Fast Semi-Supervised t-SNE for Transfer Function Enhancement in Direct Volume Rendering-Based Medical Image Visualization
Abstract
1. Introduction
2. Related Work
3. Methods
3.1. Transfer Function-Based Multi-Dimensional Feature Space
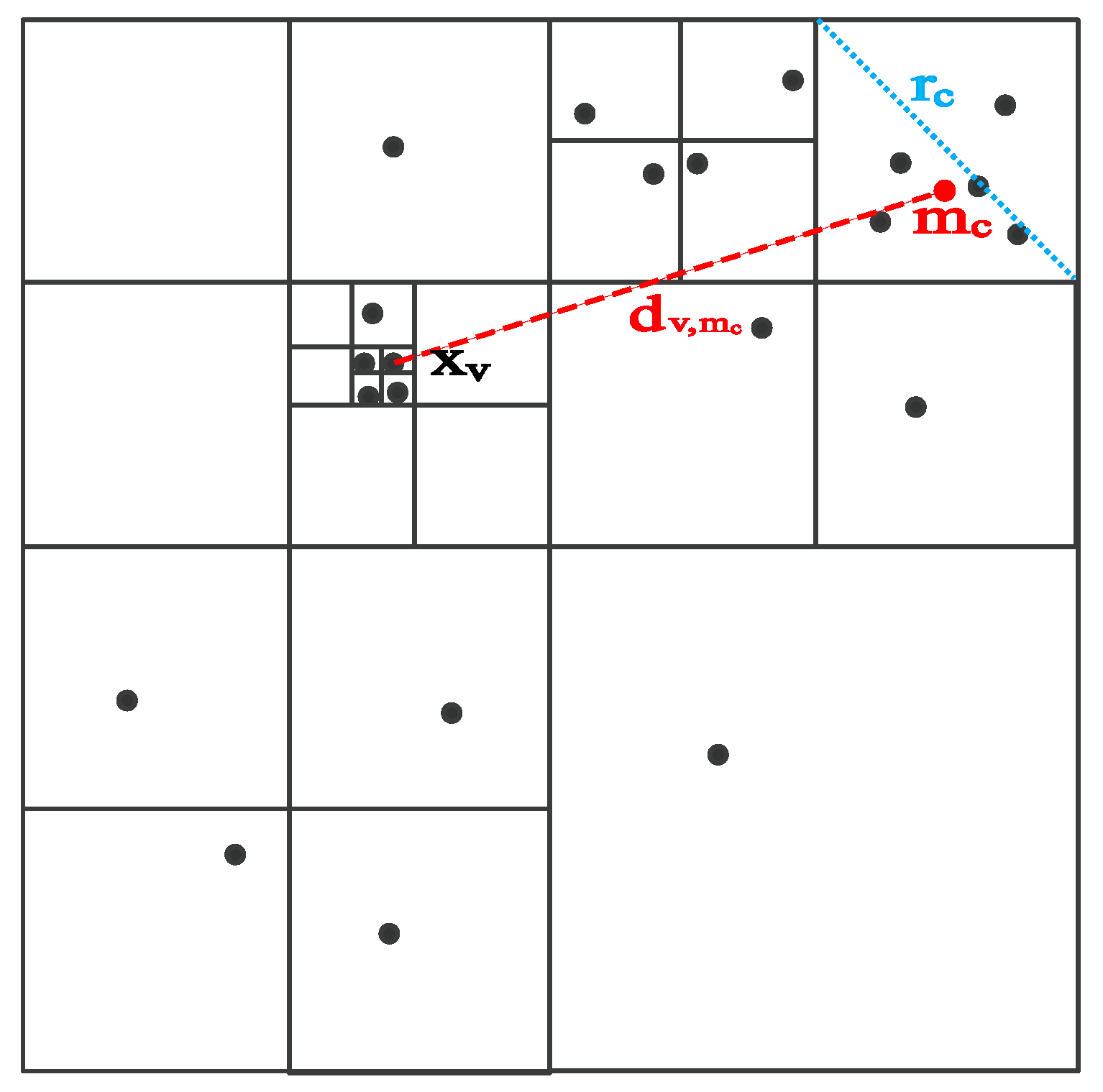
3.2. Fast Semi-Supervised t-SNE (FSS.t-SNE)
3.3. FSS.t-SNE-Based Volumetric Rendering
