Invariant Feature Learning Based on Causal Inference from Heterogeneous Environments
Abstract
1. Introduction
- We incorporate causal inference into a machine learning algorithm to identify invariant features, moving beyond a passive search through optimization.
- We eliminate redundant causal relationships by modularizing features, which significantly reduces the complexity of causal inference. This modularization makes our causal inference algorithm applicable to complex datasets.
- We introduce a statistical testing-based method for measuring causal effects, addressing the limitation of the Average Causal Effect (ACE). Our proposed method can handle scenarios where the intervention variable can take multiple values.
2. Related Works
3. Invariant Feature Detection Based on Causal Inference
3.1. Setup and Assumptions
- (a)
- Invariance property: for all , we have that holds.
- (b)
- Sufficiency property: .
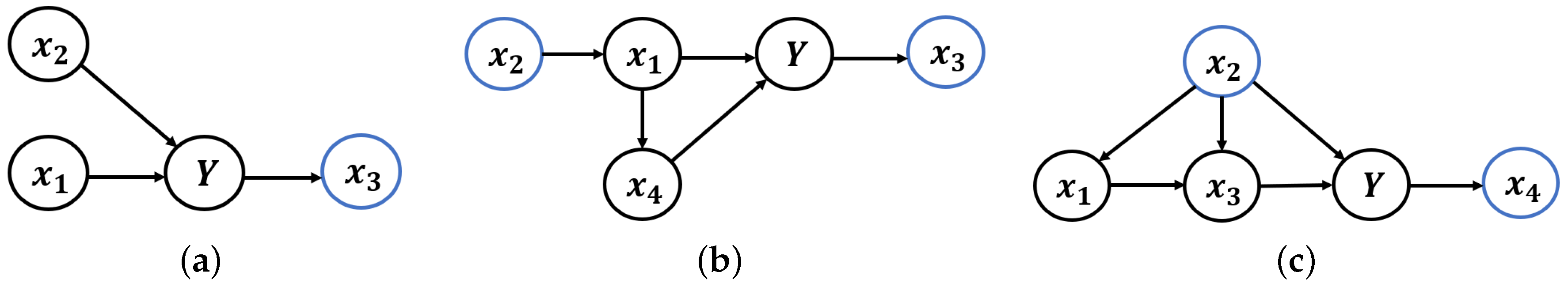
3.2. Causal Structure behind the Predict Problem
- (a)
- If a variable is the cause of Y, then is called a causal variable. All causal variables constitute the causal features.
- (b)
- If a variable is not the cause of Y and the environment e is the cause of , then is called an environmental variable. All environmental variables constitute the environmental features.
- (c)
- If a variable is in the latent features but is not the causal variable or environmental variable, then is called a redundant variable. All redundant variables constitute the redundant features.
3.3. Causal Inference with Multiple Environments
3.3.1. The Mean after Intervention
3.3.2. Analysis of Variance in Causal Inference
| Algorithm 1 Filter environmental variables |
Input: . Output: The indices of environmental variables . 1: 2: for to do 3: Calculate the mean value and the variance after intervention: ( denotes the ith dimension of z), 4: Normalization: 5: Construction of test statistics: 6: Calculate F statistics: 7: if then 8: 9: end if 10: end for 11: 12: return
|
4. Empirical Studies
4.1. Linear Simulated Data
4.2. Gaussian Mixture Model
4.3. Colored MNIST
4.4. Real-World Data
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| OOD | out of distribution |
| IFCI | invariant feature learning based on causal inference |
Appendix A. The Proof of Propositions and Theorems
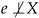 and
and 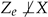 , which is contradictory.
, which is contradictory.Appendix B. Backdoor Criterion and Backdoor Adjustment Formula
- (1)
- There is no descendant node of X in ;
- (2)
- blocks every backdoor path between X and Y that passes through X.
Appendix C. Analysis of Variance
- (1)
- The dependent variable population follows a normal distribution.
- (2)
- The dependent variable population between different categories has the same variance .
- (3)
- Each observation is independent of the others.
- Suggest a hypothesis:
- 2.
- Construction of test statistics:
- (1)
- The mean of the dependent variables in each category:, where denotes the sample size in the ith category.
- (2)
- The total mean of all observations:, where n denotes the total sample size.
