Abstract
In the dynamic landscape of healthcare, decision support systems (DSS) confront continuous challenges, especially in the era of big data. Background: This study extends a Q&A-based medical DSS framework that utilizes semantic technologies for disease inference based on a patient’s symptoms. The framework inputs “evidential symptoms” (symptoms experienced by the patient) and outputs a ranked list of hypotheses, comprising an ordered pair of a disease and a characteristic symptom. Our focus is on advancing the framework by introducing ontology integration to semantically enrich its knowledgebase and refine its outcomes, offering three key advantages: Propagation, Hierarchy, and Range Expansion of symptoms. Additionally, we assessed the performance of the fully implemented framework in Python. During the evaluation, we inspected the framework’s ability to infer the patient’s disease from a subset of reported symptoms and evaluated its effectiveness in ranking it prominently among hypothesized diseases. Methods: We conducted the expansion using dedicated algorithms. For the evaluation process, we defined various metrics and applied them across our knowledge base, encompassing 410 patient records and 41 different diseases. Results: We presented the outcomes of the expansion on a toy problem, highlighting the three expansion advantages. Furthermore, the evaluation process yielded promising results: With a third of patient symptoms as evidence, the framework successfully identified the disease in 94% of cases, achieving a top-ranking accuracy of 73%. Conclusions: These results underscore the robust capabilities of the framework, and the enrichment enhances the efficiency of medical experts, enabling them to provide more precise and informed diagnostics.
Keywords:
knowledge graph; semantic reasoning; medical diagnostic; decision support systems; semantic technology MSC:
68T35
1. Introduction
The Industry 4.0 Standard incorporates automation and data exchange technologies across various domains, including cloud computing, big data, and database design. These information and communication technologies are reshaping services and production, particularly in the health domain. The integration of Internet of Things, Cloud Computing, and big data is revolutionizing eHealth, giving rise to Healthcare 4.0 [1,2]. Healthcare 4.0 addresses the significant challenges of expanding, virtualizing, and facilitating new healthcare processes such as home care, precision medicine, and personalized/remote pharmaceutical treatments [3]. Healthcare poses a critical social and economic challenge globally, with administrators, clinicians, researchers, and practitioners facing increasing pressure from rising expectations in both the public and private sectors [1]. Healthcare 4.0 reflects the trend of offering technological solutions to challenges posed by the medical realm [4]. Specifically, we are tasked with leveraging semantic technologies grounded in big data and advanced algorithms [5].
In our ongoing research [6] within the medical domain, we are concentrating on medical decision support systems (MDSS) characterized by interactions between a medical expert and a patient. The goal is to empower the medical expert to assist the patient in addressing specific issues she faces [7]. These interactions consist of a series of iterations, each involving a question posed by the medical expert and an answer provided by the patient [8,9]. Each iteration advances the medical expert closer to a decision regarding the patient’s issue, usually, the decision will be formulated as a medical diagnosis. The nature of these interactions tends to be limited, often in terms of time, which may impact the ability to provide optimal diagnoses.
We utilize semantic technology to propose a framework that supports the above process: in each iteration, it suggests a question concerning a symptom the patient is experiencing. In the final round, it provides a list of ranked hypotheses (ordered pairs of a disease and a symptom indicating it) according to the likelihood the disease is indeed the patient’s diagnosis. The framework is based on a knowledge graph (KG for short), which has gained increasing popularity as a means to represent knowledge [10]. The KG is a natural method for depicting interconnected data [11,12,13], consisting of nodes categorized as symptoms or diseases, connected by edges linking a symptom to a disease if it characterizes it. On top of the KG, we formulated a set of interactive algorithms that employ both the knowledge graph and initial input from the patient to propose pertinent questions.
In this paper, we detail an expansion of the framework by enhancing the KG with semantic knowledge extracted from an ontology of symptoms (SYMP) [14] and their relationships. Relevant elements from the ontology, especially hierarchical structures, were integrated into the KG. The enhanced KG has expanded symptom representations and a hierarchical structure, offering several benefits for the inference process. These include a broader set of recommended questions for the medical expert in each iteration with the patient and provides additional evidence of symptoms [15]. We demonstrated the extension via toy problem and highlighted the advantages it brings.
The entire framework was implemented in Python, and we conducted various tests to assess its output and its effectiveness. Primarily, we were interested in knowing to what extent the framework can infer the patient’s disease, as a function of the number of evidence symptoms (i.e., symptoms that the patient experiences). In particular, we were interested in knowing if it succeeds in inferring where the patient’s disease is located within the list of hypotheses. Perfect success is defined as inferring the patient’s illness as the first hypothesis. The results were very encouraging. For example, when we provided one third of the patient’s symptoms as evidence symptoms, in 94% of cases the framework succeeded in including the patient’s disease, and in 73% of those it was the top-ranked hypothesis. An additional evaluation test examined the number of iterations required to find a high-ranking hypothesis. As expected, a small number of evidential symptoms entails more iterations of the framework.
The rest of the paper is organized as follows: Section 2 provides background and a literature review. In Section 3, we offer a brief overview of the framework introduced in our previous study, and in Section 4, we detail the evaluation of the framework. Section 5 elaborates on KG enrichment, covering the algorithms used, their implementation, effects, and benefits. Section 6 concludes the study with a discussion that also addresses future work.
2. Background and Prior Work
In this section, we provide a concise background on ontologies and semantic technologies (Section 2.1). We also review prior related works that share similar objectives with this research and utilize similar tools and technologies.
2.1. Background
2.1.1. Ontology
An ontology, as defined by T.R. Gruber, serves as an accurate and precise specification of a conceptualization that is machine-interpretable [16]. This definition encompasses the explicit detailing of entities or concepts, their attributes, and the relationships existing between them within a particular domain. Essentially, ontology provides a shared vocabulary for both humans and machines, facilitating seamless communication and information exchange [17].
The development of ontologies holds paramount importance for several reasons [18]. Firstly, it plays a pivotal role in fostering a unified understanding of information structures among individuals and software agents [19]. By establishing a standardized framework, ontologies enable a more coherent and consistent interpretation of data across diverse contexts. Secondly, ontologies contribute significantly to the efficient reuse of domain knowledge [20]. By encapsulating core concepts, attributes, and relationships within a particular field, ontologies provide a reusable foundation, streamlining the development process and ensuring consistency and accuracy in applying domain knowledge. Lastly, ontologies are instrumental in the analysis of domain knowledge [21]. They serve as powerful tools for comprehending the intricacies of a given subject area, allowing for a deeper exploration of relationships between entities and a more nuanced understanding of the underlying structure. This analytical aspect aids researchers, developers, and practitioners in making informed decisions and advancements within their respective domains.
