range of applications as well as adaptations to the P-Spline method itself, including modifications to the penalty term, as well as the addition of secondary penalty terms to which this work also contributes. P-Splines have been applied within many different domains, including in a medical context such as with Mubarik et al. (2020) [
14] applying P-Splines to breast cancer mortality data while managing to outperform existing non-smoothing models, and also within a geospatial environment such as with Rodriguez (2018) [
15] using P-Splines to model random spatial variation within plant breeding experiments while taking advantage of properties such as a stable and fast estimate and being able to handle missing data and being able to model a non-normal response. P-Splines have also been adapted to be used within a Bayesian context by Lang and Brezger (2004) [
16] and are shown in several works to improve predictive modelling, such as with Brezger and Steiner’s (2012) [
17] work of modelling demand for brands of orange juice and also with Bremhorst and Lambert’s (2016) [
18] work using survival analysis data.
There have also been several works that have built upon the original P-Spline method to incorporate a second additional penalty parameter; this has been undertaken for a host of reasons. Aldrin (2006) introduces an additive penalty to the original Eilers and Marx P-Spline to improve the sensitivity of the smoothing curve [
19], whilst Bollaerts et al. (2006) devise a second penalty to enforce a constraint in which the assumed shape of the relationship between predictors and covariates is taken into account [
20]. Simpkin and Newell (2013) also introduce a secondary penalty, suggesting this method helps alleviate fears when derivative estimation is of concern and can also lead to an improvement in the size of errors made during estimation [
21]. Perperoglou and Eilers (2009) devise a second penalty term to capture excess variability and to explicitly estimate individual deviance effects; they use a ridge penalty to constrain these effects and the result is a very effective and more suitable model than the single penalty P-Spline [
22]. This work aims to contribute within the additionally penalized P-Spline method space; however, the second penalty we use and our reasons for taking this direction are unlike that of other authors.
3. Model and Estimation
Our data set consists of observations of response variable y and two covariates , and data set consists of observations of response variable y and single covariate x. We are interested in modelling a relationship between y and the smooth function represented using spline basis functions , which we can estimate from . However, if is small, then there is much uncertainty surrounding this relationship. We therefore look to incorporate to enhance our learning surrounding response variable y and the relationship with covariates . Provided that is large, this will provide an accurate marginal estimate which can be incorporated into our analysis. We are planning to develop three models for this relationship, with each one building upon the previous.
3.1. Model Assumptions
We assume in general that
where
is a smooth function. For simulation purposes, we take [0,1] to be the domain of each of
x and
z, and we will use B-Splines to model
. We do this in two ways:
No Interaction Between Covariates: The relationship of the response y to covariates treats each variable separately such that the model is comprised of two smooth relationships. This is expressed as .
Interaction Between Covariates: There is a single smoothing relationship that incorporates an interaction of covariates x and z with response y. This is expressed as .
Each relationship results in different ways in which the design matrix of the B-Spline basis function is created which is explained in more detail in
Appendix C.
3.2. Linear B-Spline Model
Let us firstly assume a standard linear B-Spline model, which throughout we denote with subscript ‘0’:
where
D is the design matrix,
are the corresponding coefficients, and
are
random errors as usual. We find estimated values for coefficients
by minimising:
ultimately receiving the ordinary least squares estimate:
This value can then be used to receive fitted values for the linear model:
3.3. P-Spline Estimation
Building upon the linear model and referring back to the penalty term described by Eilers and Marx in
Flexible Smoothing with B-Splines and Penalties (1996) [
6], we now apply a penalty to the B-Spline, known as a P-Spline estimation, using a fairly large number of knots to create basis matrices
and
. We denote
and
to be roughness matrices that are based upon the second-order differences in row and column directions, with
referring to covariate
x and
referring to covariate
z. The construction of roughness matrices are discussed in more detail in
Appendix D.
The least penalized squares estimate is now found through minimising:
The least penalized squares estimate is now
A proof of this is provided in
Appendix E. This value as previous can then be used to obtain the fitted value for the P-Spline estimation model:
In the P-Spline estimation model (denoted with subscript ‘1’), both roughness matrices and are regulated by the same penalty parameter . This assumes that for our case x and z are symmetrical when simulating the data and for simulation purposes keep the model complexity simple; however, in reality we would need two parameters.
3.4. New Additional Marginalisation Penalty
As of yet, we have not introduced a method of being able to take into account the vertical data set,
; we now introduce an additional second penalty term to aid with this task. Suppose
is a vector of
x values of chosen length to provide a reasonable spread across
x domains. Let
be the true marginal function at
, such that
which can be estimated from our vertical data
. We are also able to estimate these marginal values from our horizontal data
.
Let
, a vector of size .
be any element from (a scalar).
be covariates for element i within .
be a kernel function, which we take to be the probability density function of a normal distribution with mean = 0 and standard deviation = 1.
be a smoothing parameter.
A consistent estimator, i.e., converges on the true value when sample size tends to infinity, is therefore:
In vector arguments, we can write:
where
K is a matrix comprised of scaled
functions. Recalling
, therefore:
We wish for , i.e., our estimated marginal values from of x, to be as close as possible to , i.e., the true marginal values from of x. In practice, of course, would be unknown; however, we can estimate this from the vertical data using . Conversely, we have assumed since , the error in will be relatively small. Hence, for simplicity, we use the true marginal rather than the estimator for now. Our additional penalty term now added to the least penalized squares estimate takes this into account.
The new least penalized squares estimate is now found through minimising:
This is thus providing the twice least penalized squares estimate:
The proof for this is shown in
Appendix F. This value as previous again can be used to obtain the fitted value for the P-Spline estimation model now fitted with an additional marginalisation penalty to take into account
:
In this model, we note that our additional marginalisation penalty is regulated by penalty parameter , and our P-Spline smoothing penalty is regulated by as previously. Within this paper, the model in which we use the additional marginalisation penalty is denoted with a subscript ‘2’.
4. Model Testing
In this section, we test our three models upon a series of data simulations. Simulations allow for the exploration of a controlled space and also the freedom to adapt our models to a range of different parameters, including sample size, data noise, and relationships between covariates. Using data simulations also allows for the use of perfect knowledge of true values for response variable y and true marginal values of x; this provides the advantage that we are accurately able to compare our three models by comparing our fitted values of each model to the ground truth, something that is naturally unknown in real world data. The aim of these simulations is to show that our adapted model featuring the additional marginalisation penalty outperforms both the linear B-Spline method and once penalized P-Spline estimation.
4.1. Data Simulation
4.1.1. Simulating Covariates and Responses
We first of all generate some artificial
data, generate
observations, and define
x and
z within it in order to be distributed upon a regular grid spanning
. The relationship the covariates have with response variable
y depends upon whether we consider
x and
z to have independent effects (no interaction) from one another or not (interaction); therefore, there are two separate equations, one to represent each model structure. The equations for these bivariate data sets are from Wood’s
Thin Plate Regression Splines (2003) [
23]. When we assume a model structure of no interaction effect, the true value for
y, denoted
, is found through the equation:
When we assume a model structure with an interaction effect,
is found through the equation:
These equations are almost identical, the only difference being that when there is an interaction each exponent contains both
x and
z, and when there is no interaction one exponent contains just
x and the other just
z. The constants displayed in these equations are arbitrary and hold no importance to the interaction between covariates
x and
z and are there to simply create a relationship with response
y, so they could feasibly be any value not including 0. Both relationships are each evaluated at
and
. The value for
y is provided through adding artificial noise generated by
independent
random variables to the
values, which for now we evaluate at
.