- (3)
- The sum of squares of total errors:The sum of squares of intra-group errors:The sum of squares of inter-group errors:
- (4)
- Calculate the F statistics:.
Appendix D. The Structure of VAE
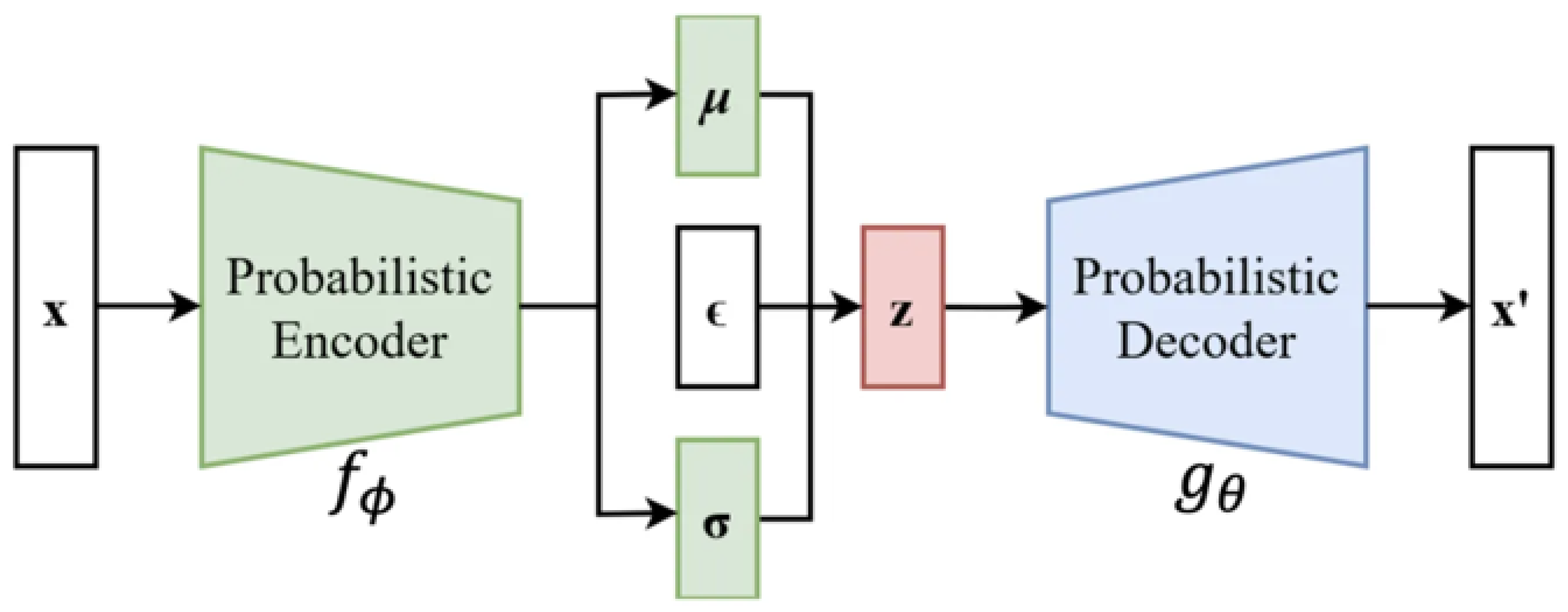
Appendix E. Model Settings
Appendix E.1. Linear Simulated Data
Appendix E.2. GMM
Appendix E.3. Colored MNIST
Appendix E.4. NICO
Appendix F. Generating Linear Simulated Data
| Case 1 | Case 2 | Case 3 |
|---|---|---|
Appendix G. Experimental Results


References
- Arjovsky, M.; Bottou, L.; Gulrajani, I.; Lopez-Paz, D. Invariant risk minimization. arXiv 2019, arXiv:1907.02893. [Google Scholar]
- Liu, J.; Hu, Z.; Cui, P.; Li, B.; Shen, Z. Heterogeneous Risk Minimization. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Volume 139, pp. 6804–6814. [Google Scholar]
- Beery, S.; Horn, G.V.; Perona, P. Recognition in terra incognita. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; Volume 16, pp. 472–489. [Google Scholar]
- Yin, M.; Wang, Y.; Blei, D.M. Optimization-based Causal Estimation from Heterogenous Environments. arXiv 2021, arXiv:2109.11990. [Google Scholar]
- Besserve, M.; Mehrjou, A.; Sun, R.; Schölkopf, B. Counterfactuals uncover the modular structure of deep generative models. In Proceedings of the Eighth International Conference on Learning Representations (ICLR 2020), Addis Ababa, Ethiopia, 26–30 April 2020. [Google Scholar]
- Pearl, J.; Glymour, M.; Jewell, N.P. Causal Inference in Statistics: A Primer; Wiley: Hoboken, NJ, USA, 2016. [Google Scholar]
- Peters, J.; Janzing, D.; Schölkopf, B. Elements of Causal Inference: Foundations and Learning Algorithms; MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Hendrycks, D.; Dietterich, T. Benchmarking Neural Network Robustness to Common Corruptions and Perturbations. arXiv 2019, arXiv:1903.12261. [Google Scholar]
- Shmueli, G. To Explain or to Predict? arXiv 2011, arXiv:1101.0891. [Google Scholar]