2.1.2. Semantic Technology and Graph Reasoning
Knowledge graphs (KG) encode information by transforming data into a coded format, specifically by organizing relationships between entities into graph structures. KGs, also referred to as semantic graphs, capture the interest of both academic and industrial researchers across a range of fields that share the common need to represent knowledge [22]. KGs possess the characteristic of delivering semantically structured information. This attribute empowers KGs to provide innovative solutions for significant tasks, including addressing queries [23], developing recommendation systems [24], and enhancing information retrieval [25]. Knowledge graphs are also regarded as holding considerable potential for advancing the capabilities of intelligent machines, representing a promising avenue for developing more sophisticated technology.
2.2. Prior Works
A range of clinical decision support systems (CDSS) have been developed to aid medical professionals in diagnosing diseases based on patients’ symptoms. Jiang et al. [26] proposed a three-layer knowledge base model that improved the accuracy of disease predictions. Silva et ai. [27] utilized a Bayesian framework to construct a web-based system with high detection accuracy for general and complex diseases. Rahaman [28] focused on diabetes diagnosis, creating a user-interactive system based on symptoms, signs, and risk factors. Dong [29] et al. suggests employing a CDSS to enhance the precision of diagnosing headache disorders, while Tandra and his colleagues [30] propose a fuzzy-neuro-based CDSS for disease diagnosis.
In recent years, with the widespread adoption of semantic technology across various content domains, there has been a natural demand to extend its application to the medical field. Researchers employ semantic technologies in the realm of medicine to offer novel solutions for diverse needs and enhance existing responses [31]. In particular, we focus on medical diagnosis/decision support systems, whose primary goal, according to Moreira and his colleagues [32], is “to provide relevant data to the medical experts where and when it is needed”. There has been a rise in new diagnostic and support systems for medical decision making, utilizing knowledge graphs and/or ontologies, or enhancing existing systems by applying semantic web techniques such as ontologies/NLP [33].
Riaño and his colleagues [34] developed an ontology-powered decision support tool that provides personalized guidelines and recommendations tailored to the specific health needs of complex chronic patients to assist doctors at the point of care. Another work [35] utilized automated reasoning with an ontology knowledge base to generate diagnostics insights and care recommendations that aid mental health professionals in making data-driven clinical decisions. Santos and colleagues [36] introduced CLINPRO—a knowledge-graph-based interpreter for clinical proteomics data. CLINPRO enables the mapping of proteomic observations to tissue and disease contexts to derive insights that explain mechanisms underlying patient disease states. According to Dissanayake and his colleagues [37], who conducted a systematic review on the utilization of clinical reasoning ontologies to enhance the capabilities of CDSS, the design and implementation of more sophisticated and context-aware CDSSs contributed to advancements in healthcare informatics. The research conducted by Shanavas et al. [38] explores the utilization of ontologies to enhance concept graphs, providing insights into how this approach can contribute to more effective medical document classification.
We selected eight studies (including our own) that share a similar objective to ours (i.e., providing assistance and recommendations for patient diagnosis) and conducted a comparative analysis (see Table 1). The table comprises 11 columns. Column 3 offers a concise description of each study, while column 4 outlines the input for the system/framework. Column 5 details the interactions between the patient and the medical expert, occasionally involving multiple iterations. Column 6 describes the main technologies employed, column 7 specifies the output, and column 8 provides insights into the implementation methods. Columns 9 and 10 present details about the sample size used for assessment, and column 11 covers the evaluation method along with the key results.

Table 1.
Comparative analysis studies aiming to assist medical experts to infer a patient’s disease.
The optimal results were observed when the system was rule-based. However, such systems are, by nature, narrower in the range of explored diseases, and lack the dynamism seen in systems like ours, which dynamically respond to each interaction between the patient and the medical expert.
To conclude, some studies leverage semantic technologies to enhance the process of medical decision making. However, the number of works utilizing knowledge graphs in conjunction with ontologies, and exploiting their integration, is still limited.
3. The Framework
In this section, we provide a brief overview of the framework introduced in our previous study [6], outlining its constituent algorithms and the interplay between them.
Recall, we aim to engage in collaborative decision making. This involves an ongoing exchange (as limited as possible) between a domain expert and an end-user, where they share questions and answers. The framework contributes by proposing questions for the expert to ask the end-user. The progression of the decision making process hinges on the responses provided by the end-user.
As we focus on the medical domain, the questions and answers should relate to symptoms and diseases. Moreover, the ultimate objective of this process is to assist the medical expert, in arriving at a diagnosis—delivering an explanation for a specific set of symptoms characterizing the end-user, who is a patient in this case, through the analysis of available data in the KG. “Does the patient display a specific symptom?” is an example of a question that may emerge during the interaction between the medical expert and the patient. The output of the framework is a list of ranked hypotheses, each hypothesis is an ordered pair of a disease and a symptom indicating it. Therefore, our jargon contains the following terms: symptoms, diseases, and hypotheses.
The framework comprises two key segments: the initial phase, known as pre-processing, executed during the framework launch, and the subsequent phase, termed processing, activated with each new request.
The pre-processing phase involves creating a knowledge graph (KG) from raw data taken from Kaggle [39]. The dataset comprises patient records, where each record represents one patient. The records include the diagnosed disease for each patient, as well as the associated symptoms reported. In total, the dataset encompasses 41 distinct diseases and 130 distinct symptoms. Certain symptoms appear only once, indicating that they characterize a single disease, while other symptoms occur multiple times, suggesting they may be characteristic of several diseases. The dataset enables the exploration of the relationships between diseases and symptoms.
The KG nodes are the symptoms and the diseases, and the KG edges are the relations between them (if a symptom indicates a disease, there is an edge between these two nodes). We then use the Louvain hierarchical clustering algorithm [40] on the KG to find clusters of diseases (named communities) that have similar symptoms (Algorithm 1 in [6]).
The processing phase is conducted whenever a new medical expert-patient interaction starts, hence, a new patient presents a set of symptoms (named evidence symptoms or ES for short). During the interaction, the framework executes a set of inference algorithms, which use the communities to determine which diseases are compatible with the symptoms reported by the patient. In particular, Algorithm 2 in [6] finds the most probable diseases (i.e., the possible diseases that are compatible with evidence symptoms); Algorithm 3 in [6] repeatedly, as needed, infers and suggests a question to the medical expert (i.e., a symptom) that indicates the most probable community to include the patient’s disease; finally, the processing phase concludes by inferring and outputting, through Algorithm 4 in [6], a list of hypotheses (i.e., ordered pairs of a disease and a symptom indicating it) that the patient might have, sorted by relevance.
