An Improved Expectation–Maximization Bayesian Algorithm for GWAS
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Model
2.2. Genome-Wide Association Analysis of the emBBI Algorithm
2.2.1. Polygenic and Residual Noise Whitening Stage
2.2.2. Variable Reduction Stage
2.2.3. EM Algorithm Stage
- Provide the initial values for the parameters.
- E-step: for each SNP, calculate and the posterior probability as shown in (10).
- M-step: updated , , , and are given according to Functions (11)–(14).
2.2.4. Likelihood Ratio (LR) Test
2.3. Comparison Algorithm
2.4. Experimental Materials
2.4.1. Simulation Datasets
2.4.2. Arabidopsis Datasets
2.5. Candidate Gene Identification and Enrichment Analysis
2.6. Tissue-Specific Expression Analysis
3. Results
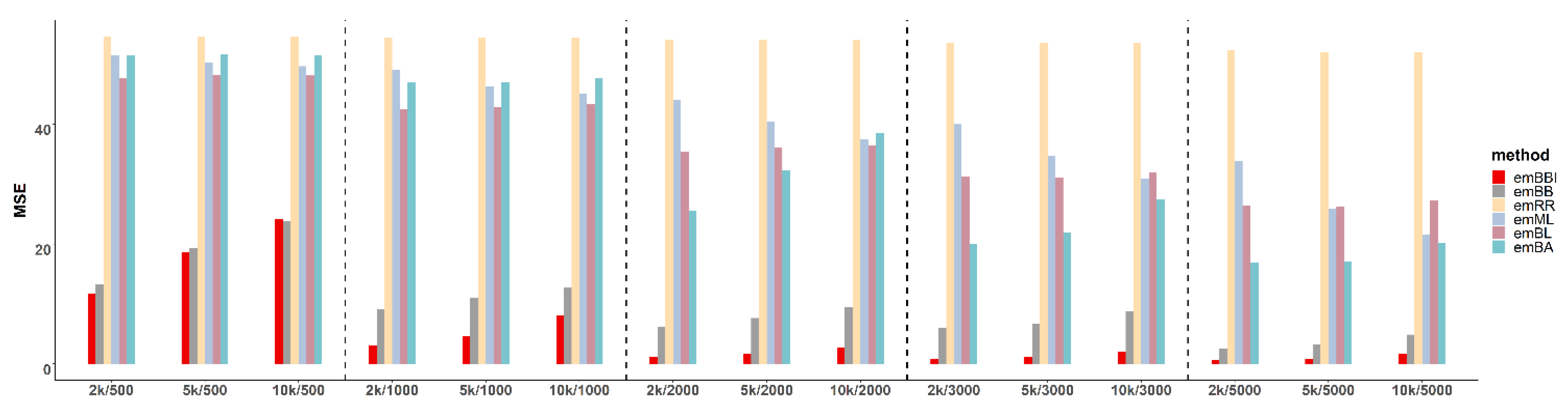
3.1. Experimental Results of Simulated Data
3.2. Results of Real Data Analysis
3.2.1. Analysis of Phenotypic Differences in Arabidopsis Data
3.2.2. GWASs and Known Genes of QTNs
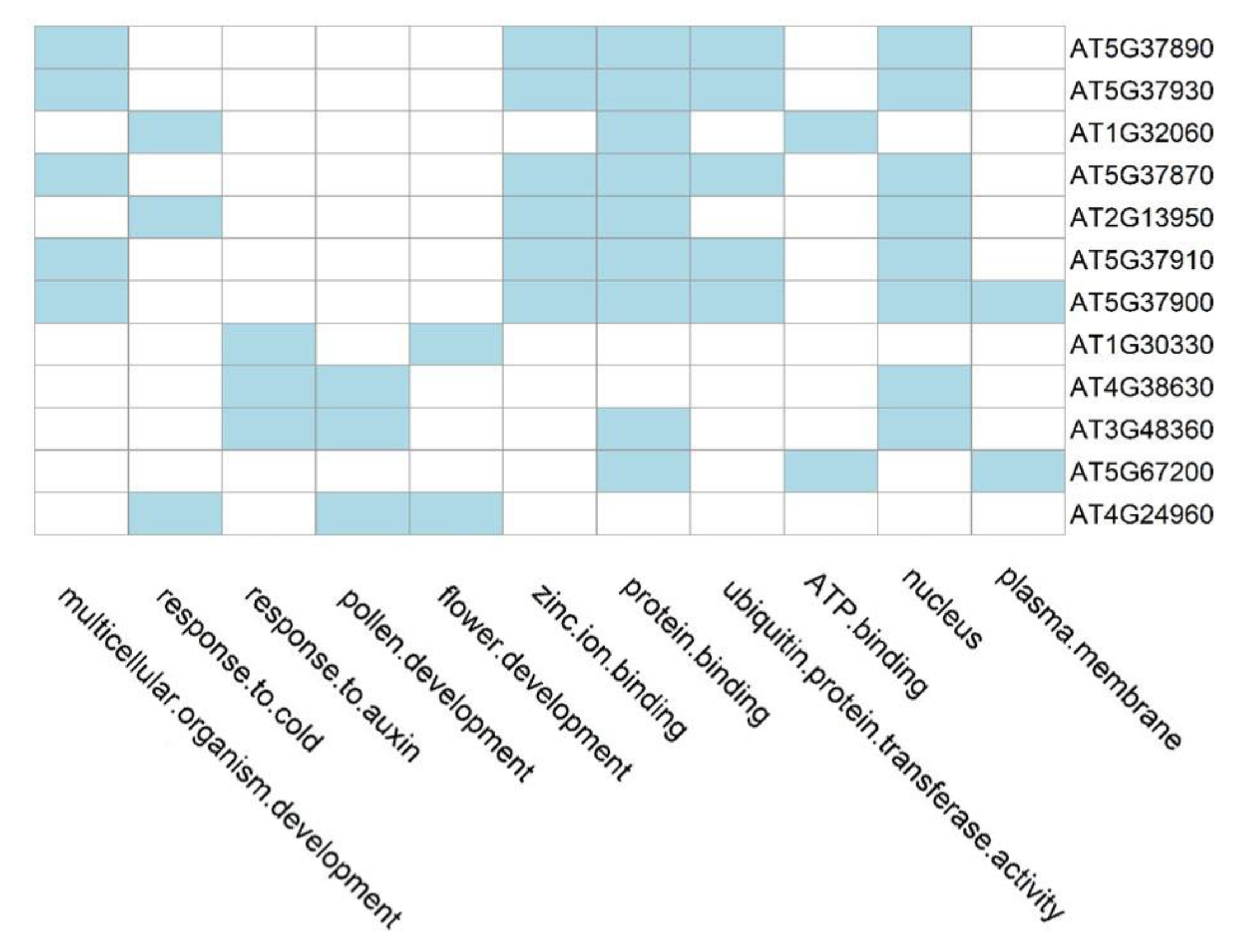
3.2.3. Functional Enrichment Analysis of Candidate Genes
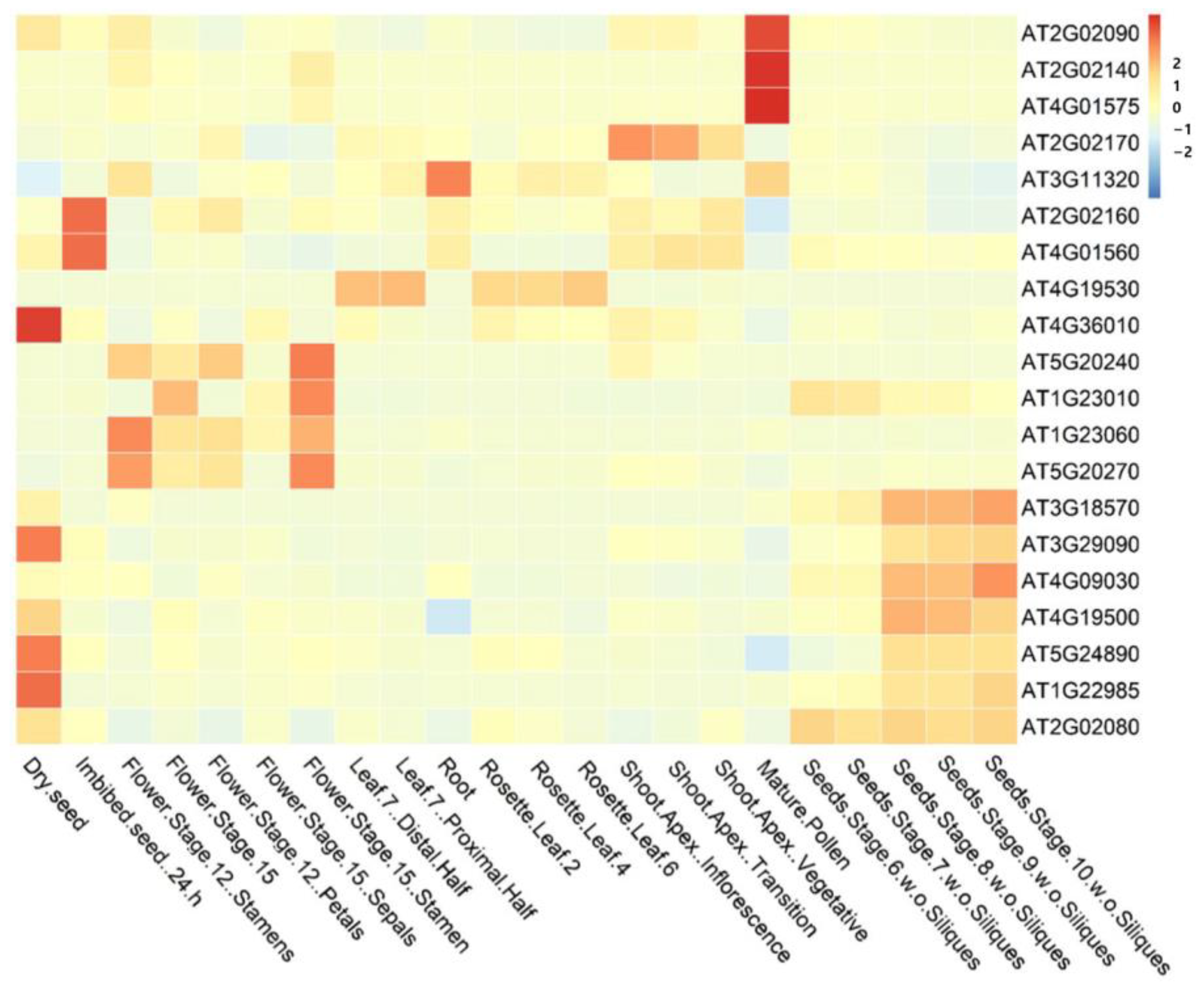
3.2.4. Expression Profiling of Candidate Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, H.; Kugathasan, S.; Annese, V.; Bradfield, J.R.; Russell, R.K.; Sleiman, P.M.A.; Imielinski, M.; Glessner, J.; Hou, C.; et al. Diverse Genome-wide Association Studies Associate the IL12/IL23 Pathway with Crohn Disease. Am. J. Hum. Genet. 2009, 84, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.W.; Yang, J.; Zhou, L.S.; Ren, J.; Liu, X.X.; Zhang, H.; Yang, B.; Zhang, Z.Y.; Ma, H.B.; Xie, X.H.; et al. A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle. PLoS Genet. 2014, 10, e1004710. [Google Scholar] [CrossRef]