4. Experimental Set-Up
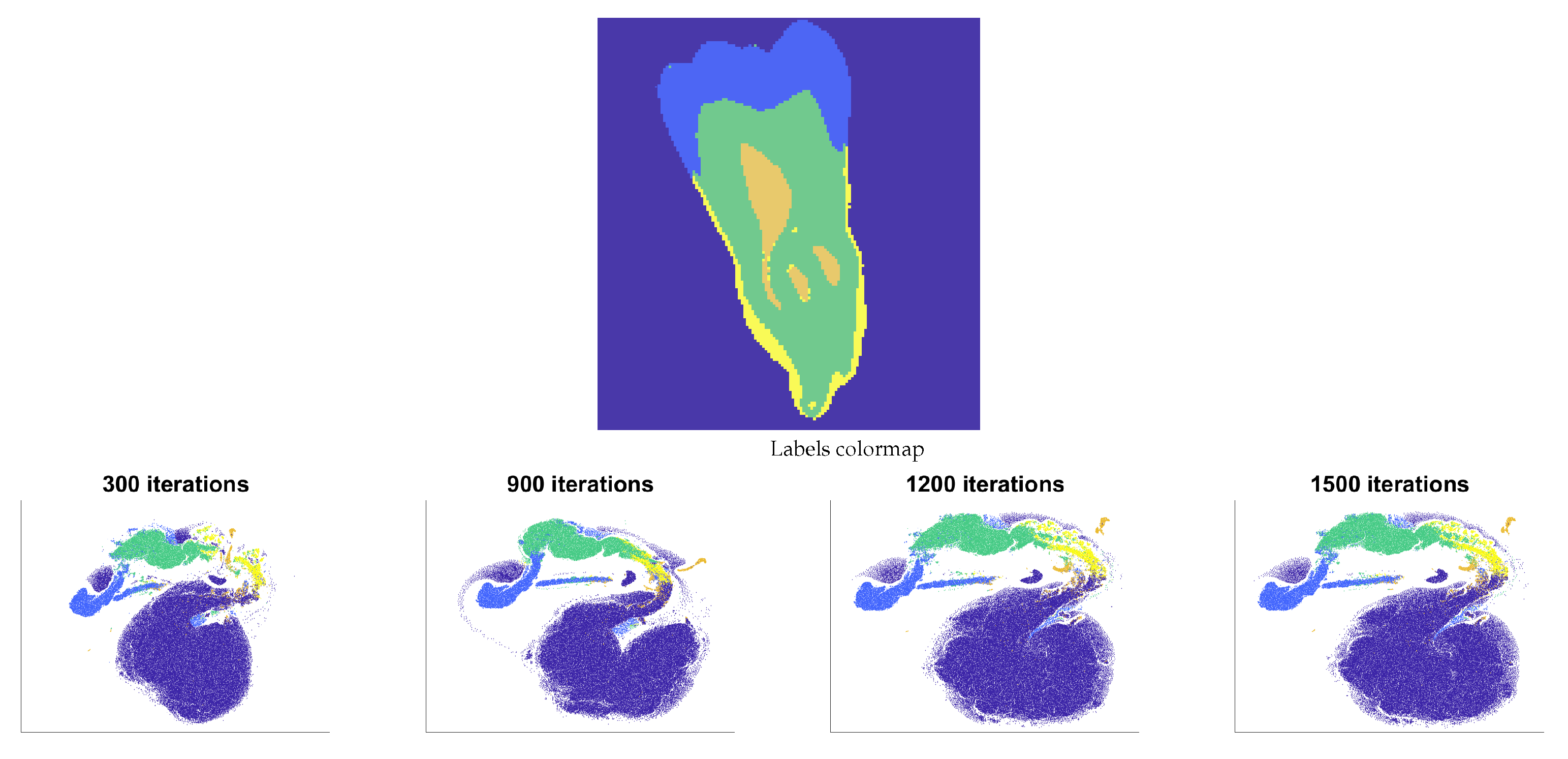
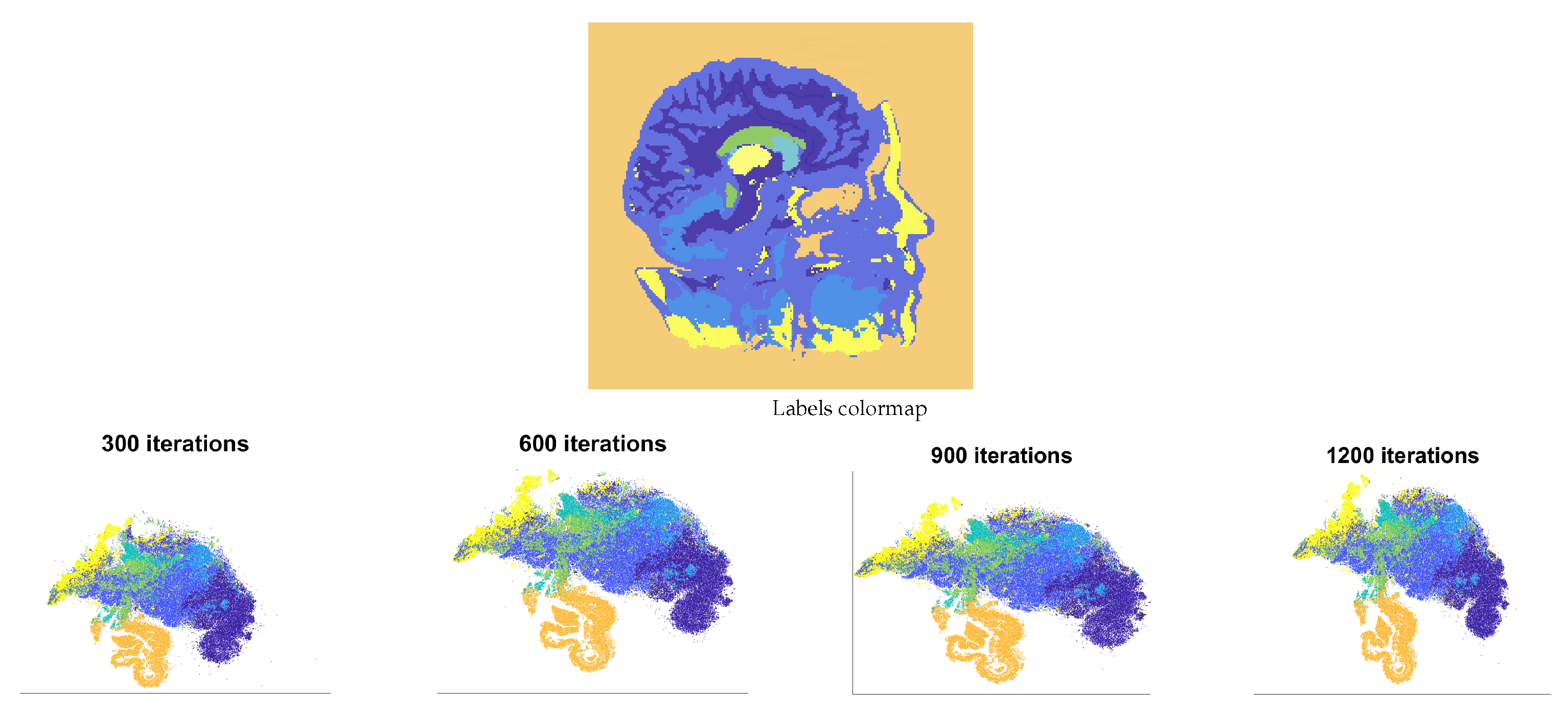
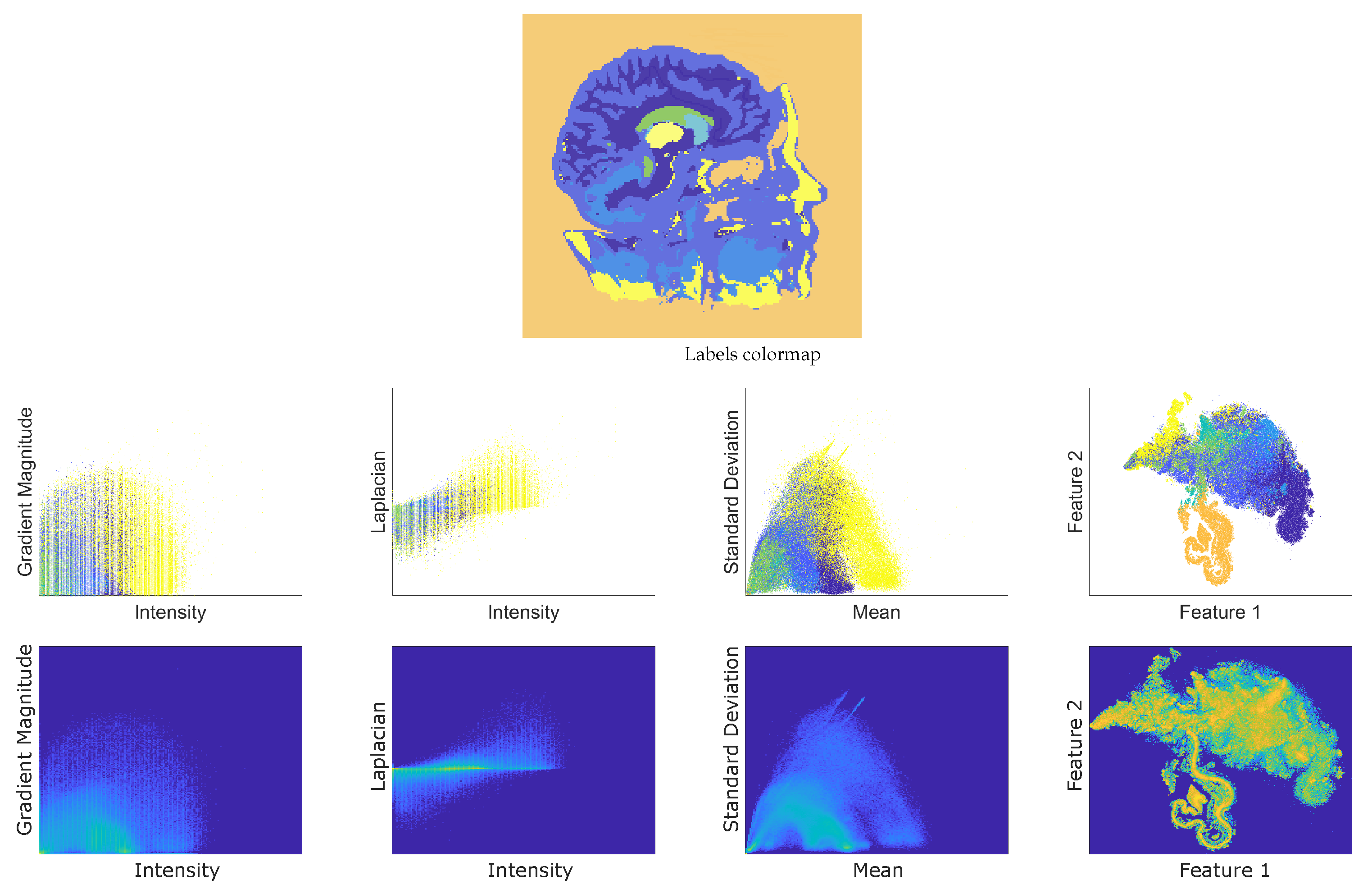
4.1. Tested Datasets
- –
- Human Tooth Computed Tomography (CT-Tooth) comprises voxels, revealing the dentine, enamel, and pulp [25]. The volume is cropped from rows 61 to 106 and columns 58 to 148 to obtain a subvolume of 46 × 91 × 161 = 673,946 voxels. The entire volume is unlabeled (supervised dataset).
- –
- T1 Magnetic Resonance of a head with skull partially removed to reveal the brain (https://graphics.stanford.edu/data/voldata/ (accessed on 1 February 2024)) (MR-Brain), of size . The volume is cropped from rows 50 to 176, the columns 75 to 173, and the slices 54 to 90 to obtain a subvolume of 565,785 voxels. On some slices, tissues of interest were hand-brushed by an amateur user. Labels are associated with the white matter, the green matter, the skull, the ventricles, the cerebellum, the basal ganglia, and the background. The number of labeled voxels corresponds to of the cropped volume (semi-supervised dataset).