Figure 1 illustrates the interactions within the framework among the patient, the medical expert, and the KG, during the processing phase.
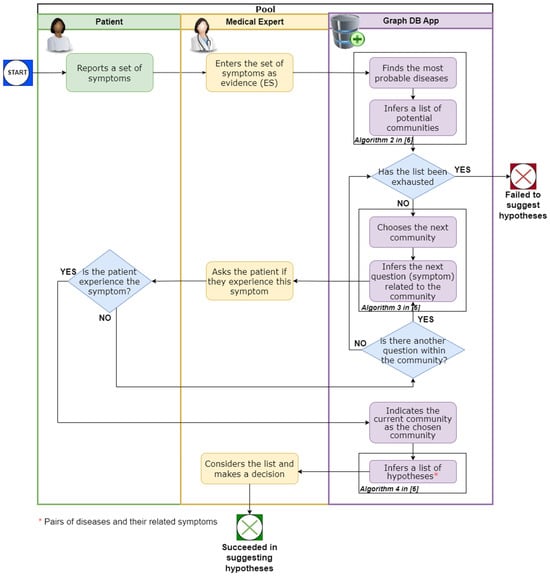
Figure 1.
The interactions within the framework among the patient, the medical expert, and the KG during the processing phase.
4. Framework Evaluation
In this section, we review the implementation details of the framework, describe the dataset we used to evaluate the framework’s capabilities, present the evaluation measures that we defined, and the outcome of running these measures on the dataset. We focus on detecting the patient’s disease, particularly on where this disease is positioned in the ranked list of possible diseases, output by the framework. The objective is for the true disease to be positioned as high as possible. We assess this capability across three different data segments, as explained below.
4.1. Implementation Details
The framework implementation includes two main parts. The first one is the KG construction, as described in Section 3. The KG was constructed using Neo4j Graph Database, Version 5. The second part includes the framework algorithms, which were developed in Python. To apply the algorithms on the KG, the Neo4j Python Driver (https://neo4j.com/developer/python/, accessed on 30 January 2024) was used.
4.2. DataSet Description
To evaluate the developed framework, we used a dataset [39] of 410 patient records. Each record referred to one patient and includes the name of the disease the patient was diagnosed with and the symptoms the patient was experiencing (between 3 and 17 symptoms).
4.3. Applying the Framework on the Dataset
For each patient record, we performed the following: we input a subset of the patient’s symptoms as evidence symptoms (i.e., the set of symptoms the patient experiences and has reported). Then, we executed the processing phase, which output a list of hypotheses. Finally, we evaluated these hypotheses against the real disease diagnosed for the patient, using its other symptoms for the interaction phase (as explained in Section 3, Algorithm 3 in [6]). The subset of the patient’s symptoms was determined using a threshold variable , which was set to be a different fraction at each run (. In this way, we were able to compare the influence of the evidence symptoms group on the diagnosis detection, and the influence on the number of iterations until reaching the right diagnostic. The next subsection describes the evaluation results, for each one of the thresholds.
4.4. Evaluation Measures
As stated, we assess the performance of the proposed framework, focusing on its capability to detect the patient’s disease—particularly its position in the ranked list of potential diseases generated by the framework. The goal is to have the true disease ranked as high as possible. We evaluate this capability in three distinct segments (i.e., , as detailed in the previous subsection.
Let H be the hypotheses output list of the framework. Recall that H is sorted by the hypothesis’s relevance to the patient. Let be the disease diagnosed for the patient (the ground truth). Let be the position of the disease in H ( = if the disease does not exist in H). Note that small values of rank indicate a better prediction of the framework. Informally, we are interested in the position of the true patient’s disease in the list of hypotheses output by the framework. The optimal scenario is when the true disease is ranked at the top of the list of hypotheses.
Consider the following example of a patient’s record, and the rank achieved by applying the framework on a of its symptoms. The patient’s record is: [Urinary tract infection; burning micturition, foul smell of urine, continuous feel of urine]. The first term (Urinary tract infection) is the disease of the patient, and the other three terms are three symptoms the patient has experienced. For the evaluation test, with a threshold of a , (i.e., of three symptoms is one symptom), we randomly chose the burning micturition symptom to serve as evidence. After inserting it as an input, the framework outputs the following ranked list of six hypotheses: {(Urinary tract infection, continuous feel of urine); (Urinary tract infection, bladder discomfort); (Drug Reaction, stomach pain); (Drug Reaction, spotting urination); (Drug Reaction, itching), (Drug Reaction, skin rash)}. Note that in this test case, the patient’s true disease is urinary tract infection, which is positioned as the first hypothesis in the list, i.e., the rank is defined as 1, (the top rank), which is the best rank.
Our assessments include several steps. First, for every patient , we checked where the patient’s diagnosis () is located within this list of hypotheses (that is, weather ). Out of 410 patients, by setting (that is, having only a third of the patient symptoms as evidence symptoms) for only 23 patients, the list of hypotheses did not include the patient’s diagnosis, i.e., in 94% of cases, the framework succeeded in inferring and including the patient’s true disease in the output ranked list. With a softer threshold (), for only 10 patients out of 410, the list of hypotheses did not include the patient’s diagnosis (i.e., 98% success). Then, we examined how many patients there are for each rank (that is, a histogram of the patients’ rank). The results are presented in Figure 2. Note that even with a of the patient’s symptoms, the framework infers the patient’s true disease as the top ranked hypothesis in more than 73% of the patients.
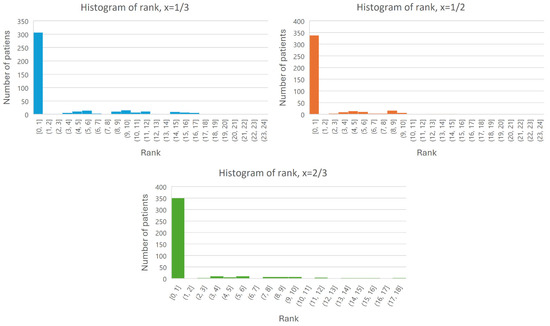
Figure 2.
Histogram of the rank of each patient (the rank is on the X-axis, and the number of patients is on the Y-axis).
Figure 3 presents the rank of each patient, for all three thresholds (). As expected, when applying a smaller threshold, it is harder to detect the patient’s disease, and the rank of the patient is smaller overall. In practice, it means that receiving the same diagnosis will take longer.