- Fan, Q.C.; Wu, P.F.; Dai, G.J.; Zhang, G.X.; Zhang, T.; Xue, Q.; Shi, H.Q.; Wang, J.Y. Identification of 19 loci for reproductive traits in a local Chinese chicken by genome-wide study. Genet. Mol. Res. 2017, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Demars, J.; Fabre, S.; Sarry, J.; Rossetti, R.; Gilbert, H.; Persani, L.; Tosser-Klopp, G.; Mulsant, P.; Nowak, Z.; Drobik, W.; et al. Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep. PLoS Genet. 2013, 9, e1003482. [Google Scholar] [CrossRef]
- Lin, H.; Zhou, Z.; Zhao, J.; Zhou, T.; Bai, H.; Ke, Q.; Pu, F.; Zheng, W.; Xu, P. Genome-Wide Association Study Identifies Genomic Loci of Sex Determination and Gonadosomatic Index Traits in Large Yellow Croaker (Larimichthys crocea). Mar. Biotechnol. 2021, 23, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Huang, X.H.; Wei, X.H.; Sang, T.; Zhao, Q.A.; Feng, Q.; Zhao, Y.; Li, C.Y.; Zhu, C.R.; Lu, T.T.; Zhang, Z.W.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–976. [Google Scholar] [CrossRef]
- Li, H.; Peng, Z.Y.; Yang, X.H.; Wang, W.D.; Fu, J.J.; Wang, J.H.; Han, Y.J.; Chai, Y.C.; Guo, T.T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Chao, Z.F.; Chen, Y.Y.; Ji, C.; Wang, Y.L.; Huang, X.; Zhang, C.Y.; Yang, J.; Song, T.; Wu, J.C.; Guo, L.X.; et al. A genome-wide association study identifies a transporter for zinc uploading to maize kernels. EMBO Rep. 2023, 24, e55542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, M.; Wen, Y.; Zhang, Y.; Lu, Y.; Wang, S.; Chen, J. A Fast Multi-Locus Ridge Regression Algorithm for High-Dimensional Genome-Wide Association Studies. Front. Genet. 2021, 12, 649196. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.W.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.M.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Lippert, C.; Listgarten, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef]