- Wang, R.; Yi, M.; Chen, Z.; Zhu, S. Out-of-distribution Generalization with Causal Invariant Transformations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 375–385. [Google Scholar]
- Recht, B.; Roelofs, R.; Schmidt, L.; Shankar, V. Do ImageNet classifiers generalize to ImageNet? In Proceedings of the 36th International Conference on Machine Learning, ICML, Long Beach, CA, USA, 9–15 June 2019; pp. 9413–9424. [Google Scholar]
- Yi, M.; Wang, R.; Sun, J.; Li, Z.; Ma, Z.-M. Improved OOD Generalization via Conditional Invariant Regularizer. arXiv 2022, arXiv:2207.06687. [Google Scholar]
- Schneider, S.; Rusak, E.; Eck, L.; Bringmann, O.; Brendel, W.; Bethge, M. Improving robustness against common corruptions by covariate shift adaptation. In Advances in Neural Information Processing Systems 33; NeurIPS: La Jolla, CA, USA, 2020. [Google Scholar]
- Tu, L.; Lalwani, G.; Gella, S.; He, H. An empirical study on robustness to spurious correlations using pre-trained language models. Trans. Assoc. Comput. Linguist. 2020, 8, 621–633. [Google Scholar] [CrossRef]
- Muandet, K.; Balduzzi, D.; Schölkopf, B. Domain generalization via invariant feature representation. In Proceedings of the 30th International Conference on Machine Learning, ICML, Atlanta, GA, USA, 17–19 June 2013; PART 1. pp. 10–18. [Google Scholar]
- Su, H.; Wang, W. An Out-of-Distribution Generalization Framework Based on Variational Backdoor Adjustment. Mathematics 2024, 12, 85. [Google Scholar] [CrossRef]
- Scholkopf, B.; Locatello, F.; Bauer, S.; Ke, N.R.; Goyal, A.; Bengio, Y.; Kalchbrenner, N. Toward Causal Representation Learning. Proc. IEEE 2021, 109, 612–634. [Google Scholar] [CrossRef]
- Sinha, A.; Namkoong, H.; Duchi, J. Certifying some distributional robustness with principled adversarial training. In Proceedings of the 6th International Conference on Learning Representations, ICLR, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Sagawa, S.; Koh, P.W.; Hashimoto, T.B.; Liang, P. Distributionally Robust Neural Networks for Group Shifts. arXiv 2019, arXiv:1911.08731. [Google Scholar]