- –
- Abdominal Computed Tomography (CT-Abdomen) with manual annotations of the lung, bones, liver, kidneys, and bladder, labeled by an expert (https://www.cancerimagingarchive.net/collections/ (accessed on 1 February 2024)) [44]. The volume size is and is cropped from rows 14 to 150, columns 30 to 185, obtaining a subvolume of 751,080 voxels. Original manual annotations label of the voxels in the subvolume by clinicians (semi-supervised dataset).
4.2. Training Details and Method Comparison
5. Results and Discussion
5.1. FSS.t-SNE Embedding Results
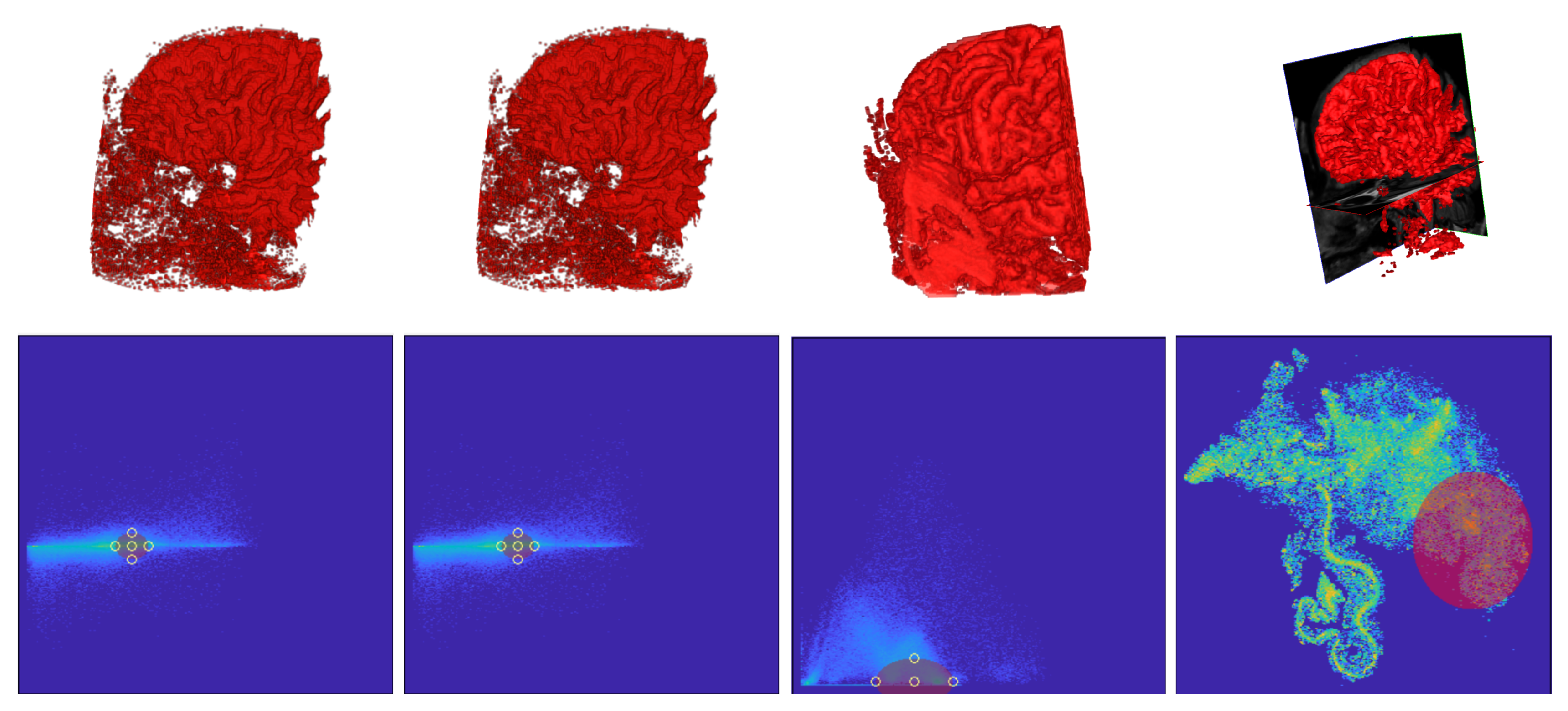
5.2. DVR Visual Inspection Results
5.3. Segmentation Results
5.4. Limitations
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mijwil, M.M.; Al-Mistarehi, A.H.; Abotaleb, M.; El-kenawy, E.S.M.; Ibrahim, A.; Abdelhamid, A.A.; Eid, M.M. From Pixels to Diagnoses: Deep Learning’s Impact on Medical Image Processing-A Survey. Wasit J. Comput. Math. Sci. 2023, 2, 9–15. [Google Scholar] [CrossRef]
- Magadza, T.; Viriri, S. Deep learning for brain tumor segmentation: A survey of state-of-the-art. J. Imaging 2021, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhuang, Y.; Luo, Y.; Zhu, F.; Zhao, W.; Zeng, H. Deep learning-based automated lesion segmentation on pediatric focal cortical dysplasia II preoperative MRI: A reliable approach. Insights Imaging 2024, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Cheng, H.C.; Cardone, A.; Jain, S.; Krokos, E.; Narayan, K.; Subramaniam, S.; Varshney, A. Deep-learning-assisted volume visualization. IEEE Trans. Vis. Comput. Graph. 2018, 25, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Han, J. Dl4scivis: A state-of-the-art survey on deep learning for scientific visualization. IEEE Trans. Vis. Comput. Graph. 2022, 29, 3714–3733. [Google Scholar] [CrossRef] [PubMed]
- Ljung, P.; Krüger, J.; Groller, E.; Hadwiger, M.; Hansen, C.D.; Ynnerman, A. State of the art in transfer functions for direct volume rendering. Comput. Graph. Forum 2016, 35, 669–691. [Google Scholar] [CrossRef]
- Besançon, L.; Ynnerman, A.; Keefe, D.F.; Yu, L.; Isenberg, T. The state of the art of spatial interfaces for 3d visualization. Comput. Graph. Forum 2021, 40, 293–326. [Google Scholar] [CrossRef]
- Suganyadevi, S.; Seethalakshmi, V.; Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retr. 2022, 11, 19–38. [Google Scholar] [CrossRef]
- Ruijters, D. Common Artifacts in Volume Rendering. arXiv 2021, arXiv:2109.13704. [Google Scholar]
- Berisha, V.; Krantsevich, C.; Hahn, P.R.; Hahn, S.; Dasarathy, G.; Turaga, P.; Liss, J. Digital medicine and the curse of dimensionality. NPJ Digit. Med. 2021, 4, 153. [Google Scholar] [CrossRef]