Figure 5 displays the two relationships between covariates and responses:
4.1.2. Estimating Marginal Effects
We next need to find
in order to form our second penalty term. In our simulations, take
to be an equidistant sequence of 100 values between [0,1] to provide a reasonable spread across the
x domain whilst remaining low in dimension. We estimate
by calculating
using either Equations (
34) or (
35) as appropriate, at each value of
using 10,000 values of
z equidistant between [0,1] and then averaging. The value of 10,000 was selected to again provide a good spread across the
z domain and also to be large enough to provide an accurate true estimate. In this way, we estimate
under the assumption that
z is uniformly distributed. For other distributions, we would need a weighted average.
Figure 6 is an illustration of the true marginal values
for both relationships outlined in
Section 4.1.1.
4.1.3. Assessing Model Fit
To assess model fit, we compare the fitted marginal of attained by our models with the true marginal of x found in , , and we also compare the fitted values of each model with the true values for y, . The comparison for each case is in the form of sum of squares (SS), i.e., . The desired value is for this sum to be as close to zero as possible, as this will suggest a better fit. In practice, and would be unknown; however, for model testing/simulation purposes we assume that we have perfect knowledge.
4.2. Model Fit Comparison
We will first of all fit our three models to a single simulated data set with a predetermined number of observations, level of noise, and relationship between covariates x and z using the sum of squares of fitted values and sum of squares of marginal values as a means of comparison. Following this, we will then vary our simulated data’s parameters and increase the number of simulations for each varying parameter combination.
Single Data Set
The following three model fits are applied to a simulated data set evaluated at
and
. The data follows a structure where there is an interaction between
x and
z for now, i.e., the effects are not independent. The number of knots for each covariate is set at the highest even number they can be at
, noting that the restriction for this is that
in order to create a valid design matrix. Finally, penalty parameters are given default values of
and
for now—we will investigate optimal values later. Recall as shown in
Section 3 that the fitted values for each model are defined as follows:
in which:
And the fitted marginal values of
x for each model are defined as:
in which
with
K defined in
Section 3.4.
We evaluate each model by finding the sum of squares for the fitted values (
) for all
i’s and the sum of squares for the marginal values (
) for
x in
. The closer both of these sums are to zero, the better the model fit is to the data. In
Table 1, we illustrate the sum of squares for each fit:
We see here that the sum of squares for the fitted values and the marginal values are both at their lowest for Fit2, the fit which incorporates the additional penalty term. Fit1 is also a better fit to the data than Fit0 in terms of sum of squares of fitted values; however, Fit0 does display a better fit when comparing the sum of squares of marginal values. We can illustrate the three fits upon the simulated data along with the true function in 3D plots in
Figure 7:
We see that the linear model with B-Spline basis functions produces a very jagged fit between response
y and covariates
. When the penalty parameter
is introduced in Fit1, the fit becomes far smoother. It is difficult to spot any real difference between the model fit of Fit2 to Fit1 in the above 3D plots. In
Figure 8, we show the estimated marginal functions of
x,
found from each model fit against the true marginal fit
, recalling that in practice this would not be known.
There is very little difference between the estimated marginal function in Fit0 and Fit1; however, Fit2 does offer an improved fit to the true marginal function of
x. This is highlighted in
Table 1 where Fit2 has a lower sum of squares of the marginal values than that of Fit0 and Fit1. We have shown that for one particular data set where
, in which there is an interaction between covariates, that Fit2 in which we use our novel additional penalty to take into account the marginal value for
x thus outperforms standard linear B-Spline methods and penalized P-Spline estimations in terms of model fit.
4.3. Varying Size, Noise, and Structure
Having observed that Fit2 provides a better model fit to Fit0 and Fit1 upon one simulation of a data set with specific parameters, it is now necessary to investigate across many simulations where parameters now vary. This includes the number of observations within the horizontal data set
, the level of noise in the data set
(recalling that noise is determined via
independent random variables being added to the
values), and also the structure of the data set, i.e., whether or not there is an interaction between covariates
x and
z. We therefore alter
to be = 100 or 400,
, and to represent the relationship the two covariates have with one another. We repeat each combination of these three parameters in 100 simulations and report the mean average sum of squares of fitted values and marginal values for each model fit across each parameter set and report our results in
Table 2. It is important to note the number of knots,
for when
and
for when
, recalling that
must hold. Penalty parameters
and
are at their optimal values for each sample size, noise, and covariate relationship combination—we will explore in
Section 4.4 how these values are evaluated.
The average sum of square values for both the fitted values and the marginal values for x are such that Fit2 is the lowest for every model structure, sample size, and noise combination across 100 simulations. It is also the case that for all mean average sum of squares for fitted values that Fit1 outperforms Fit0; however, when looking at the mean average sum of squares for marginal values, Fit1 does not always perform better than Fit0; this is not particularly surprising as there is nothing within the single penalty P-Spline that pulls the estimated marginal towards the true marginal. It is worth mentioning that comparing model fits between different combinations of parameters and structures is unwise. The key purpose of this exercise was to illustrate that when we take into account the marginal value of x through the use of an additional penalty term, that this offers an improved model fit than that of existing linear and penalized regression methods.
4.4. Approximating Penalty Parameters
In P-Splines, the larger is, the more penalized the curvature of the fit is; therefore, it is less sensitive to the data providing lower variance and higher bias. As , bias is low and variance is high. Typically, we would want as . Our problem is more complex to solve as previous literature offers solutions when there is only a single penalty parameter; in our case with Fit2 and our additional penalty term, we require the selection of two penalty terms, and .
Focusing initially upon one data set which considers an interaction between covariates x and z, and model parameters and , we elect to treat the penalty parameter differently within Fit1 and Fit2 such that determines Fit1 only and determines Fit2 along with . Within Fit1, we aim to find the value of that minimises the value of the sum of squares between the fitted values, , and the true values of the response, . We require values to be non-negative; therefore, we select an appropriate range of [0, 1, 2, …, 100] for . Note that in practice this is not possible as is unknown (this is discussed later). We find that a value of provides the lowest value of SS(Fitted) for Fit1 upon this particular data set. Fixing for now , we now define to be along the range [0, 0.5, 1.0, …, 50.0] and find each SS value corresponding to each value. The minimum value for for this data set. We repeat this process now fixing to alter of which we find the approximate optimal value for .
Naturally, these penalty parameter values will not hold as data set parameters are altered; similarly, these values may even vary from simulation to simulation. The optimum penalty parameters across 100 simulations of specified parameters are demonstrated within
Appendix G.1. We accept that this method is ad hoc; however, within simulations this method of selecting penalty parameters is valid as the sum of squares is a comparison between the fitted and true values of the response. In reality, this is unknown and a new method of optimising
is required which is discussed in
Section 6.
5. Logistic Regression for a Binary Response
Several adaptations to our models are required to take into account a binary response variable.
Let us now assume that
in which
is a smooth yet unknown function of probabilities. We can calculate the marginal effect of
x via
As previously, we estimate the smooth function from our horizontal data . We modify our estimated smooth function so that the marginal estimate of x from the horizontal data is close to the more accurate marginal found from the vertical data, . For now, we assume that is so large that the uncertainty which comes from is so small that we may as well use the true marginal instead. We reiterate that in reality this would not be possible as we would not know the true marginal; however, for simulation purposes it is useful as we try to achieve a marginal estimate as close to the truth as possible.
5.1. Data Creation
For data creation in simulations, we allow the true probabilities to be equal to the standard logistic function, also known as the expit:
in which
is a scaled form of the smooth function we used previously in
Section 4 for the linear model simulations. In simulated binary response data,
y is created through generating random samples from a uniform distribution. We find the true marginal
in our simulations through the same way as previously.
In the linear model, we will begin with a B-Spline approximation to create design matrix
D for the horizontal data
; however, now we will take the logistic model rather than the linear. Therefore, for any case
i:
in which
is row
i in design matrix
D.
We can now fit our three models once again, highlighting several differences that occur from the linear approach.