- Li, Y.; Tian, X.; Gong, M.; Liu, Y.; Liu, T.; Zhang, K.; Tao, D. Deep Domain Generalization via Conditional Invariant Adversarial Networks. In Proceedings of the 15th European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar]
- Chang, S.; Zhang, Y.; Yu, M.; Jaakkola, T. Invariant rationalization. In Proceedings of the 37th International Conference on Machine Learning, ICML, Virtual, 13–18 July 2020. [Google Scholar]
- Rojas-Carulla, M.; Schölkopf, B.; Turner, R.; Peters, J. Invariant models for causal transfer learning. J. Mach. Learn. Res. 2018, 19, 1309–1342. [Google Scholar]
- Shen, Z.; Cui, P.; Zhang, T.; Kunag, K. Stable Learning via Sample Reweighting. In Proceedings of the 34th AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 5692–5699. [Google Scholar]
- Schölkopf, B. Causality for Machine Learning. arXiv 2019, arXiv:1911.10500. [Google Scholar]
- Kuang, K.; Cui, P.; Athey, S.; Xiong, R.; Li, B. Stable prediction across unknown environments. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, London, UK, 19–23 August 2018; pp. 1617–1626. [Google Scholar]
- Peters, J.; Bühlmann, P.; Meinshausen, N. Causal inference by using invariant prediction: Identification and confidence intervals. J. R. Stat. Soc. Ser. Stat. Methodol. 2016, 78, 947–1012. [Google Scholar] [CrossRef]
- Cui, P.; Athey, S. Stable learning establishes some common ground between causal inference and machine learning. Nat. Mach. Intell. 2022, 4, 110–115. [Google Scholar] [CrossRef]
- Rosenfeld, E.; Ravikumar, P.; Risteski, A. The Risks of Invariant Risk Minimization. arXiv 2020, arXiv:2010.05761. [Google Scholar]
- Kamath, P.; Tangella, A.; Sutherland, D.J.; Srebro, N. Does Invariant Risk Minimization Capture Invariance? arXiv 2021, arXiv:2101.01134. [Google Scholar]
- Rubin, D.B. Causal Inference Using Potential Outcomes: Design, Modeling, Decisions. J. Am. Stat. Assoc. 2005, 469, 322–331. [Google Scholar] [CrossRef]