- de Moura Pinto, F.; Freitas, C.M. Design of multi-dimensional transfer functions using dimensional reduction. In Proceedings of the 9th Joint Eurographics/IEEE VGTC Conference on Visualization, Norrköping, Sweden, 23–25 May 2007; Eurographics Association: Goslar, Germany, 2007; pp. 131–138. [Google Scholar]
- Kim, H.S.; Schulze, J.P.; Cone, A.C.; Sosinsky, G.E.; Martone, M.E. Dimensionality reduction on multi-dimensional transfer functions for multi-channel volume data sets. Inf. Vis. 2010, 9, 167–180. [Google Scholar] [CrossRef]
- de Bodt, C.; Mulders, D.; Vserleysen, M.; Lee, J.A. Fast Multiscale Neighbor Embedding. IEEE Trans. Neural Netw. Learn. Syst. 2020, 34, 1546–1560. [Google Scholar] [CrossRef]
- Zhu, R.; Dornaika, F.; Ruichek, Y. Semi-supervised elastic manifold embedding with deep learning architecture. Pattern Recognit. 2020, 107, 107425. [Google Scholar] [CrossRef]
- Zheng, J.; Qiu, H.; Xu, X.; Wang, W.; Huang, Q. Fast Discriminative Stochastic Neighbor Embedding Analysis. Comput. Math. Methods Med. 2013, 2013, 106867. [Google Scholar] [CrossRef]
- de Bodt, C.; Mulders, D.; López-Sánchez, D.; Verleysen, M.; Lee, J.A. Class-aware t-SNE: cat-SNE; ESANN: Bruges, Belgium, 2019; pp. 409–414. [Google Scholar]
- Sheikhpour, R.; Sarram, M.A.; Gharaghani, S.; Chahooki, M.A.Z. A survey on semi-supervised feature selection methods. Pattern Recognit. 2017, 64, 141–158. [Google Scholar] [CrossRef]
- Zhu, T.; Pimentel, M.A.; Clifford, G.D.; Clifton, D.A. Unsupervised bayesian inference to fuse biosignal sensory estimates for personalizing care. IEEE J. Biomed. Health Inform. 2018, 23, 47–58. [Google Scholar] [CrossRef]
- Huang, S.; Elgammal, A.; Huangfu, L.; Yang, D.; Zhang, X. Globality-locality preserving projections for biometric data dimensionality reduction. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014; pp. 15–20. [Google Scholar]
- Huang, R.; Zhang, G.; Chen, J. Semi-supervised discriminant Isomap with application to visualization, image retrieval and classification. Int. J. Mach. Learn. Cybern. 2019, 10, 1269–1278. [Google Scholar] [CrossRef]
- Engel, K.; Hadwiger, M.; Kniss, J.M.; Lefohn, A.E.; Salama, C.R.; Weiskopf, D. Real-Time Volume Graphics. In Proceedings of the SIGGRAPH ’04: ACM SIGGRAPH 2004 Course Notes, Los Angeles, CA, USA, 8–12 August 2004; Association for Computing Machinery: New York, NY, USA, 2004; p. 29-es. [Google Scholar] [CrossRef]
- Serna-Serna, W.; de Bodt, C.; Alvarez-Meza, A.M.; Lee, J.A.; Verleysen, M.; Orozco-Gutierrez, A.A. Semi-supervised t-SNE with multi-scale neighborhood preservation. Neurocomputing 2023, 550, 126496. [Google Scholar] [CrossRef]