5.2. No Penalty—B-Spline Logistic Regression (Fit0)
The first difference in this approach is that we estimate our coefficient values
using maximum likelihood estimation rather than through least squares. Allowing
for
, the likelihood is
and the log-likelihood is
Unfortunately, there is no simple closed form for
which maximises
, so we therefore require a numerical method. As the first and second derivatives can be easily obtained, the obvious choice is the Newton–Raphson method [
24].
Let us assume that design matrix
D is
in dimension, and so
are a
vector of first derivatives and
matrix of second derivatives, respectively. Iteratively, we start with an initial coefficient estimate guess of
and then create a sequence
until the sequence has converged, or is adjudged to have converged, sufficiently.
Allowing the current estimate to be
, then the next estimate according to the Newton–Raphson method is defined as follows:
If the absolute differences between and are below some predefined tolerance threshold, convergence can be declared and we decide we have obtained the estimated coefficients . Alternatively, if the algorithm fails to converge, we set a maximum number of iterations to prevent an infinite loop.
We derive the first and second derivatives of the likelihood function with respect to
to be
and
A full proof of this is found in
Appendix H.1. The Newton–Raphson method for finding
that maximises the log-likelihood for logistic regression with no additional penalty can therefore be expressed as follows:
Convergence problems can, however, still exist. This occurs when the number of parameters is large compared with the information in the data—as a result two different errors can occur, either the algorithm does not converge or it does converge but some of the estimated coefficients are very high. There are two solutions that we can implement to avoid these errors: replicate the data several times (denoted nrep) or reduce the number of parameters to be estimated by reducing the number of knots, and/or . The selection of the number of replications and the numbers of knots is not explored in this work, but as a result of these errors the default simulation setup is with two replicates of each observation, using knots when fitting a B-Spline estimate to achieve the design matrix D.
5.3. Single Penalty—P-Spline Estimation (Fit1)
As was the case for the linear model, we penalize using P-Spline estimations, selecting
to maximise:
in which
and
are the same row/column roughness matrices used previously in the linear model to prevent overfitting. A key difference from the previous single penalty usage, however, is that we are now trying to maximise the objective likelihood function rather than minimise the least squares objective function—therefore, the penalty is now subtracted rather than added.
We find the first and second derivatives of the P-Spline estimation to be used within the Newton–Raphson method (of which a proof is provided in
Appendix H.2) such that
and:
We note that the penalty term for the first derivative is a vector and a matrix for the second derivative, and hence the
j and
subscripts, respectively. We use these values within our Newton–Raphson approximation as outlined previously in Equation (
47) to find the estimated
coefficients.
5.4. Double Penalty—Marginal Penalization (Fit2)
We now wish to add the novel second penalty taking into account the discrepancies between the marginal estimated from the horizontal data and vertical data . To reiterate, for simulation purposes we use the true marginal instead of the estimate .
We use a kernel smoothing method to estimate
from the fitted
such that
with
being a kernel function,
being a smoothing parameter,
being any element from
(a scalar), and
representing covariates for element
i within
. We can express this in vector form:
where
K is a
matrix of weights that have been suitably scaled. These weights do not contain
and so therefore
K is a fixed constant in the optimisation of the maximum log-likelihood objective function. The objective function now contains two penalties, evaluated at test vector
. The objective function is as follows:
As before and for simplicity, we define the marginal penalty (
MP) as follows:
and for simplicity:
We find the first derivatives of the marginal penalty as follows:
And the second derivatives as follows:
A proof for these derivatives is found in the
Appendix H.3. We use these values within our Newton–Raphson approximation as outlined previously in Equation (
47) to find the estimated
coefficients.
5.5. Measure of Fit
As we have undertaken previously, we will measure the fit of these three approaches using the sum of squares, comparing the estimated probabilities
; however, we now take into account heterogeneous variances that may exist within
x and
z; thus, we use a weighted sum of squares as an alternative:
Varying sample size
, number of knots
, and the relationship between covariates
x and
z are recorded. Displayed in
Table 3 are the mean average sums of squares for fitted and marginal values from 100 simulations of each data set with these varying combinations. The penalty parameters
and
are at their approximate optimal values and are illustrated in
Appendix G.2.
6. Application
6.1. Data Set
We now wish to test whether the double penalty method yields similarly better fitting results upon real data. The data we use within this Application section is the LITMUS (Liver Investigation: Testing Marker Utility in Steatohepatitis) Metacohort, making up a part of the European Non-Alcoholic Fatty Liver Disease (NAFLD) Registry [
25]. We utilise 19 covariates that are easily attained through a blood test or a routine GP appointment, whereby this includes information such as age, gender, BMI, and pre-existing health conditions, in order to form our vertical data set
. We focus upon a binary response variable of ‘At-Risk MASH’—a key stage in the MASLD natural progression between benign steatosis and more serious fibrosis and cirrhosis—with ‘1’ representing an individual being positive At-Risk MASH, and ‘0’ representing negative. There are approximately 6000 individuals that have an At-Risk MASH response and have had the 19 covariates that make up the vertical data set
measured. The horizontal data set
includes approximately 1500 individuals who have had the 19 core features of
measured, alongside further genomic sequencing data collected. This includes a further 37 additional covariates within the horizontal data set
.
A summary of the
and
data sets is illustrated in
Table 4:
6.2. Adaptations from Simulated Models
6.2.1. Dimensionality Reduction
Data simulations have been limited to where the number of covariates is equal to two. In principle, the methods we have created would work for more than two covariates, but the number of parameters would become very large and the fits therefore unstable. Instead, we will adopt three dimensionality reduction techniques to obtain the best linear combinations of the 19 and 37 covariates: linear predictor following the fit of a GLM; principle component analysis (PCA); and t-distributed stochastic neighbour embedding (tSNE). These will be taken as
x and
z, respectively. We discuss this issue of dimensionality reduction in
Section 7.
The first method of dimensionality reduction applied on the real data set is using the linear predictor fitted on the link scale following the fit of a generalised linear model (GLM) of the covariates to the response [
26]. GLMs are formed using three components: a linear predictor—a linear combination of covariates and coefficients; a probability distribution—used to generate the response variable; and a link function—simply a function that ‘links’ together the linear predictors and probability distribution parameter. By fitting a GLM to
upon the 19 covariates that also exist within
with the corresponding binary response
y for these observations, we take the linear predictor fitted on the link scale and return a single vector that represents
x. In the same way, we fit a GLM upon the 37 additional covariates that exist only within
with the corresponding binary response
y for these observations and take the linear predictor fitted on the link scale to return another single vector, this time representing
z. This technique is common in prognostic modelling within medical domains where the linear predictor is often used as a prognostic index, i.e., a measure of future risk), for patients [
27,
28].
The second method used in this section is principal component analysis (PCA). Developed by Karl Pearson [
29], PCA is one of the most common methods of dimensionality reduction. In a nutshell, supposing we have
p covariates, PCA transforms
p variables
called principal components, each of which are linear combinations of the original covariates
. We select coefficients for each covariate so that the first principal component
explains the most variation within the data, and then the second principal component
(uncorrelated with
) explains the next most variation, and so on. For our purpose, we use the first principal component when performing a PCA on the 19 and then the 37 covariates, thus providing
x and
z as single vectors we can use within our analysis.
The final method of dimensionality reduction we use is t-distributed stochastic neighbour embedding (tSNE). Based upon the van der Maaten t-distributed variant of stochastic neighbour embedding, developed by Hinton and Roweis [
30,
31], tSNE, unlike the linear predictor and PCA methods, is a non-linear technique that aims to preserve pairwise similarities between data points in a low-dimensional space. The tSNE method calculates the pairwise similarity of data points within high and low dimensional space and assigns high and low probabilities to data points that are close and far away from a selected data point, respectively. It then maps the higher dimensional data onto a lower dimensional space whilst minimizing the divergence in the probability distributions of data points within the higher and lower dimensional data. This mapping then provides a vector which can be used within our methods to represent both
x and
z variables. The greatest difference between the PCA and tSNE methods are that PCA aims to preserve the variance of the data whereas tSNE aims to preserve the relationship between the data points.