- Dawid, A.P. Causal Inference Without Counterfactuals. J. Am. Stat. Assoc. 2000, 95, 407–424. [Google Scholar] [CrossRef]
- Robins, J.M.; Hernón, M.Á.; Brumback, B. Marginal Structural Models and Causal Inference in Epidemiology. Epidemiology 2000, 11, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Pearl, J. Causality: Models, Reasoning, and Inference; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Greenl, S.; Pearl, J.; Robins, J.M. Causal Diagrams for Epidemiologic Research. Epidemiology 1999, 10, 37–48. [Google Scholar] [CrossRef]
- Spirtes, P. Single World Intervention Graphs (SWIGs): A Unification of the Counterfactual and Graphical Approaches to Causality; Working Paper Number 128; Center for Statistics and the Social Sciences University of Washington: Seattle, WA, USA, 2013. [Google Scholar]
- Richardson, T.; Robins, J.M. Causation, Prediction, and Search. MIT Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Yao, L.; Chu, Z.; Li, S.; Li, Y.; Gao, J.; Zhang, A. A Survey on Causal Inference. Assoc. Comput. Mach. 2021, 15, 1–46. [Google Scholar] [CrossRef]
- Pearl, J. Causal inference in statistics: An overview. Stat. Surv. 2009, 3, 96–146. [Google Scholar] [CrossRef]
- Brand, J.E.; Zhou, X.; Xie, Y. Recent Developments in Causal Inference and Machine Learning. Annu. Rev. Sociol. 2023, 49, 81–110. [Google Scholar] [CrossRef]
- Hair, J.F., Jr.; Sarstedt, M. Data, measurement, and causal inferences in machine learning: Opportunities and challenges for marketing. J. Mark. Theory Pract. 2021, 29, 65–77. [Google Scholar] [CrossRef]
- Higgins, I.; Amos, D.; Pfau, D.; Racaniere, S.; Matthey, L.; Rezende, D.; Lerchner, A. Towards a Definition of Disentangled Representations. arXiv 2023, arXiv:1812.02230. [Google Scholar]
- Wang, X.; Chen, H.; Tang, S.; Wu, Z.; Zhu, W. Disentangled Representation Learning. arXiv 2023, arXiv:2211.11695. [Google Scholar]
- Kuang, K.; Xiong, R.; Cui, P.; Athey, S.; Li, B. Stable prediction with model misspecification and agnostic distribution shift. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 4485–4492. [Google Scholar]