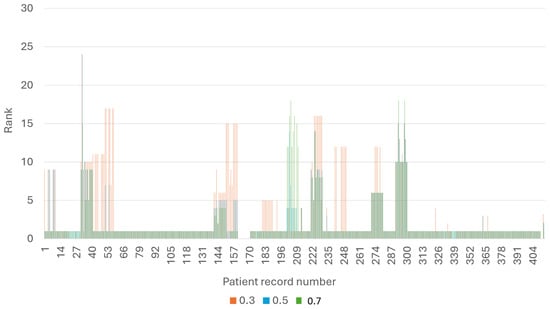
Figure 3.
The rank of each patient, for every threshold.
However, as illustrated in Figure 3, this is not the case for patients 201–211. Upon closer examination, we discovered that these patients have Hepatitis B, while the framework outputs Hepatitis E as the primary diagnosis. Upon a more in-depth analysis of the knowledge graph (refer to Figure 4) for these two diseases, it becomes evident that examining them is challenging due to the shared symptoms. In such cases, additional knowledge is necessary, such as ranking the symptoms based on their level of indication, if this information is available.
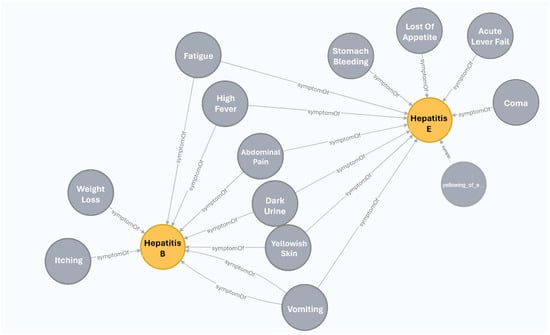
Figure 4.
A subgraph of the KG including hepatitis B and hepatitis E diseases, and their symptoms.
We also examined the number of hypotheses received in each of the three thresholds. The results are presented in Figure 5. Note that the number of hypotheses for each patient is similar when applying different thresholds, and as expected there are more hypotheses when using fewer evidence symptoms (ES) since there are more symptoms left as unknowns.
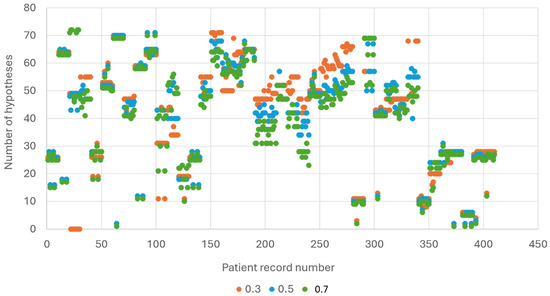
Figure 5.
Number of hypotheses received in each of the three thresholds.
Finally, we examined the number of iterations required to find a high-ranking hypothesis, for each threshold. As seen in Figure 6, and as expected, when inputting a small number of evidence symptoms to the framework, achieving improved ranking results demands more iterations.
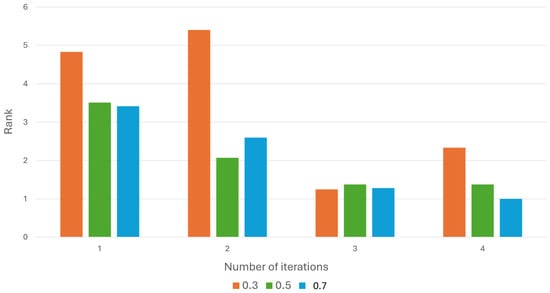
Figure 6.
The average rank obtained with every number of iterations.
5. Enriching the Framework with Semantic Technology
In this section, we introduce semantic expansion, its associated algorithms, and the additional value it offers. Specifically, Section 5.1 delves into the Enrichment Algorithms, while Section 5.2 provide the implementation details of these algorithms. In Section 5.3, we proceed to explore the added value of KG enrichment, leading to the creation of the enhanced KG, and conclude with a description of a toy problem to illustrate the points raised.
Let be a directed graph, which is defined as follows. Let be the set of nodes, where is the set of diseases and S is the set of symptoms. The edges of the graph are defined as follows: E = , that is, there is an edge from a symptom s to disease d if s might indicate d.
The KG was constructed based on a historical examination of relevant domain experts, and it serves as data-driven knowledge (Figure 7A illustrates an example of such a KG). The knowledge contained within the KG is limited in nature and lacks the classic hierarchical structure of symptoms and diseases. Integrating ontology elements into the KG expands it and semantically enriches the KG. This integration facilitates the inference of new symptoms as evidence, leading to the deduction of additional relevant diseases for the domain expert.
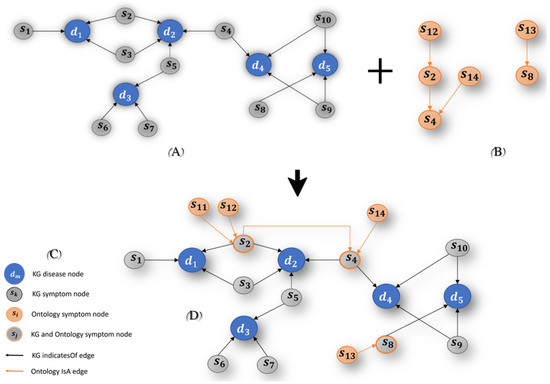
Figure 7.
The Ontology integration. (A) illustrates an example of a KG, (B) illustrates an example of a structured hierarchically Ontology, and (C) A illustrates an example of an enhanced KG.
5.1. Knowledge Graph Enhancement
The first step for enhancing the KG is to explore ontologies with relevant domains, and then integrate them. The second step is to use the enhanced KG to improve the inference in it and by that improve the framework. In this research, we chose to use the Symptoms Ontology (SYMP) [14] since it contains terms that are relevant to the KG domain and can naturally enrich it. The SYMP ontology is structured hierarchically, comprising nodes and edges. Each node represents a symptom, and each edge signifies the inheritance relation (ISA) between two symptoms (refer to Figure 7B for an example).
The SYMP ontology was integrated into the KG according to the following algorithms. The first Algorithm 1 adds the relevant symptom nodes to the KG. The second Algorithm 2 adds ISA relations to the KG.
| Algorithm 1: Add Symptom Nodes to the KG |
| Input: KG, SYMP |
| Output: KG |
| Algorithm: |
| For all edges ) in SYMP, such that and |
| Add as a symptom node to KG. |
| Algorithm 2: Add ISA Relations between Symptoms in the KG, according to the Ontology |
| Input: KG, SYMP |
| Output: enhanced KG |
| Algorithm: |
| For all edges ) in and |
| Add the edge to KG, labeled ISA. |
Figure 7 illustrates an example of integrating an ontology (B) into an existing KG (A), generating a semantic technology platform (C), which we named enhanced KG. Note that Figure 7D defines the legend for the symbols used in Figure 7A–C.