- Wang, Q.S.; Tian, F.; Pan, Y.C.; Buckler, E.S.; Zhang, Z.W. A SUPER Powerful Method for Genome Wide Association Study. PLoS ONE 2014, 9, e107684. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Jiang, L.; Zheng, Z.; Qi, T.; Kemper, K.E.; Wray, N.R.; Visscher, P.M.; Yang, J. A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet. 2019, 51, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.J.; Zhang, H.; Ni, Y.L.; Huang, B.; Zhang, J.; Feng, J.Y.; Wang, S.B.; Dunwell, J.M.; Zhang, Y.M.; Wu, R. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 2018, 19, 700–712. [Google Scholar] [CrossRef]
- Iwata, H.; Uga, Y.; Yoshioka, Y.; Ebana, K.; Hayashi, T. Bayesian association mapping of multiple quantitative trait loci and its application to the analysis of genetic variation among L. germplasms. Theor. Appl. Genet. 2007, 114, 1437–1449. [Google Scholar] [CrossRef]
- Zhang, J.; Yue, C.; Zhang, Y.M. Bias correction for estimated QTL effects using the penalized maximum likelihood method. Heredity 2012, 108, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Moser, G.; Lee, S.H.; Hayes, B.J.; Goddard, M.E.; Wray, N.R.; Visscher, P.M. Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model. PLoS Genet. 2015, 11, e1004969. [Google Scholar] [CrossRef]
- Shepherd, R.K.; Meuwissen, T.H.; Woolliams, J.A. Genomic selection and complex trait prediction using a fast EM algorithm applied to genome-wide markers. BMC Bioinform. 2010, 11, 529. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Iwata, H. EM algorithm for Bayesian estimation of genomic breeding values. BMC Genet. 