- George, C.; Roger, L.B. Statistical Inference; Duxbury Press: London, UK, 2001. [Google Scholar]
- Krueger, D.; Caballero, E.; Jacobsen, J.; Zhang, A.; Binas, J.; Zhang, D.; Priol, R.L.; Courville, A. Out-of-Distribution Generalization via Risk Extrapolation (REx). arXiv 2020, arXiv:2003.00688. [Google Scholar]
- Xie, C.; Chen, F.; Liu, Y.; Li, Z. Risk variance penalization: From distributional robustness to causality. arXiv 2020, arXiv:2006.07544. [Google Scholar]
- Higgins, I.; Matthey, L.; Pal, A.; Burgess, C.; Glorot, X.; Botvinick, M.; Mohamed, S.; Lerchner, A. beta-VAE: Learning Basic Visual Concepts with a Constrained Variational Framework. In Proceedings of the International Conference on Learning Representations, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. In Proceedings of the International Conference on Learning Representations, ICLR, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]
- He, Y.; Shen, Z.; Cui, P. Towards non-iid Image Classification: A Dataset and Baselines. In Proceedings of the Computer Vision and Pattern Recognition, CVPR, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]

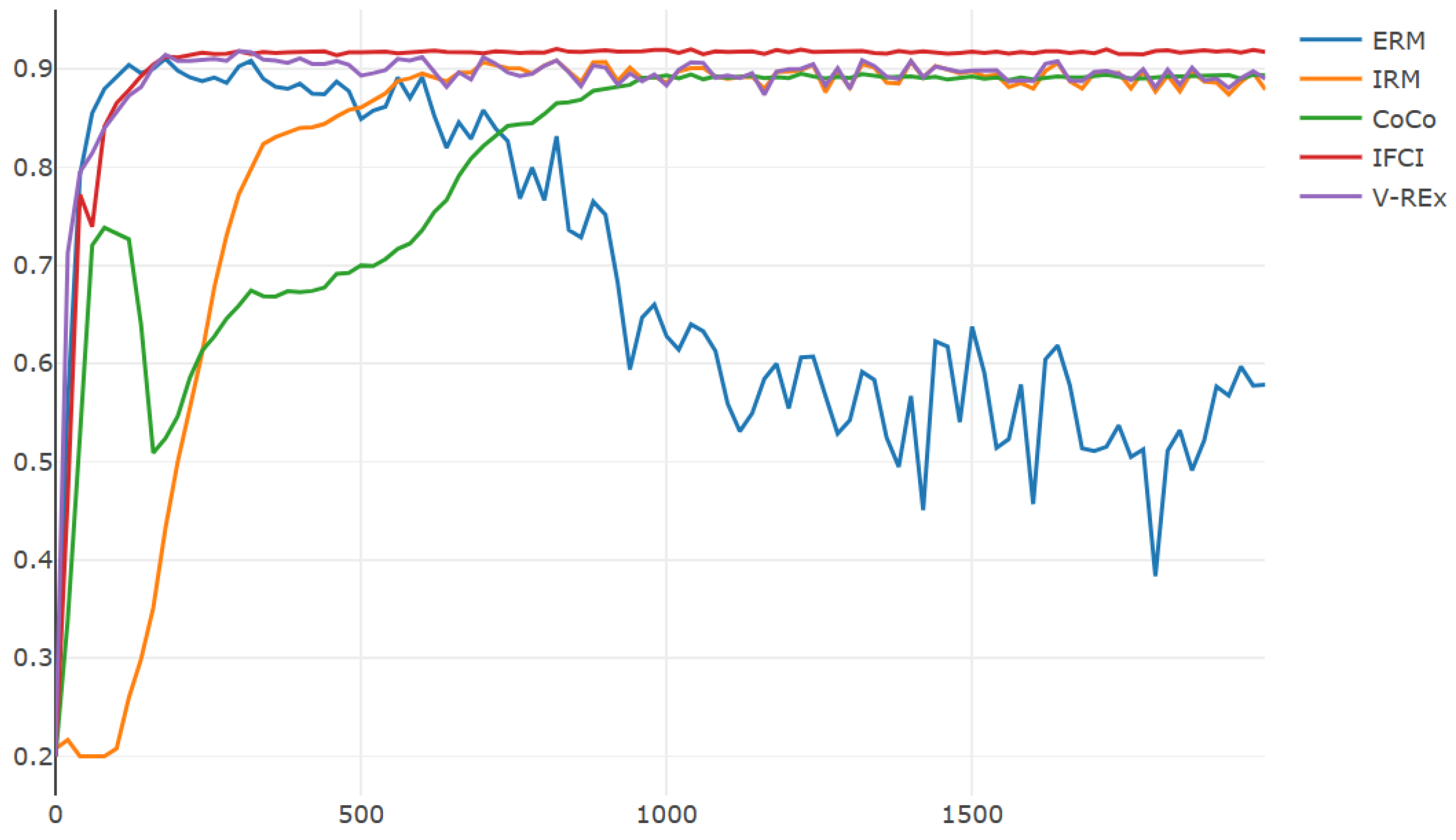
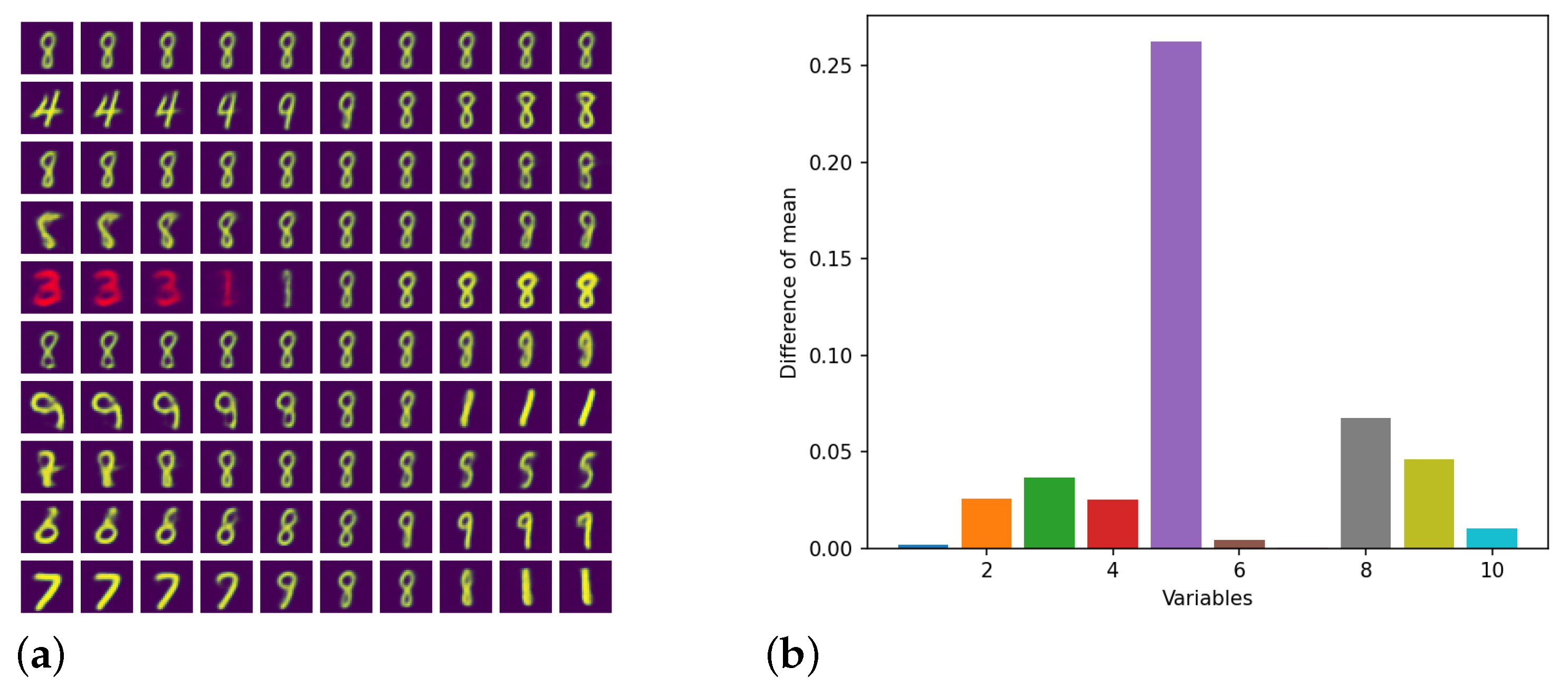
| Algorithm | Training Accuracy | Testing Accuracy | Training Time |
|---|---|---|---|
| ERM | 92.8 | 80.7 | 18.85 s |
| IRM | 90.2 | 83.9 | 17.72 s |
| V-REx | 90.0 | 86.8 | 17.70 s |
| RVP | 91.1 | 87.0 | 23.60 s |
| CoCo | 85.1 | 84.5 | 20.1 s |
| ICP | 93.6 | 92.3 | 19.69 s |
| IFCI | 93.6 | 92.3 | 18.03 s |
| Algorithm | Training Accuracy | Testing Accuracy | Training Time |
|---|---|---|---|
| ERM | 96.4 | 82.0 | 16.83 s |
| IRM | 94.4 | 83.6 | 17.36 s |
| V-REx | 93.0 | 77.2 | 34.50 s |
| RVP | 88.1 | 74.2 | 34.53 s |
| CoCo | 86.1 | 81.8 | 42.90 s |
| ICP | - | - | - |
| IFCI | 94.2 | 91.5 | 16.96 s |
| Algorithm | Training Accuracy | Testing Accuracy | Training Time |
|---|---|---|---|