6.2.2. Estimating
Recall that for a binary response variable
y
where
is a smooth but unknown function of all fitted probabilities. The associated marginal is given as follows:
in which
is the conditional probability density function of
z given
x.
As we have undertaken previously, we are planning to estimate from but modify our estimate to make sure that , i.e., the estimated marginal x attained from our horizontal data, is close to the more accurate marginal from our vertical data, . In simulations we used the true marginal ; however, it is not possible to calculate in real data, so our first task is therefore to estimate .
6.2.3. Marginal in the Second Penalty
In our simulations to use the second marginalisation penalty, we compare the marginal from our fit using
x and
z in our horizontal data, and compare this with the true marginal values. Naturally, the true marginal is unknown in our real data, so we therefore use an accurate estimate from
. As mentioned in
Section 3, we use a predefined vector
to calculate the marginal from
. In our real data, we can now simply use
, including the observed
x-values from
, to calculate the marginal. The advantage of this is that we use all values in
x from
rather than just unique values, allowing the second penalty term to have greater weight for more common values of
x. Another advantage is the marginal from
, whereby
for
is produced as a part of the fitting procedure.
6.2.4. Means of Comparison
In our real data as mentioned we now do not know the true smooth function
; therefore, a comparison by means of the sum of squares of fitted values as undertaken in simulations is now redundant. By allowing the estimated marginal of
x from
to now replace the true marginal
which is now also unknown, our only means of comparison now is through the sum of squares of the marginal values, with values closer to zero indicating a greater fit. In
Section 7, we mention possible future work of evaluating model fits upon data in which we do not have perfect knowledge.
6.2.5. Cross-Validation for Approximating Penalty Parameters
One final adaptation from modelling upon real data to simulations is that it is now feasible to undertake a
k-fold cross-validation method in order to determine the smoothing parameter
. Recall in simulations how
was selected through comparing the sum of squares values when fitting the simulated data across a grid of
values. This sum of squares value was found through comparing model fit values to the truth; however, in our real data application where ground truth is unknown,
k-fold cross-validation [
32] is now required.
Setting number of folds and allowing for a 90:10 train/test split, each train set data is fitted using a P-Spline approximation whilst iterating through a grid of values. The coefficients of each of these fits are then multiplied with the design matrix created from the test set and then put into an expit function to give the estimated fitted probabilities . We use three different metrics to compare and the values of y within the test set: sum of squares (SS), log-likelihood (LL), and area under curve (AUC). For each value, the median value for each metric across the folds is found. The ‘best’ value is therefore the median value that is either the smallest SS or the greatest LL or AUC value. We then use all three supposed ‘best’ values according to these metrics to find . This is simply found through using these values and scanning through a grid of values, until an acceptable improvement in fit from using the additional marginalisation penalty over the single penalty P-Spline approximation is found, whereby in our case this acceptable improvement is a 50% reduction in the sum of squares in marginal values. We can also select as simply the value that provides the lowest sum of squares of marginal values when using the additional marginalisation penalty.
We accept that our method of selecting
is ad hoc and discuss potential future work options to select
values in
Section 7. Increasing
values will take
values ever closer to
at the expense of a poorer and more biased estimate of
. In practice, we would like
to be just close enough to
to consider a realistic and feasible estimate of the underlying true
. This depends upon the level of noise that exists in
and to a lesser extent the noise in
. Selecting
based upon a 50% reduction in the SS of marginal values is therefore preferable rather than the outright best SS value—this is because we accept that there is a level of noise in
and that it would not be exactly the same as
even if we had perfect knowledge on the correct marginal, so we just expect these values to be close. By increasing
, we force these values to be closer together, leading to more bias within
.
6.3. Results
Following the dimensionality reduction of the real data set, x and z are now single vectors. Errors frequently arose at two points during the modelling process for the application data. The first occasion is when fitting the three model types upon the newly scaled data, and the second occasion is when cross-validating upon the data to find optimal values for . In the second instance, errors occur in particular for values of high . For both occasions, this is due to the algorithm for fitting a generalised linear model either not converging or producing coefficient ’s that are ridiculously high. This error happens when the fitted probabilities are extremely close to 0 or 1, occurring when the predictor variable x is able to perfectly separate the response variable. The consequence of this is that maximum likelihood estimates of the coefficients do not exist, and therefore the algorithm fails to converge. These errors can be alleviated by trimming the scaled values for x and z by removing extreme values at either end of the range. Following an extensive search of altering the minimum and maximum values of x and z, the number of data points that are removed without causing either a fitting or cross-validation error was 24 for both the interaction and non-interaction data sets when using PCA as a form of dimensionality reduction. This compares with 46 data points removed in the non-interaction data set and 52 data points removed in the interaction data set when using the Linear Predictor as a means of dimensionality reduction, and no data points removed for both the non-interaction and interaction data sets when using tSNE.
As mentioned within
Section 6.2.1, we perform three methods of dimensionality reduction (linear predictor, PCA, and tSNE) and we have throughout two different relationship covariates
x and
z with one another (interaction or no interaction). Along with having three methods of determining
as mentioned in
Section 6.2.5 (AUC, SS, and Log-likelihood) and two methods of determining
(outright best SS of marginal values and lowest value that offers a 50% improvement in SS of marginal values compared to Fit1), we simulate all of these combinations of possible changes to our modelling.
Table 5 lists all results of these calculations. We can report that for every combination of dimensionality reduction, choice of
and
, interaction or no interaction between covariates, Fit2 always provides a notable enhancement in marginal fit of on average 65%; this is because after all setting
would at worst reduce Fit2 to Fit1. Generally for interaction data sets, the additional penalty model offers less of an improvement in comparison to non-interaction data sets—in some cases such as the modelling upon an interaction data set using linear predictor as a dimensionality reduction method and using AUC as the
determination method, there is no value of
that offers a greater than 50% improvement in Fit2 over Fit1. This is the case for four other instances, as shown in
Table 5. This is arbitrary, however, as it is clear from our results that Fit2 always provides an improvement in fit compared to Fit1 and Fit0. It is also notable that the P-Spline approximation method does not always offer an improvement upon the standard linear model. Generally, log-likelihood and sum of squares methods select the same values for
; however, there is more variation when AUC is the method of determination for
. Values for
are almost always identical regardless of
determination method, typically offering
approximately equal to 2 when selecting Fit2 purely on best fit, and
approximately equal to 1 when selecting
based upon a 50% improvement in fit for Fit2 over Fit1.
In
Figure 9, we graphically compare the marginal estimates from
,
, which we receive from each model fit as outlined in
Section 6.2.3 with our estimated marginal of
x from
,
(which we receive as outlined in
Section 6.2.2), for each model fit and each dimensionality reduction method— note that the relationship between covariates in this case is one of which there is an interaction. The red lines in each plot represent
and blue lines represent
. The top row of plots are obtained through using the Linear Predictor as a means of dimensionality reduction; the middle row via PCA; and bottom row through tSNE. Each graph on the left of the plot illustrates the fitted marginal probabilities achieved from Fit0; the middle via Fit1; and the right hand side via Fit2. Better model fit is demonstrated the closer the blue and red lines are to one another, and as we can see for Fit2
(red) is closest to
(blue) for all dimensionality reduction methods. Fit2 is therefore a better fit for our applications data compared to Fit0 and Fit1.