The resulting KG, that is, the enhanced KG, contains three symptom nodes:
- (i)
- An “original” KG symptom node, named as KG symptom node: these nodes appeared in the KG before the enhancement, and are directly connected to disease nodes, via relation (for example, see node s1 in Figure 7C).
- (ii)
- New ontological symptom node, named ontology symptom node: these are SYMP ontology nodes, which were added by Algorithm 1. These nodes are directly connected to the KG symptom node via relation, according to Algorithm 2 rules (for example, see node s11 in Figure 7C).
- (iii)
- A node that is both “original” and ontological, named a hybrid symptom node: these nodes are directly connected to a KG disease node (via relation) and to some other hybrid node or ontology symptom node (via relation). For example, see node s2 in Figure 7C.
Similarly, the enhanced KG has two types of relations:
- (i)
- An edge between a KG node to a disease node it indicates, named as KG edge.
- (ii)
- An edge between the ontology symptom node or hybrid node to its parent node (which can be an ontology symptom node or hybrid node), named an ontology edge.
Figure 7C ilustrures both types.
5.2. Implementing Algorithms 1 and 2
In this subsection, we present an overview of the stage involving the identification of matching symptoms, a crucial step for implementing Algorithms 1 and 2 (as discussed in the previous subsection) responsible for generating the enhanced KG.
5.2.1. Overview
A necessary step in implementing Algorithm 1 and Algorithm 2 is identifying the symptoms, denoted as s, which exist in both the KG and SYMP (as detailed in the next subsection). Subsequently, the sub-tree rooted by s is integrated into the KG. For instance, the KG symptom also exists in SYMP (as ). Consequently, the subtree rooted by was integrated into the KG (nodes integrated by Algorithm 1, and edges by Algorithm 2). Figure 8, which is a Neo4j screenshot, illustrates the creation of the symptom node along with its edge pointing to GERD disease and with its descendant (e.g., ontology node), which is connected via edge to its parent. Note that ontology node has a descendant as well, namely the ontology node. The associated Neo4j commands establishing these nodes and edges can be found in Appendix A.
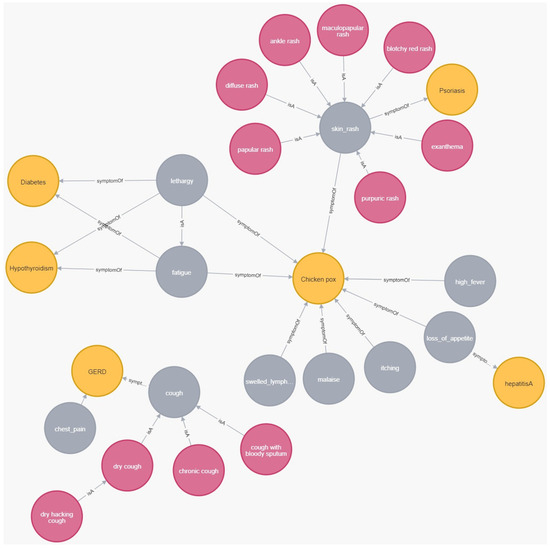
Figure 8.
An example of integrating a hierarchical tree of symptoms into the KG. Disease nodes are represented in gray, symptom nodes in yellow, and ontology nodes in red.
5.2.2. Identifying Matching Symptoms
This step arises when natural language is involved since there is more than one way to describe a symptom. To detect matching symptoms, we examined all 128 symptoms within the KG, such that for each symptom, we conducted a search using different similarity methods (substring, Levenshtein distance, exploring synonymous) in the SYMP ontology. For some instances, the match was one-to-one, meaning that in both the KG and the SYMP, the symptom had the same name (e.g., in the KG, the symptom was named identical to the ontology symptom named ). In other cases, the symptom names were similar but not identical (e.g., in the KG and in the ontology). More rarely, the names were entirely different but represented synonymous concepts, which were found by using synonymous terms for the relevant symptoms (e.g., in the ontology, and in the KG).
Out of the 128 symptoms in the KG, 51 symptoms had no corresponding match in the SYMP ontology. Additionally, 44 symptoms had a match, but no hierarchical tree was rooted under them. Another 4 symptoms had a match, but it was already assigned to a different hierarchical tree. Finally, 26 symptoms had a match with a hierarchical tree rooted under them, and these trees were integrated into the KG. Overall, 21 new ontological nodes were added to the KG, along with 22 ontological edges (i.e., ISA edges).
5.3. Inference in the Enhanced KG: Demonstrating via a Toy Problem
In this subsection, we highlight the advantages of this approach and demonstrate the process of inference within the Enhanced KG using a toy problem.
The enhanced KG expands in both symptom representation and hierarchical structure, providing several advantages for the inference process:
- (i)
- Evidence Propagation: Evidence symptoms (ES) can propagate through the edges of the graph, providing additional evidence, hence increasing the number of ES. This process has the potential to discover new diseases and expand the number of possible diseases for the patient.
- (ii)
- Symptoms Hierarchy Impact: Incorporating the symptoms hierarchy, along with the given ES, can indicate which community is more likely to be considered, especially in cases where multiple communities have equal LIND (LIND (=Local-in-Degree) of a given community c, is defined by the number of edges that point to diseases of c, by ES) scores.
- (iii)
- Expansion of Symptoms Range:
- ◯
- Increasing the number of hypotheses presented to the medical expert.
- ◯
- Facilitating a broader coverage of potential patient symptoms through the utilization of natural language processing (NLP) techniques (see further details in Section 6 where we discuss future work).
To demonstrate these capabilities, consider the following toy problem scenario exhibited in Figure 9. The figure presents the enhanced KG that was previously presented in Figure 7. The enhanced KG consists of 5 disease nodes (d1, d2, d3, d4, d5), and 14 symptom nodes, part of which are KG symptom nodes (s1, s3, s5, s6, s7, s9, s10), ontology symptoms nodes (s11, s11, s13, s14), and hybrid symptom nodes (s2, s4, s8). After executing the clustering algorithm (i.e., Algorithm 1 in [6]), three communities were created: C1, C2, and C3 as presented in the figure.
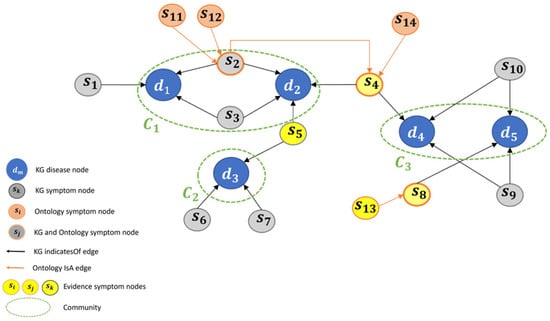
Figure 9.