- Levoy, M. Display of surfaces from volume data. IEEE Comput. Graph. Appl. 1988, 8, 29–37. [Google Scholar] [CrossRef]
- Kniss, J.; Kindlmann, G.; Hansen, C. Multidimensional transfer functions for interactive volume rendering. IEEE Trans. Vis. Comput. Graph. 2002, 8, 270–285. [Google Scholar] [CrossRef]
- Sereda, P.; Bartroli, A.V.; Serlie, I.W.; Gerritsen, F.A. Visualization of boundaries in volumetric data sets using LH histograms. IEEE Trans. Vis. Comput. Graph. 2006, 12, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Haidacher, M.; Patel, D.; Bruckner, S.; Kanitsar, A.; Gröller, M.E. Volume visualization based on statistical transfer-function spaces. In Proceedings of the IEEE PacificVis, Taipei, Taiwan, 2–5 March 2010. [Google Scholar]
- Correa, C.; Ma, K.L. Size-based transfer functions: A new volume exploration technique. IEEE Trans. Vis. Comput. Graph. 2008, 14, 1380–1387. [Google Scholar] [CrossRef] [PubMed]
- Kindlmann, G.; Whitaker, R.; Tasdizen, T.; Moller, T. Curvature-based transfer functions for direct volume rendering: Methods and applications. In Proceedings of the IEEE Visualization (VIS 2003), Seattle, WA, USA, 19–24 October 2003; IEEE: Piscataway, NJ, USA, 2003; pp. 513–520. [Google Scholar]
- Totsuka, T.; Levoy, M. Frequency domain volume rendering. In Proceedings of the 20th Annual Conference on Computer Graphics and Interactive Techniques, Anaheim, CA, USA, 2–6 August 1993; pp. 271–278. [Google Scholar]
- Roettger, S.; Bauer, M.; Stamminger, M. Spatialized transfer functions. In Proceedings of the EuroVis, Leeds, UK, 1–3 June 2005; pp. 271–278. [Google Scholar]
- Caban, J.J.; Rheingans, P. Texture-based transfer functions for direct volume rendering. IEEE Trans. Vis. Comput. Graph. 2008, 14, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, F.Y.; Ma, K.L. A cluster-space visual interface for arbitrary dimensional classification of volume data. In Proceedings of the IEEE TCVG Conference on Visualization, Konstanz, Germany, 19–21 May 2004. [Google Scholar]
- Nguyen, B.P.; Tay, W.L.; Chui, C.K.; Ong, S.H. A clustering-based system to automate transfer function design for medical image visualization. Vis. Comput. 2012, 28, 181–191. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, W.; Zhang, J.; Dong, T.; Shan, G.; Chi, X. Efficient volume exploration using the gaussian mixture model. IEEE Trans. Vis. Comput. Graph. 2011, 17, 1560–1573. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, K.P.; Schultz, T. Learning probabilistic transfer functions: A comparative study of classifiers. Comput. Graph. Forum 2015, 34, 111–120. [Google Scholar] [CrossRef]