7. Discussion and Future Work
To our knowledge, this is the first work to propose additional marginal penalties in a flexible regression. There are, however, a number of areas for future development. The first is that we were unable to develop a succinct method of selecting penalty parameter relating to the discrepancies between marginal values of x; we relied upon cross-validation to select ; however, this method is not possible in selection for —we therefore relied upon manual scanning across a range of values to select . We are prepared to accept a slightly worse fit to the data in if a more realistic marginal when compared with that from is obtained. This means we are not trying to optimise the fit but we desire as good a fit as possible subject to the marginal estimate being consistent with . Future work would therefore include the development of a concise method to choose . We know that as , ; therefore, one possibility for future work would be to gradually increase until a consistent estimator is reached. Another method would include a computationally intense method of iterating through a grid of values, this time fitting our model to a sample of values within for each value. For each iteration, we can receive the marginal values from these samples as well as a percentage confidence band for . If also lies within this band, then we accept this value as the ‘optimal’, and if not then we try the next value in our defined grid. An alternative solution could be to withhold some of to assess fit, through dividing into parts similar to a train/test split, using the train set to determine penalty parameter values and and using the test set to evaluate model performance with these selected values.
Secondly, we are reliant upon the sum of squares of the marginal values to be our sole measurement of fit for modelling within our application to the real data section. As seen in simulations, we also used the sum of squares of fitted values as a means for comparison between different model fits; however, with the true fitted values now unknown, the method we utilised within simulations is now infeasible. Future work would therefore include the development of other methods of evaluating the performance of our model when the ground truth is unknown. One possible method of achieving this objective would be to use two thirds of to cross-validate and calculate optimal penalty parameter values and while using the remaining third of to assess fit. However, this is not entirely necessary as our aim for this work was not to model as well as possible but rather to integrate the smaller cohort and larger cohort into a predictive analysis without the requirement of imputation. We would therefore consider accepting a slightly worse model fit for to ensure a marginal that is closer to .
Furthermore, we have developed our method for the case of
x and
z being scalars in
Section 6.2.1. We used dimensionality reduction methods to reduce multivariate covariates to scalar summaries. These techniques naturally come with their own disadvantages. For example, if data is strongly non-linear, then dimensionality techniques such as PCA can struggle to fully capture covariate relationships, potentially resulting in a loss of information. Dimensionality reduction can also result in difficult to interpret transformations of covariates and are not always easy to visualize. Future work therefore includes being able to develop our methods and additional marginalisation penalties to work upon data sets without the need for transforming
covariates into single
x and
z vectors. Finally, we have only considered non-parametric models for representing both
and
. We mentioned in
Section 2 and
Appendix A our motivation and the suitability for non-parametric modelling, noting that if
takes a parametric modelling form, then it is unlikely
could also be of the same parametric form. However, it is possible for one of
or
to be modelled parametrically provided the other is modelled non-parametrically. This is therefore another potential avenue for future work.
8. Conclusions
Referring back to the purpose of this research, it is a common issue within data science of how to maximise the level of information that can be attained from asymmetric overlapping data sets. In a medical context, we have highlighted how particular subjects may have more information available to utilise within predictive analysis than the more common baseline information, such as specialist testing. Common solutions to this problem involve missing data imputation or simply two separate predictive models, one using baseline information only on a large number of individuals and one using baseline plus specialist testing information on a select number of individuals. The issue with missing data imputation is that it is infeasible and bad practice to impute large levels of missing data, particularly if the cohort with larger levels of information available is substantially smaller than that of the larger cohort with less information. Utilising two separate predictive models for each cohort limits analysis and what we can learn from both the response variable and its interaction with covariates.
In this work, we propose a method to integrate the smaller cohort, named horizontal data (), and the larger cohort, named vertical data (), without the requirement for data imputation or data deletion. Simplifying the number of covariates down to two, x and z, in which x represents covariates every individual has recorded, and z represents the added covariates only individuals within have recorded, we are motivated by non-parametric models for modelling each cohort. We find that utilising flexible smoothing via B-Splines offers opportunities to take into account both cohorts into our analysis. Flexible smoothing models provide more robustness and flexibility to model complexly distributed data points where linear and polynomial regression models are unsatisfactory. Smoothness can be controlled by the introduction of a penalty term to B-Splines, also known as P-Splines—these penalties are desirable to prevent over/under-fitting to data. By looking at discrepancies between the marginal value of x obtained from , denoted , with the marginal value of x obtained from , denoted , we introduce a second penalty term to be able to model whilst taking into account .
Through a series of data simulations, penalty parameter tunings, and model adaptations to take into account both a continuous and binary response, we found that the model with the additional marginalisation penalty appended to a P-Spline approximation method outperformed both the linear B-Spline method and the standard P-Spline approximation method utilising the single smoothing penalty. Applying the model to a real life healthcare data set of the LITMUS Metacohort with binary response relating to an individual’s risk of developing MASH (metabolic dysfunction associated steatohepatitis), we let represent individuals who had a routine blood test taken, and represent individuals who had further specialist genomic sequencing data collected. We found similar results in that this model with the additional marginalisation penalty fitted the marginal values of the data better than both the linear B-Spline model and the single penalty P-Spline approximation.
Areas for future work include the development of a succinct method to select penalty parameter and the finding of a measurement to take into account overall model fit when applying models to a real world data set. In this work we omitted this, as our overall aim was to develop a method in which we could integrate asymmetric data sets into a predictive analysis upon a binary target, and therefore we had less of a focus on model fit. Future work will also include adapting our method to not require dimensionality reduction and also to consider parametric modelling for one of the and data sets. We have shown in this work that the novel additional marginalisation penalty improved the fit of the models as opposed to standard B-Spline and P-Splines approximation methods. These results are encouraging and illustrate a novel technique of how it is possible to integrate asymmetric data sets that share common levels of information without the need for data imputation or separate predictive modelling.
Author Contributions
Conceptualization, M.M. and R.H.; methodology, M.M. and R.H.; software, M.M. and R.H.; validation, M.M.; formal analysis, M.M.; investigation, M.M. and R.H.; resources, Q.M.A.; data curation, Q.M.A.; writing—original draft preparation, M.M.; writing—review and editing, M.M, R.H. and P.M.; visualization, M.M.; supervision, R.H. and P.M.; project administration, R.H. and P.M.; funding acquisition, Q.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Newcastle University and Red Hat UK. This work has been supported by the LITMUS project, which has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 777377. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. QMA is an NIHR Senior Investigator and is supported by the Newcastle NIHR Biomedical Research Centre. This communication reflects the view of the authors and neither IMI nor the European Union and EFPIA are liable for any use that may be made of the information contained herein.
Institutional Review Board Statement
This study utilised data drawn from the LITMUS Metacohort from patients participating in the European NAFLD Registry (NCT04442334), an international cohort of NAFLD patients prospectively recruited following standardized procedures and monitoring; see Hardy and Wonders et al. for details [
25]. Patients were required to provide informed consent prior to inclusion. Studies contributing to the Registry were approved by the relevant Ethical Committees in the participating countries and conform to the guidelines of the Declaration of Helsinki.
Data Availability Statement
Data underpinning this study are not publicly available. The European NAFLD Registry protocol has been published in [
25], including details of sample handing and processing, and the network of recruitment sites. Patient level data will not be made available due to the various constraints imposed by ethics panels across all the different countries from which patients were recruited and the need to maintain patient confidentiality. The point of contact for any inquiries regarding the European NAFLD Registry is Quentin M. Anstee via email:
NAFLD.Registry@newcastle.ac.uk.
Conflicts of Interest
Quentin M. Anstee has received research grant funding from AstraZeneca, Boehringer Ingelheim, and Intercept Pharmaceuticals, Inc.; has served as a consultant on behalf of Newcastle University for Alimentiv, Akero, AstraZeneca, Axcella, 89bio, Boehringer Ingelheim, Bristol Myers Squibb, Galmed, Genfit, Genentech, Gilead, GSK, Hanmi, HistoIndex, Intercept Pharmaceuticals, Inc., Inventiva, Ionis, IQVIA, Janssen, Madrigal, Medpace, Merck, NGM Bio, Novartis, Novo Nordisk, PathAI, Pfizer, Poxel, Resolution Therapeutics, Roche, Ridgeline Therapeutics, RTI, Shionogi, and Terns; has served as a speaker for Fishawack, Integritas Communications, Kenes, Novo Nordisk, Madrigal, Medscape, and Springer Healthcare; and receives royalties from Elsevier Ltd.