Enhanced KG: toy problem illustration.
Recall, the creation of the enhanced KG and its related communities is part of the pre-processing phase. The processing part starts each time for each patient. Let us assume that the patient in our scenario reports on the symptoms: s4, s5, and s13. Thus, these are the evidence symptoms (ES). In addition, due to the ISA relationship (s13 ISA s8), s8 becomes ES as well because of evidence propagation, as mentioned in point #1. Thus, the final set of ES includes {s4, s5, s8 and s13}. As a result, the pool of possible diseases expands to include {d2, d3, d4, d5}, as s5 d2 and d3, s4 d4, and s8 indicates d5 (first part of Algorithm 2 in [6]).
Each community is then assessed along with its LIND. Consequently, both communities C1 and C3 exhibit an identical LIND of 2, whereas the LIND of C2 is 1. This implies that C1 and C3 are more likely to encompass the patient’s disease. At this stage, the algorithm chooses randomly between C1 and C3. However, this random choice now changes since the enhanced KG contains additional new information, regarding the interconnections between symptoms. In particular, according to point #2, the hierarchy of symptoms of s4, which is an evidence symptom, increases the possibility that one of its descendant symptoms (i.e., s2, s11, s12, and s14) is also an evidence symptom (ES). This knowledge must be considered; thus, the impact of these four symptoms on both communities is examined. It appears that community C1 is strengthened more than community C2, since these four symptoms (s2, s11, s12, s14) point to C1, while only symptom s4 points to community C2. To conclude, C1 is the most probable community to include the patient’s disease (last part of Algorithm 2 in [6]).
At this stage (Algorithm 3 in [6]), the framework infers and suggests to the medical expert a question (=symptom) that will strengthen the choice of C1. The algorithm will choose one of the three symptoms s2, s11, and s12, since, as mentioned, they point to C1 and are descendants of an evidence symptom (s4). It can be seen that enriching the KG increased the range of symptoms that can be suggested to the medical expert.
The rest of the interaction depends on the patient’s answer: if the patient exhibits the suggested symptom, the next step of the framework involves exploring hypotheses that include diseases from C1, along with additional symptoms indicating them. These are then suggested to the medical expert, ranked by relevance (Algorithm 4 in [6]). With the integration of ontology elements, the expanded pool of symptoms allows for an increased number of hypotheses, as we have stated in point #3. Otherwise, if the patient does not exhibit the suggested symptom, the framework recommends the next possible symptom within the current community (C1), or if no possible symptoms remain within C1, select the next community to explore (C2), as demonstrated in Figure 1.
Let us recap the description of the toy problem and underscore the added value of ontology enrichment:
When a patient meets with a medical expert and presents 3 symptoms, the framework will increase the number of ES to 4 (compared to 3 in the framework without enrichment), as per Evidence Propagation. Consequently, the number of possible diseases increases to 4 (as opposed to 3 without enrichment).
In the subsequent stage, the framework chooses the community with the highest likelihood of encompassing the patient’s disease. Without enrichment, communities C1 and C3 receive identical scores. However, with enrichment (attributed to the hierarchy of symptoms), community C1 gains precedence over community C2. Next, the framework will present the medical expert with symptoms not previously in the KG but now integrated into the added hierarchic structures.
Finally, the framework will furnish the specialist (according to Expansion of Symptoms Range) with an expanded list of hypotheses (more extensive than without enrichment), encompassing diseases and their indicated symptoms, potentially specifying the patient’s disease.
6. Discussion and Future Work
Over the last decade, researchers have explored the utilization of big data analytics in biomedicine and healthcare, with a focus on areas such as public health, the medical Internet of Things, personalized medicine, medical training, and clinical decision making [41,42]. Focusing on the last, our primary objective centers on interaction-driven decision-making processes, where dynamic interplay occurs between a medical expert and a patient. In this process, the two parties interact with each other through a series of questions and answers to address a disease faced by the patient. To assist the medical expert in formulating a medical diagnosis, we proposed a framework that suggests a list of hypotheses, all inferred from the medical–patient interaction of questions and answers. The framework utilizes a knowledge graph (KG) and a set of algorithms to create hypotheses, where each hypothesis comprises an ordered pair of a disease and an associated symptom.
In the current follow-up study, similar to other researchers [15], we evaluated our framework and examined its applicability and effectiveness via several defined measures, after we fully implemented it. As other researchers clarified [43,44], there is a challenge in the implementation, assessment, and integration of medical frameworks utilizing AI tools. As our framework shares a common objective with other systems (i.e., focusing on assisting and providing recommendations for patient diagnosis), a comparison is required (see Section 3). However, our output differs somewhat from other systems, as we do not provide probabilities for the presence of specific diseases. Instead, our framework suggests that the medical expert explores the ranked list of hypotheses. Still, the results of the comparison are encouraging.
Additional contribution presented in the paper, is the enrichment of the KG, generated by integrating ontological elements (taken from a symptoms ontology [14]) and creating hierarchical structures within the knowledge graph. The KG enrichment strengthens the framework on three dimensions: (1) Evidence Propagation, which expands the set of evidence symptoms, thereby enabling the framework to suggest more hypotheses; (2) Symptoms Hierarchy Impact integration can refine the list of communities with the highest likelihood of containing the patient’s illness, especially in cases where there are several communities with the same score; and (3) Expansion of Symptoms Range to increase the number of hypotheses suggested to the medical expert.
The contribution of the current work is manifested as follows: (1) feasibility testing of the framework presented in our previous work. We believe that our framework is innovative because of its usage of semantic technologies along with advanced algorithms to enable the inference of big data; (2) expanding the framework through the use of additional semantic technologies to generate extra value for the expert and thereby the patient.
Our upcoming challenge involves integrating Natural Language Processing (NLP) into our algorithmic toolbox, particularly for the semantic-similarity process. This integration aims to aid medical experts in identifying additional symptoms, thereby providing further enhancement to the framework across the three dimensions previously outlined. The adoption of NLP in the medical field is on the rise, with researchers exploring these techniques to enrich the representation of clinical information in healthcare applications [45]. In addition, we wish our framework to generate an output that includes a graph illustrating the decision-making process, aiding the medical expert in following the process. In professional terms, we provide Explainable Artificial Intelligence (XAI), which has become necessary when using AI technologies [46,47]. Lastly, as mentioned in our earlier paper [6], we intend to explore the incorporation of weighted edges in the knowledge graph to signify the cost associated with each hypothesis.