2010, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.; Muir, W.M.; Rainey, K.M. bWGR: Bayesian whole-genome regression. Bioinformatics 2020, 36, 1957–1959. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B-Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Park, T.; Casella, G. The Bayesian Lasso. J. Am. Stat. Assoc. 2008, 103, 681–686. [Google Scholar] [CrossRef]
- Swallow, W.H.; Monahan, J.F. Monte Carlo Comparison of ANOVA, MIVQUE, REML, and ML Estimators of Variance Components. Technometrics 1984, 26, 47–57. [Google Scholar] [CrossRef]
- da Silva, F.A.; Viana, A.P.; Correa, C.C.G.; Santos, E.A.; de Oliveira, J.A.V.S.; Andrade, J.D.G.; Ribeiro, R.M.; Glória, L.S. Bayesian ridge regression shows the best fit for SSR markers in Psidium guajava among Bayesian models. Sci. Rep. 2021, 11, 13639. [Google Scholar] [CrossRef] [PubMed]
- Yi, N.J.; Xu, S.H. Bayesian LASSO for quantitative trait loci mapping. Genetics 2008, 179, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Feng, J.Y.; Ni, Y.L.; Wen, Y.J.; Niu, Y.; Tamba, C.L.; Yue, C.; Song, Q.; Zhang, Y.M. pLARmEB: Integration of least angle regression with empirical Bayes for multilocus genome-wide association studies. Heredity 2017, 118, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.J.; Zhang, Y.W.; Zhang, J.; Feng, J.Y.; Zhang, Y.M. The improved FASTmrEMMA and GCIM algorithms for genome-wide association and linkage studies in large mapping populations. Crop J. 2020, 8, 723–732. [Google Scholar] [CrossRef]
- Wen, Y.J.; Zhang, Y.W.; Zhang, J.; Feng, J.Y.; Dunwell, J.M.; Zhang, Y.M. An efficient multi-locus mixed model framework for the detection of small and linked QTLs in F2. Brief. Bioinform. 2019, 20, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wu, Q.; Shen, D.; Wen, Y.; Liu, F.; Gao, Y.; Ding, J.; Zhang, J. TSLRF: Two-Stage Algorithm Based on Least Angle Regression and Random Forest in genome-wide association studies. Sci. Rep. 2019, 9, 18034. [Google Scholar] [CrossRef] [PubMed]