Appendix A. Motivation for Non-Parametric Models
Considering a response
y and two vector covariates
x and
z, we denote
as a generic notation for probability functions, whether for discrete or continuous random variables. We are particularly interested in the conditional probability functions
and
, with the former being the marginal after integrating out
z of the latter. In general, the relationship is as follows:
When determining the suitability of either a parametric or non-parametric approach to modelling both probability functions, we note that if takes a parametric modelling form, then it is not usually possible for to also be the same parametric form.
Example: Assume
y is binary and suppose the full conditional is logistic:
where
. Take
z to be a binary scalar that is independent of
x with
, then
Hence is not of logistic form. We are therefore motivated to look at non-parametric models to take into account conditional models.
Appendix B. Data Generation for B-Splines Example
Data for
Figure 2 and
Figure 3 was created within R by generating 400 random samples from the uniform distribution for each covariate
x and
z. The relationship the covariates
x and
z have with the true value for the response, denoted
, is found through an example arbitrary equation:
Noise is then added to to give values for y which are then plotted, as shown by the blue crosses.
Appendix C. Construction of Design Matrices
As outlined in
Section 3.1, there are two relationships the response
y has with covariates
. The first instance of the smoothing relationship relating to there being no interaction between response
y and covariates
, suggests that given a predefined number of knots
, a B-Spline basis is fitted to covariate
x to provide the B-Spline basis matrix
, which has dimensions
, i.e., the number of observations within the
data set by the number of knots
. This matrix represents the list of basis functions across all predefined knots, evaluated at each observation within
. Similarly, fitting a B-Spline basis function to covariate
z with predefined number of knots,
, basis matrix
with dimensions
is outputted. The design matrix
D is then constructed by appending
to
and then adding an intercept term. The design matrix therefore has dimensions
.
For the second instance of the smoothing relationships, in this case where response y has an interaction with covariates , the design matrix D is constructed differently. Matrices and are both constructed in the same way as before; however, D is now achieved through taking all products of a column in and a column in and then adding an intercept term. This therefore provides the design matrix D with the dimensions .
Appendix D. Construction of Roughness Matrices
Section 3.3 introduces P-Spline estimation as a means of penalizing the B-Spline, achieved through the creation of penalty roughness matrices
and
. The way
and
are constructed depends upon the relationship between response
y and covariates
. When there is an interaction,
is found through the product between the identity matrix,
I, of dimensions
, and the difference matrix of dimensions
, plus an intercept term, thus giving
the dimensions of
. Similarly,
is found in the exact same way, using number of splines
this time.
therefore has the dimensions of
.
When there is no interaction between the response and the covariates, roughness matrices and have identical dimensions. In this case, and take the dimensions of , whereby this is simply the two difference matrices applied to covariates x and z appended together, with an added intercept term.
Appendix E. Proof of Least Penalized Squares Estimate with Single Penalty
Proof. Let
. The penalized sum of squares is
Differentiating
leading to
provided the inverse exists. □
Appendix F. Proof of Least Penalized Squares Estimate with Additional Penalty
Proof. Let
. The twice penalized sum of squares is
Differentiating
leading to
provided the inverse exists. □
Appendix G. Approximate Optimum Penalty Parameters
Appendix G.1. Continuous Response
Rounded to sensible values, we display the optimum
values for each model structure and data set parameter combination in
Table A1.
Table A1.
Penalty parameter values for each model structure and parameter combinations.
Table A1.
Penalty parameter values for each model structure and parameter combinations.
| Interaction | | | | | |
|---|
| Yes | 100 | 0.2 | 0.1 | 0.1 | 0.2 |
| 0.5 | 0.3 | 0.3 | 0.6 |
| 1.0 | 0.9 | 0.9 | 1.8 |
| Yes | 400 | 0.2 | 2 | 2 | 2.3 |
| 0.5 | 6 | 6 | 7 |
| 1.0 | 18 | 18 | 21 |
| No | 100 | 0.2 | 0.1 | 0.1 | 0.5 |
| 0.5 | 0.3 | 0.3 | 1.5 |
| 1.0 | 0.9 | 0.9 | 4.5 |
| No | 400 | 0.2 | 4.3 | 6 | 1 |
| 0.5 | 13 | 18 | 3 |
| 1.0 | 36 | 54 | 9 |
We see generally that as and increase, the size of each penalty parameter also increases. For data sets with a covariate interaction, the optimum values for typically follow ; however, for non-interaction data sets and differ when is larger.
Appendix G.2. Binary Response
We see in
Table A2 that generally as
increases, the size of each penalty parameter also increases. For both interaction and non-interaction data sets, the optimum values for
typically follow
; however, the value of these penalty parameters greatly increases when the number of knots
increases significantly to 18. Values for
also tend to increase as sample size increases, albeit from a far greater initial value.
Table A2.
Optimum penalty parameter values for each model structure and parameter combinations.
Table A2.
Optimum penalty parameter values for each model structure and parameter combinations.
| Interaction | | | nrep | | | |
|---|
| No | 100 | 4 | 4 | 0.06 | 0.23 | 8.94 |
| 100 | 8 | 4 | 0.21 | 0.22 | 8.92 |
| 400 | 8 | 2 | 0.28 | 0.30 | 18.86 |
| 400 | 18 | 2 | 6.34 | 7.06 | 18.98 |
| 900 | 8 | 1 | 0.25 | 0.33 | 20.82 |
| Yes | 100 | 8 | 8 | 0.71 | 0.67 | 13.06 |
| 400 | 8 | 2 | 0.62 | 0.59 | 15.98 |
| 900 | 8 | 1 | 0.57 | 0.59 | 18.46 |
Appendix H. Newton–Raphson Method for Finding
Appendix H.1. No Penalties—Fit0
Let us consider a single term within the the log-likelihood (no penalty):
Therefore
and also:
which are both scalars.
Recall that
and so for
, we have
and
We can now derive the first and second derivatives of the likelihood function with respect to
to be
and
Appendix H.2. Single Penalty—Fit1
Defining this as the roughness penalty (RP):
we are able to find the first and second derivatives that are to be added to the terms we found in the previous chapter when using Newton–Raphson upon the no penalty method, such that
and
Thus giving the overall first derivative term of the P-Spline Estimation using the Newton–Raphson method:
and second derivative:
Appendix H.3. Double Penalty—Fit2
Defining the additional marginal penalty (MP) as follows:
and for simplicity:
Only
depends upon
. Differentiating the
i-th term of
with respect to
:
and then collecting these into an
vector
, we find the first derivatives of the marginal penalty as follows:
Now differentiating the
i-th term of
again, this time with respect to
:
and then collecting these into an
vector,
, we obtain the second derivatives of the marginal penalty as follows:
These values are then used within our Newton–Raphson approximation as outlined previously to find the estimated coefficients.