| ERM | 89.0 | 70.9 | 23.47 s |
| IRM | 89.6 | 80.9 | 16.33 s |
| V-REx | 86.8 | 74.6 | 34.47 s |
| RVP | 85.9 | 71.7 | 35.37 s |
| CoCo | 79.0 | 64.0 | 20.1 s |
| ICP | 89.0 | 70.9 | 19.69 s |
| IFCI | 91.2 | 88.3 | 15.03 s |
| Algorithm | Training Accuracy | Testing Accuracy | Training Time |
|---|---|---|---|
| ERM | 99.0 | 50.5 | 21.9 s |
| IRM | 93.1 | 88.0 | 32.24 s |
| V-REx | 92.7 | 85.9 | 17.70 s |
| RVP | 93.1 | 88.6 | 18.16 s |
| CoCo | 89.3 | 89.3 | 65.92 s |
| ICP | - | - | - |
| IFCI | 91.8 | 91.7 | 16.96 s |
| 1.19 | 0.27 | 0.23 | 0.12 | 0.80 | 3561.62 | 485.23 | 589.32 |
| Algorithm | Training Accuracy | Testing Accuracy | Training Time |
|---|---|---|---|
| ERM | 99.1 | 47.4 | 30.30 s |
| IRM | 96.4 | 70.3 | 35.38 s |
| V-REx | 98.9 | 49.5 | 30.86 s |
| RVP | 98.5 | 56.9 | 33.92 s |
| CoCo | 93.5 | 88.7 | 92.24 s |
| ICP | 85.3 | 82.1 | 35.44 s |
| IFCI | 93.8 | 91.9 | 33.12 s |
| Algorithm | Training Accuracy | Testing Accuracy | Training Time |
|---|---|---|---|
| ERM | 96.9 | 54.0 | 1648.6 s |
| IRM | 85.3 | 73.4 | 2351.2 s |
| V-REx | 95.6 | 69.1 | 2169.3 s |
| RVP | 92.1 | 73.7 | 2571.3 s |
| CoCo | 81.9 | 79.5 | 4687.2 s |
| ICP | 75.1 | 72.6 | 1990.5 s |
| IFCI | 83.1 | 81.7 | 1953.0 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, H.; Wang, W. Invariant Feature Learning Based on Causal Inference from Heterogeneous Environments. Mathematics 2024, 12, 696. https://doi.org/10.3390/math12050696
Su H, Wang W. Invariant Feature Learning Based on Causal Inference from Heterogeneous Environments. Mathematics. 2024; 12(5):696. https://doi.org/10.3390/math12050696
Chicago/Turabian StyleSu, Hang, and Wei Wang. 2024. "Invariant Feature Learning Based on Causal Inference from Heterogeneous Environments" Mathematics 12, no. 5: 696. https://doi.org/10.3390/math12050696
APA StyleSu, H., & Wang, W. (2024). Invariant Feature Learning Based on Causal Inference from Heterogeneous Environments. Mathematics, 12(5), 696. https://doi.org/10.3390/math12050696