Author Contributions
Conceptualization, D.B. and S.A.-K.; Formal analysis, D.B. and S.A.-K.; Investigation, D.B. and S.A.-K.; Methodology, D.B. and S.A.-K.; Software, S.A.-K.; Validation, S.A.-K.; Writing—original draft, D.B.; Writing—review and editing, D.B. and S.A.-K. All authors have read and agreed to the published version of the manuscript.
Funding
The authors declare no funding was received.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
Figure A1A illustrates the hierarchical structure of the symptom as it appears in the SYMP ontology [14]. The hierarchical tree rooted in encompasses three sub-symptoms (~children), and one sub-sub-symptom (~grandchild). Figure A1B exhibits the Neo4j creation commands of these sub-symptoms (Algorithm 1). The subsequent step involves creating the ISA edges in the KG (according to Algorithm 2) are displayed in Figure A1C.

Figure A1.
An example of integrating a hierarchical tree of symptoms into the KG. (A): illustrates the hierarchical structure of the cough symptom as it appears in the SYMP ontology, (B): exhibits the commands for creating sub-symptoms in the KG, (C): exhibits the commands for creating the ISA edges in the KG.
References
- Aceto, G.; Persico, V.; Pescapé, A. The role of Information and Communication Technologies in healthcare: Taxonomies, perspectives, and challenges. J. Netw. Comput. Appl. 2018, 107, 125–154. [Google Scholar] [CrossRef]
- Aceto, G.; Persico, V.; Pescapé, A. Industry 4.0 and health: Internet of things, big data, and cloud computing for healthcare 4.0. J. Ind. Inf. Integr. 2020, 18, 100129. [Google Scholar] [CrossRef]
- Estrela, V.V.; Monteiro AC, B.; França, R.P.; Iano, Y.; Khelassi, A.; Razmjooy, N. Health 4.0: Applications, management, technologies and review: Array. Med. Technol. J. 2018, 2, 262–276. [Google Scholar]
- Magrabi, F.; Ammenwerth, E.; McNair, J.B.; De Keizer, N.F.; Hyppönen, H.; Nykänen, P.; Georgiou, A. Artificial intelligence in clinical decision support: Challenges for evaluating AI and practical implications. Yearb. Med. Inform. 2019, 28, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Lan, K.; Wang, D.T.; Fong, S.; Liu, L.S.; Wong, K.K.; Dey, N. A survey of data mining and deep learning in bioinformatics. J. Med. Syst. 2018, 42, 139. [Google Scholar] [CrossRef] [PubMed]
- Albagli-Kim, S.; Beimel, D. Knowledge Graph-Based Framework for Decision Making Process with Limited Interaction. Mathematics 2022, 10, 3981. [Google Scholar] [CrossRef]
- Denecke, K. How to Design Successful Conversations in Conversational Agents in Healthcare? In Proceedings of the International Conference on Human-Computer Interaction, Copenhagen, Denmark, 23–28 July 2023; Springer Nature: Cham, Switzerland, 2023; pp. 39–45. [Google Scholar]
- Hakimov, S.; Tunc, H.; Akimaliev, M.; Dogdu, E. Semantic question answering system over linked data using relational patterns. In Proceedings of the Joint EDBT/ICDT 2013 Workshops, Genoa, Italy, 18–22 March 2013; pp. 83–88. [Google Scholar]
- Goodwin, T.R.; Harabagiu, S.M. Medical question answering for clinical decision support. In Proceedings of the 25th ACM International conference on Information and Knowledge Management, Indianapolis, IN, USA, 24–28 October 2016; pp. 297–306. [Google Scholar]
- Davis, R.; Shrobe, H.; Szolovits, P. What is a Knowledge Representation? AI Mag. 1993, 14, 17–33. [Google Scholar]
- Webber, R.J.; Eifrem, E. Graph Databases: New Opportunities for Connected Data; O’Reilly Media, Inc.: Middlesex County, MA, USA, 2015. [Google Scholar]
- Wang, Q.; Mao, Z.; Wang, B.; Guo, L. Knowledge graph embedding: A survey of approaches and applications. IEEE Trans. Knowl. Data Eng. 2020, 29, 2724–2743. [Google Scholar] [CrossRef]
- Rajabi, E.; Kafaie, S. Knowledge graphs and explainable ai in healthcare. Information 2022, 13, 459. [Google Scholar] [CrossRef]
- Symptom Ontology (SYMP), Ontology Lookup Service (OLS). Available online: https://www.ebi.ac.uk/ols/ontologies/symp (accessed on 30 January 2024).
- Bonner, S.; Barrett, I.P.; Ye, C.; Swiers, R.; Engkvist, O.; Hoyt, C.T.; Hamilton, W.L. Understanding the performance of knowledge graph embeddings in drug discovery. Artif. Intell. Life Sci. 2022, 2, 100036. [Google Scholar] [CrossRef]
- Gruber, T.R. Toward Principles for the Design of Ontologies Used for Knowledge Sharing. Intl. J. Hum. Comput. Stud. 1995, 43, 907–928. [Google Scholar] [CrossRef]
- Smith, B.; Ashburner, M.; Rosse, C.; Bard, J.; Bug, W.; Ceusters, W.; Goldberg, L.J.; Eilbeck, K.; Ireland, A.; Mungall, C.J. The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2004, 25, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Noy, N.F.; McGuinness, D.L. Ontology Development 101: A Guide to Creating Your First Ontology. Stanford Medical Informatics Technical Report SMI-2001-0880. 2001. Available online: http://protege.stanford.edu/publications/ontology_development/ontology101.pdf (accessed on 30 January 2024).