References
- Kang, H. The prevention and handling of the missing data. Korean J. Anesthesiol. 2013, 64, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate imputation by chained equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Lee, J.H.; Huber, J.C., Jr. Evaluation of multiple imputation with large proportions of missing data: How much is too much? Iran. J. Public Health 2021, 50, 1372. [Google Scholar] [PubMed]
- Schafer, J.L. Multiple imputation: A primer. Stat. Methods Med. Res. 1999, 8, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.A. How can I deal with missing data in my study? Aust. N. Z. J. Public Health 2001, 25, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Eilers, P.H.; Marx, B.D. Flexible smoothing with B-splines and penalties. Stat. Sci. 1996, 11, 89–121. [Google Scholar] [CrossRef]
- Hastie, T.J.; Tibshirani, R.J. Generalized Additive Models; CRC Press: Boca Raton, FL, USA, 1990; Volume 43. [Google Scholar]
- Perperoglou, A.; Sauerbrei, W.; Abrahamowicz, M.; Schmid, M. A review of spline function procedures in R. BMC Med. Res. Methodol. 2019, 19, 46. [Google Scholar] [CrossRef]
- Schoenberg, I.J. Contributions to the problem of approximation of equidistant data by analytic functions. Part B. On the problem of osculatory interpolation. A second class of analytic approximation formulae. Q. Appl. Math. 1946, 4, 112–141. [Google Scholar] [CrossRef]
- De Boor, C. On calculating with B-splines. J. Approx. Theory 1972, 6, 50–62. [Google Scholar] [CrossRef]
- Cox, M. The numerical evaluation of a spline from its B-spline representation. IMA J. Appl. Math. 1978, 21, 135–143. [Google Scholar] [CrossRef]
- O’sullivan, F.; Yandell, B.S.; Raynor, W.J., Jr. Automatic smoothing of regression functions in generalized linear models. J. Am. Stat. Assoc. 1986, 81, 96–103. [Google Scholar] [CrossRef]
- Currie, I.D.; Durban, M. Flexible smoothing with P-splines: A unified approach. Stat. Model. 2002, 2, 333–349. [Google Scholar] [CrossRef]
- Mubarik, S.; Hu, Y.; Yu, C. A multi-country comparison of stochastic models of breast cancer mortality with P-splines smoothing approach. BMC Med. Res. Methodol. 2020, 20, 299. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Alvarez, M.X.; Boer, M.P.; van Eeuwijk, F.A.; Eilers, P.H. Correcting for spatial heterogeneity in plant breeding experiments with P-splines. Spat. Stat. 2018, 23, 52–71. [Google Scholar] [CrossRef]
- Lang, S.; Brezger, A. Bayesian P-splines. J. Comput. Graph. Stat. 2004, 13, 183–212. [Google Scholar] [CrossRef]
- Brezger, A.; Steiner, W.J. Monotonic regression based on bayesian p–splines: An application to estimating price response functions from store-level scanner data. J. Bus. Econ. Stat. 2008, 26, 90–104. [Google Scholar] [CrossRef]
- Bremhorst, V.; Lambert, P. Flexible estimation in cure survival models using Bayesian P-splines. Comput. Stat. Data Anal. 2016, 93, 270–284. [Google Scholar] [CrossRef][Green Version]
- Aldrin, M. Improved predictions penalizing both slope and curvature in additive models. Comput. Stat. Data Anal. 2006, 50, 267–284. [Google Scholar] [CrossRef]
- Bollaerts, K.; Eilers, P.H.; Van Mechelen, I. Simple and multiple P-splines regression with shape constraints. Br. J. Math. Stat. Psychol. 2006, 59, 451–469. [Google Scholar] [CrossRef]
- Simpkin, A.; Newell, J. An additive penalty P-Spline approach to derivative estimation. Comput. Stat. Data Anal. 2013, 68, 30–43. [Google Scholar] [CrossRef]
- Perperoglou, A.; Eilers, P.H. Penalized regression with individual deviance effects. Comput. Stat. 2010, 25, 341–361. [Google Scholar] [CrossRef]
- Wood, S.N. Thin plate regression splines. J. R. Stat. Soc. Ser. B Stat. Methodol. 2003, 65, 95–114. [Google Scholar] [CrossRef]
- Ypma, T.J. Historical development of the Newton–Raphson method. SIAM Rev. 1995, 37, 531–551. [Google Scholar] [CrossRef]
- Hardy, T.; Wonders, K.; Younes, R.; Aithal, G.P.; Aller, R.; Allison, M.; Bedossa, P.; Betsou, F.; Boursier, J.; Brosnan, M.J.; et al. The European NAFLD Registry: A real-world longitudinal cohort study of nonalcoholic fatty liver disease. Contemp. Clin. Trials 2020, 98, 106175. [Google Scholar] [CrossRef] [PubMed]
- Nelder, J.A.; Wedderburn, R.W. Generalized linear models. J. R. Stat. Soc. Ser. A Stat. Soc. 1972, 135, 370–384. [Google Scholar] [CrossRef]
- Ramspek, C.L.; Jager, K.J.; Dekker, F.W.; Zoccali, C.; van Diepen, M. External validation of prognostic models: What, why, how, when and where? Clin. Kidney J. 2021, 14, 49–58. [Google Scholar] [CrossRef]
- Al-Taie, W.A.; Farrow, M. Bayes Linear Bayes Networks with an Application to Prognostic Indices. Bayesian Anal. 2023, 18, 437–463. [Google Scholar] [CrossRef]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9. [Google Scholar]
- Hinton, G.E.; Roweis, S. Stochastic neighbor embedding. Adv. Neural Inf. Process. Syst. 2002, 15. [Google Scholar]
- Larson, S.C. The shrinkage of the coefficient of multiple correlation. J. Educ. Psychol. 1931, 22, 45. [Google Scholar] [CrossRef]
Figure 1.
Vertical Data, and Horizontal Data, .
Figure 1.
Vertical Data, and Horizontal Data, .
Figure 2.
Comparison between linear, polynomial, and spline fits to true function upon bivariate data. Green = linear function/red = polynomial function/black = B-Spline function/orange = true function.
Figure 2.
Comparison between linear, polynomial, and spline fits to true function upon bivariate data. Green = linear function/red = polynomial function/black = B-Spline function/orange = true function.
Figure 3.
B-Splines fitted with varying number of knots. Orange line represents true function, black line represents fitted B-Spline.
Figure 3.
B-Splines fitted with varying number of knots. Orange line represents true function, black line represents fitted B-Spline.
Figure 4.
P-Splines fitted with varying magnitude of penalty term. Orange line represents true function, black line represents fitted P-Spline.
Figure 4.
P-Splines fitted with varying magnitude of penalty term. Orange line represents true function, black line represents fitted P-Spline.
Figure 5.
Perspective plot of fitted relationships between and y as model structure varies. (Left): (interaction), (Right): (no interaction).
Figure 5.
Perspective plot of fitted relationships between and y as model structure varies. (Left): (interaction), (Right): (no interaction).
Figure 6.
True marginal for the two model structures. (Left): (interaction), (Right): (no interaction).
Figure 6.
True marginal for the two model structures. (Left): (interaction), (Right): (no interaction).
Figure 7.
3D plot of all three model fits and the true function. From left to right: True function, Fit0, Fit1, Fit2.
Figure 7.
3D plot of all three model fits and the true function. From left to right: True function, Fit0, Fit1, Fit2.
Figure 8.
Blue = estimated marginal /Red = true marginal . From left to right: Fit0, Fit1, Fit2.
Figure 8.
Blue = estimated marginal /Red = true marginal . From left to right: Fit0, Fit1, Fit2.
Figure 9.
Comparison between (Red) and (Blue) for each model fit and each dimensionality reduction: Linear Predictor (top row)/PCA (middle row)/tSNE (bottom row).
Figure 9.
Comparison between (Red) and (Blue) for each model fit and each dimensionality reduction: Linear Predictor (top row)/PCA (middle row)/tSNE (bottom row).
Table 1.
Sum of squares for model fits upon a single data set where and the model structure is such that there is an interaction between x and z. Bold indicates best value for each row.
Table 1.
Sum of squares for model fits upon a single data set where and the model structure is such that there is an interaction between x and z. Bold indicates best value for each row.
| | Fit0 | Fit1 | Fit2 |
|---|
| SS Fitted Values | 21.72 | 8.98 | 8.79 |
| SS Marginal Values | 0.50 | 0.52 | 0.27 |
Table 2.
Mean average sum of squares of fitted and marginal values for each model fit as model structure, sample size, and noise are varied (100 simulations). Bold indicates best value for each row and SS metric.
Table 2.