- Tamba, C.L.; Ni, Y.L.; Zhang, Y.M. Iterative sure independence screening EM-Bayesian LASSO algorithm for multi-locus genome-wide association studies. PLoS Comput. Biol. 2017, 13, e1005357. [Google Scholar] [CrossRef]
- Kao, C.H.; Zeng, Z.B.; Teasdale, R.D. Multiple interval mapping for quantitative trait loci. Genetics 1999, 152, 1203–1216. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Guo, W.; Zhang, Y.M.; Zhang, T. QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor. Appl. Genet. 2008, 117, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Atwell, S.; Huang, Y.S.; Vilhjalmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Akond, Z.; Ahsan, M.A.; Alam, M.; Mollah, M.N.H. Robustification of GWAS to explore effective SNPs addressing the challenges of hidden population stratification and polygenic effects. Sci. Rep. 2021, 11, 13060. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]

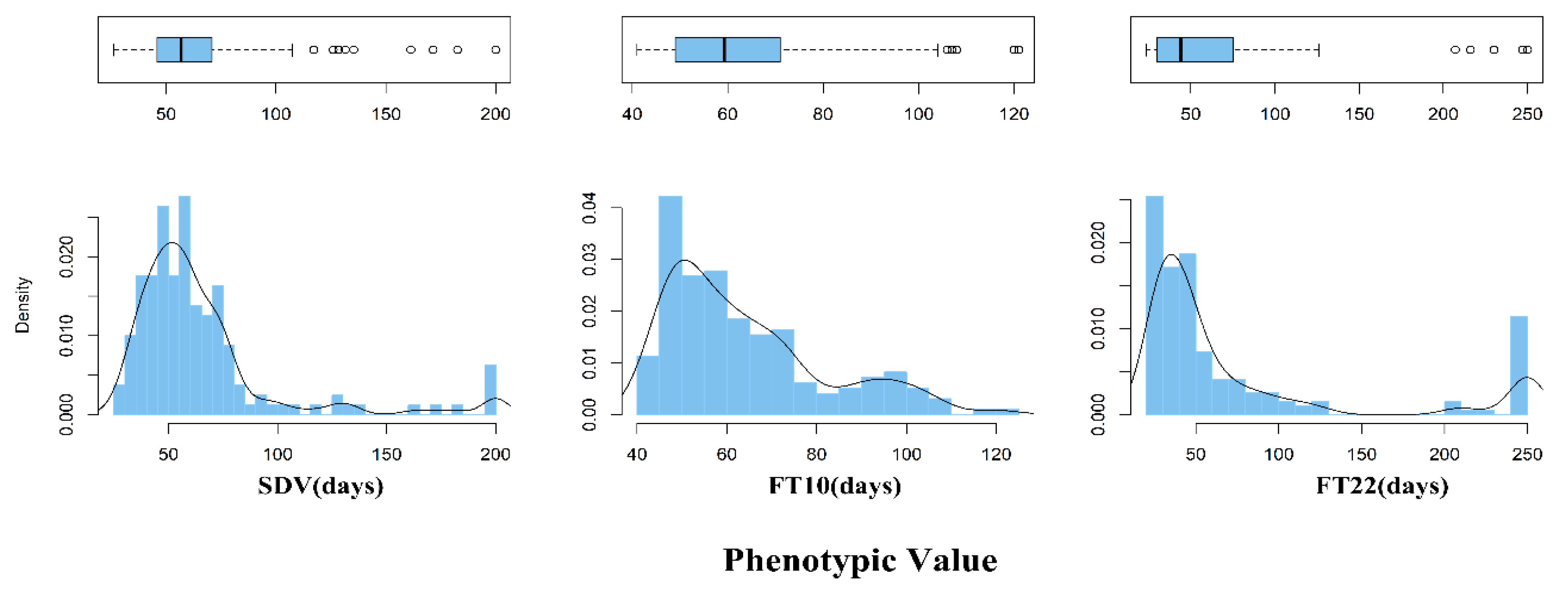
| Trait | Mean | SD | CV | Min | Max | Range |
|---|---|---|---|---|---|---|
| SDV | 64.86 | 35.07 | 0.54 | 26.33 | 200.00 | 173.67 |
| FT10 | 63.97 | 17.82 | 0.28 | 41.00 | 121.00 | 80.00 |
| FT22 | 74.72 | 71.75 | 0.96 | 23.30 | 250.00 | 226.70 |
| Chr. | Position | Gene | Method | Chr. | Position | Gene | Method |
|---|---|---|---|---|---|---|---|
| 1 | 2747091 | AT1G08660 | emRR, emBB | 2 | 13862690 | AT2G32700 | emRR |
| 2752442 | emRR | 2 | 10555534 | AT2G24790 | emBBI | ||
| 1 | 2779526 | AT1G08730 | emRR | 2 | 14696964 | AT2G34880 | emBBI |
| 1 | 6470743 | AT1G18750 | emBB | 2 | 16020457 | AT2G38185 | emBBI |