- Guarino, N. (Ed.) Formal ontology in information systems. In Proceedings of the First International Conference (FOIS’98), Trento, Italy, 6–8 June 1998; IOS Press: Amsterdam, The Netherlands, 1998; Volume 46. [Google Scholar]
- Fernández-López, M.; Gómez-Pérez, A.; Juristo, N. Methontology: From Ontological Art Towards Ontological Engineering; Association for the Advancement of Artificial Intelligence: Washington, DC, USA, 1997. [Google Scholar]
- Studer, R.; Benjamins, V.R.; Fensel, D. Knowledge engineering: Principles and methods. Data Knowl. Eng. 1998, 25, 161–197. [Google Scholar] [CrossRef]
- Zou, X. A survey on application of knowledge graph. J. Phys. Conf. Ser. 2020, 1487, 012016. [Google Scholar] [CrossRef]
- Gashkov, A.; Perevalov, A.; Eltsova, M.; Both, A. Improving Question Answering Quality through Language Feature-Based SPARQL Query Candidate Validation. The Semantic Web. ESWC 2022. Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2022; Volume 13261. [Google Scholar] [CrossRef]
- Guo, Q.; Zhuang, F.; Qin, C.; Zhu, H.; Xie, X.; Xiong, H.; He, Q. A survey on knowledge graph-based recommender systems. IEEE Trans. Knowl. Data Eng. 2020, 34, 3549–3568. [Google Scholar] [CrossRef]
- Dietz, L.; Kotov, A.; Meij, E. Utilizing knowledge graphs for text-centric information retrieval. In Proceedings of the 41st International ACM SIGIR Conference on Research & Development in Information Retrieval, Ann Arbor, MI, USA, 8–12 July 2018; pp. 1387–1390. [Google Scholar]
- Jiang, Y.; Qiu, B.; Xu, C.; Li, C. The research of clinical decision support system based on three-layer knowledge base model. J. Healthc. Eng. 2017, 2017, 6535286. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.T.; Jayamanne, D.J. Computer-aided medical diagnosis using bayesian classifier-decision support system for medical diagnosis. Int. J. Multidiscip. Stud. 2016, 3, 2. [Google Scholar] [CrossRef]
- Rahaman, S. Diabetes diagnosis decision support system based on symptoms, signs and risk factor using special computational algorithm by rule base. In Proceedings of the 2012 15th International Conference on Computer and Information Technology (ICCIT), Chittagong, Bangladesh, 22–24 December 2012; pp. 65–71. [Google Scholar]
- Dong, Z.; Yin, Z.; He, M.; Chen, X.; Lv, X.; Yu, S. Validation of a guideline-based decision support system for the diagnosis of primary headache disorders based on ICHD-3 beta. J. Headache Pain 2014, 15, 40. [Google Scholar] [CrossRef]
- Tandra, S.; Gupta, D.; Amudha, J.; Sharma, K. A fuzzy-neuro-based clinical decision support system for disease diagnosis using symptom severity. In Soft Computing and Signal Processing, Proceedings of the 2nd ICSCSP, Hyderabad, India, 21–22 June 2019; Springer: Singapore, 2020; pp. 81–98. [Google Scholar]
- Belciug, S.; Gorunescu, F. Intelligent Decision Support Systems-A Journey to Smarter Healthcare; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 130–137. [Google Scholar]
- Moreira, M.W.; Rodrigues, J.J.; Korotaev, V.; Al-Muhtadi, J.; Kumar, N. A comprehensive review on smart decision support systems for health care. IEEE Syst. J. 2019, 13, 3536–3545. [Google Scholar] [CrossRef]
- Li, L.; Wang, P.; Yan, J.; Wang, Y.; Li, S.; Jiang, J.; Liu, Y. Real-world data medical knowledge graph: Construction and applications. Artif. Intell. Med. 2020, 103, 101817. [Google Scholar] [CrossRef]
- Riaño, D.; Real, F.; López-Vallverdú, J.A.; Campana, F.; Ercolani, S.; Mecocci, P.; Caltagirone, C. An ontology-based personalization of health-care knowledge to support clinical decisions for chronically ill patients. J. Biomed. Inform. 2012, 45, 429–446. [Google Scholar] [CrossRef]
- Yamada, D.B.; Bernardi, F.A.; Miyoshi NS, B.; de Lima, I.B.; Vinci, A.L.T.; Yoshiura, V.T.; Alves, D. Ontology-based inference for supporting clinical decisions in mental health. In Proceedings of the Computational Science–ICCS 2020: 20th International Conference, Amsterdam, The Netherlands, 3–5 June 2020; Proceedings, Part IV 20. Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 363–375. [Google Scholar]
- Santos, A.; Colaço, A.R.; Nielsen, A.B.; Niu, L.; Strauss, M.; Geyer, P.E.; Mann, M. A knowledge graph to interpret clinical proteomics data. Nat. Biotechnol. 2022, 40, 692–702. [Google Scholar] [CrossRef]
- Dissanayake, P.I.; Colicchio, T.K.; Cimino, J.J. Using clinical reasoning ontologies to make smarter clinical decision support systems: A systematic review and data synthesis. J. Am. Med. Inform. Assoc. 2020, 27, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Shanavas, N.; Wang, H.; Lin, Z.; Hawe, G. Ontology-based enriched concept graphs for medical document classification. Inf. Sci. 2020, 525, 172–181. [Google Scholar] [CrossRef]
- Kaggle. Available online: https://www.kaggle.com (accessed on 30 January 2024).
- Hao, L.; Halappanavar, M.; Kalyanaraman, A. Parallel heuristics for scalable community detection. Parallel Comput. 2015, 47, 19–37. [Google Scholar]
- Luo, J.; Wu, M.; Gopukumar, D.; Zhao, Y. Big data application in biomedical research and health care: A literature review. Biomed. Inform. Insights 2016, 8, BII-S31559. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Jyan, H.W.; Chien, S.C.; Jen, H.H.; Hsu, C.Y.; Lee, P.C.; Chan, C.C. Containing COVID-19 among 627,386 persons in contact with the diamond princess cruise ship passengers who disembarked in Taiwan: Big data analytics. J. Med. Internet Res. 2020, 22, e19540. [Google Scholar] [CrossRef]
- Svedberg, P.; Reed, J.; Nilsen, P.; Barlow, J.; Macrae, C.; Nygren, J. Toward successful implementation of artificial intelligence in health care practice: Protocol for a research program. JMIR Res. Protoc. 2022, 11, e34920. [Google Scholar] [CrossRef]
- van der Vegt, A.H.; Scott, I.A.; Dermawan, K.; Schnetler, R.J.; Kalke, V.R.; Lane, P.J. Implementation frameworks for end-to-end clinical AI: Derivation of the SALIENT framework. J. Am. Med. Inform. Assoc. 2023, 30, 1503–1515. [Google Scholar] [CrossRef]
- Thukral, A.; Dhiman, S.; Meher, R.; Bedi, P. Knowledge graph enrichment from clinical narratives using NLP, NER, and biomedical ontologies for healthcare applications. Int. J. Inf. Technol. 2023, 15, 53–65. [Google Scholar] [CrossRef]
- Malik, P.; Pathania, M.; Rathaur, V.K. Overview of artificial intelligence in medicine. J. Fam. Med. Prim. Care 2019, 8, 2328. [Google Scholar]
- Chaddad, A.; Peng, J.; Xu, J.; Bouridane, A. Survey of explainable AI techniques in healthcare. Sensors 2023, 23, 634. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).