Mean average sum of squares of fitted and marginal values for each model fit as model structure, sample size, and noise are varied (100 simulations). Bold indicates best value for each row and SS metric.
| Interaction | | | Fit0 SS(Fitted) | Fit1 SS(Fitted) | Fit2 SS(Fitted) | Fit0 SS(Marg) | Fit1 SS(Marg) | Fit2 SS(Marg) |
|---|
| Yes | 100 | 0.2 | 6.55 | 4.74 | 4.71 | 0.94 | 0.98 | 0.73 |
| 0.5 | 19.95 | 7.73 | 7.34 | 1.71 | 1.51 | 0.74 |
| 1.0 | 70.26 | 15.41 | 13.20 | 4.93 | 3.53 | 0.76 |
| Yes | 400 | 0.2 | 20.64 | 9.07 | 8.99 | 0.32 | 0.40 | 0.22 |
| 0.5 | 88.66 | 13.18 | 12.53 | 0.56 | 0.67 | 0.20 |
| 1.0 | 333.85 | 23.46 | 20.32 | 1.34 | 1.22 | 0.16 |
| No | 100 | 0.2 | 0.68 | 0.54 | 0.45 | 0.81 | 0.95 | 0.53 |
| 0.5 | 4.23 | 3.04 | 2.08 | 1.89 | 2.13 | 0.58 |
| 1.0 | 16.84 | 10.47 | 5.88 | 4.77 | 4.73 | 0.48 |
| No | 400 | 0.2 | 1.56 | 0.75 | 0.68 | 0.75 | 0.83 | 0.67 |
| 0.5 | 8.98 | 3.43 | 2.67 | 0.90 | 1.10 | 0.64 |
| 1.0 | 37.76 | 11.65 | 8.19 | 1.95 | 2.35 | 0.62 |
Table 3.
Mean average weighted sum of squares of fitted values for each model fit as model structure, sample size, and number of knots are varied (100 simulations). Bold indicates best value for each row and SS metric.
Table 3.
Mean average weighted sum of squares of fitted values for each model fit as model structure, sample size, and number of knots are varied (100 simulations). Bold indicates best value for each row and SS metric.
| Interaction | | | nrep | Fit0 WSS(Fitted) | Fit1 WSS(Fitted) | Fit2 WSS(Fitted) | Fit0 WSS(Marg) | Fit1 WSS(Marg) | Fit2 WSS(Marg) |
|---|
| No | 100 | 4 | 4 | 16.76 | 11.86 | 9.22 | 1.37 | 1.32 | 0.99 |
| 100 | 8 | 4 | 16.50 | 10.34 | 8.66 | 1.14 | 1.21 | 0.83 |
| 400 | 8 | 2 | 17.75 | 12.85 | 9.26 | 0.85 | 1.00 | 0.49 |
| 400 | 18 | 2 | 37.99 | 12.32 | 9.13 | 0.77 | 0.90 | 0.47 |
| 900 | 8 | 1 | 17.94 | 12.62 | 9.14 | 0.76 | 0.88 | 0.46 |
| Yes | 100 | 8 | 8 | 83.63 | 32.44 | 30.79 | 0.85 | 0.72 | 0.34 |
| 400 | 8 | 2 | 74.55 | 22.98 | 20.71 | 0.65 | 0.56 | 0.24 |
| 900 | 8 | 1 | 73.20 | 22.48 | 20.18 | 0.54 | 0.45 | 0.21 |
Table 4.
Characteristics of and data sets within the LITMUS Metacohort.
Table 4.
Characteristics of and data sets within the LITMUS Metacohort.
| Dataset | N | p | Y = 0 | Y = 1 |
|---|
| 6024 | 19 | 4014 | 2010 |
| 1456 | 19 + 37 | 860 | 596 |
Table 5.
Complete results for modelling upon the LITMUS Metacohort. Bold indicates best results for each row.
Table 5.
Complete results for modelling upon the LITMUS Metacohort. Bold indicates best results for each row.
| Dim. Reduction | Interaction | Determination | Determination | | | Fit 0 SS(Marg) | Fit 1 SS(Marg) | Fit 2 SS(Marg) |
|---|
| Linear Predictor | No | SS | 50% Improvement | 4 | 1.2 | 23.80 | 22.94 | 11.32 |
| SS | Best Fit2 | 4 | 2.2 | 23.80 | 22.94 | 7.66 |
| Log-likelihood | 50% Improvement | 3 | 1.2 | 23.80 | 22.80 | 11.12 |
| Log-likelihood | Best Fit2 | 3 | 2.2 | 23.80 | 22.80 | 7.39 |
| AUC | 50% Improvement | 6 | 1.3 | 23.80 | 23.16 | 11.11 |
| AUC | Best Fit2 | 6 | 2.3 | 23.80 | 23.16 | 7.70 |
| Linear Predictor | Yes | SS | 50% Improvement | 4 | NA | - | - | - |
| SS | Best Fit2 | 4 | 2.4 | 24.69 | 25.83 | 13.42 |
| Log-likelihood | 50% Improvement | 3 | NA | - | - | - |
| Log-likelihood | Best Fit2 | 3 | 2.4 | 24.69 | 25.66 | 13.09 |
| AUC | 50% Improvement | 6 | NA | - | - | - |
| AUC | Best Fit2 | 6 | 2.4 | 24.69 | 26.03 | 13.76 |
| PCA | No | SS | 50% Improvement | 2 | 1.0 | 42.37 | 33.85 | 16.30 |
| SS | Best Fit2 | 2 | 2.0 | 42.37 | 33.85 | 9.81 |
| Log-likelihood | 50% Improvement | 2 | 1.0 | 42.37 | 33.85 | 16.30 |
| Log-likelihood | Best Fit2 | 2 | 2.0 | 42.37 | 33.85 | 9.81 |
| AUC | 50% Improvement | 2 | 1.0 | 42.37 | 33.85 | 16.30 |
| AUC | Best Fit2 | 2 | 2.0 | 42.37 | 33.85 | 9.81 |
| PCA | Yes | SS | 50% Improvement | 3 | 1.1 | 36.20 | 35.07 | 17.49 |
| SS | Best Fit2 | 3 | 2.0 | 36.20 | 35.07 | 12.12 |
| Log-likelihood | 50% Improvement | 2 | 1.1 | 36.20 | 34.85 | 17.28 |
| Log-likelihood | Best Fit2 | 2 | 2.0 | 36.20 | 34.85 | 11.87 |
| AUC | 50% Improvement | 1 | 1.1 | 36.20 | 34.55 | 16.91 |
| AUC | Best Fit2 | 1 | 2.0 | 36.20 | 34.55 | 11.48 |
| tSNE | No | SS | 50% Improvement | 3 | 1.4 | 46.66 | 36.29 | 17.54 |
| SS | Best Fit2 | 3 | 2.0 | 46.66 | 36.29 | 14.24 |
| Log-likelihood | 50% Improvement | 3 | 1.4 | 46.66 | 36.29 | 17.54 |
| Log-likelihood | Best Fit2 | 3 | 2.0 | 46.66 | 36.29 | 14.24 |
| AUC | 50% Improvement | 8 | 1.5 | 46.66 | 34.25 | 16.74 |
| AUC | Best Fit2 | 8 | 2.0 | 46.66 | 34.35 | 14.26 |
| tSNE | Yes | SS | 50% Improvement | 5 | NA | - | - | - |
| SS | Best Fit2 | 5 | 2.0 | 48.59 | 53.35 | 30.19 |
| Log-likelihood | 50% Improvement | 5 | NA | - | - | - |
| Log-likelihood | Best Fit2 | 5 | 2.0 | 48.59 | 53.35 | 30.19 |
| AUC | 50% Improvement | 0 | 2.1 | 48.59 | 48.59 | 21.00 |
| AUC | Best Fit2 | 0 | 2.1 | 48.59 | 48.59 | 21.00 |
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).