| 1 | 4345034 | AT1G12790 | emBL | AT2G38195 | |||
| 1 | 4953573 | AT1G14440 | emBL | AT2G38220 | |||
| 1 | 2779077 | AT1G08660 | emML | 3 | 18923922 | AT3G50870 | emRR |
| AT1G08730 | 4 | 17263477 | AT4G36620 | emRR | |||
| 1 | 3180545 | AT1G09780 | emML | 4 | 13976417 | AT4G28190 | emBBI |
| 1 | 3849924 | AT1G11410 | emML | 5 | 13923880 | AT5G35750 | emRR |
| 1 | 9053189 | AT1G26220 | emML | 5 | 14296026 | AT5G36260 | emBA |
| 9054289 | 5 | 14296180 | AT5G36260 | emBA | |||
| 9055133 | 5 | 3163806 | AT5G10140 | emBBI | |||
| 9072307 | 5 | 6851199 | AT5G20280 | emBBI | |||
| 9073534 | 5 | 8609688 | AT5G24930 | emBBI | |||
| 9082795 | 5 | 19938064 | AT5G49150 | emBBI | |||
| 1 | 9483525 | AT1G27320 | emML | 5 | 26786046 | AT5G67180 | emBBI |
| 9486308 | 5 | 26793959 | |||||
| 9488653 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Zhao, J.; Wang, J.; Lin, G.; Li, L.; Ban, F.; Zhu, M.; Wen, Y.; Zhang, J. An Improved Expectation–Maximization Bayesian Algorithm for GWAS. Mathematics 2024, 12, 1944. https://doi.org/10.3390/math12131944
Zhang G, Zhao J, Wang J, Lin G, Li L, Ban F, Zhu M, Wen Y, Zhang J. An Improved Expectation–Maximization Bayesian Algorithm for GWAS. Mathematics. 2024; 12(13):1944. https://doi.org/10.3390/math12131944
Chicago/Turabian StyleZhang, Ganwen, Jianini Zhao, Jieru Wang, Guo Lin, Lin Li, Fengfei Ban, Meiting Zhu, Yangjun Wen, and Jin Zhang. 2024. "An Improved Expectation–Maximization Bayesian Algorithm for GWAS" Mathematics 12, no. 13: 1944. https://doi.org/10.3390/math12131944
APA StyleZhang, G., Zhao, J., Wang, J., Lin, G., Li, L., Ban, F., Zhu, M., Wen, Y., & Zhang, J. (2024). An Improved Expectation–Maximization Bayesian Algorithm for GWAS. Mathematics, 12(13), 1944. https://doi.org/10.3390/math12131944