- Matrakas, M.D.; Scheer, S. Three-Dimensional Representation of a Multidimensional Data Set. Appl. Math. Sci. 2016, 10, 959–971. [Google Scholar] [CrossRef]
- Ponciano, D.; Seefelder, M.; Marroquim, R. Graph-based interactive volume exploration. Comput. Graph. 2016, 60, 55–65. [Google Scholar] [CrossRef]
- Berger, M.; Li, J.; Levine, J.A. A generative model for volume rendering. IEEE Trans. Vis. Comput. Graph. 2018, 25, 1636–1650. [Google Scholar] [CrossRef]
- Engel, D.; Ropinski, T. Deep volumetric ambient occlusion. IEEE Trans. Vis. Comput. Graph. 2020, 27, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.; Bohak, C.; Engel, D.; Mindek, P.; Strnad, O.; Wonka, P.; Li, S.; Ropinski, T.; Viola, I. Finding Nano-Ötzi: Cryo-Electron Tomography Visualization Guided by Learned Segmentation. IEEE Trans. Vis. Comput. Graph. 2022, 29, 4198–4214. [Google Scholar] [CrossRef] [PubMed]
- Van Der Maaten, L. Barnes-hut-sne. arXiv 2013, arXiv:1301.3342. [Google Scholar]
- Max, N. Optical models for direct volume rendering. IEEE Trans. Vis. Comput. Graph. 1995, 1, 99–108. [Google Scholar] [CrossRef]
- Rister, B.; Yi, D.; Shivakumar, K.; Nobashi, T.; Rubin, D.L. CT-ORG, a new dataset for multiple organ segmentation in computed tomography. Scientific Data 2020, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive machine learning for (bio) image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Duran, A.H.; Duran, M.N.; Masood, I.; Maciolek, L.M.; Hussain, H. The additional diagnostic value of the three-dimensional volume rendering imaging in routine radiology practice. Cureus 2019, 11, e5579. [Google Scholar] [CrossRef]
- Bai, S.; Ma, C.; Wang, X.; Zhou, S.; Jiang, H.; Ma, L.; Jiang, H. Application of Medical Image 3D Visualization Web Platform in Auxiliary Diagnosis and Preoperative Planning. J. Image Graph. 2023, 11, 32–39. [Google Scholar] [CrossRef]
- Chen, R.; Ran, Y.; Wu, Y.; Xu, H.; Niu, J.; Zhang, Y.; Cheng, J. The value of the cinematic volume rendering technique: Magnetic resonance imaging in diagnosing tumors associated with the brachial plexus. Eur. J. Med. Res. 2023, 28, 569. [Google Scholar] [CrossRef]
- Chen, R.; Ran, Y.; Xu, H.; Niu, J.; Wang, M.; Wu, Y.; Zhang, Y.; Cheng, J. The guiding value of the cinematic volume rendering technique in the preoperative diagnosis of brachial plexus schwannoma. Front. Oncol. 2023, 13, 1278386. [Google Scholar] [CrossRef]
- Ardakani, A.A.; Mohammadi, A.; Faeghi, F.; Acharya, U.R. Performance evaluation of 67 denoising filters in ultrasound images: A systematic comparison analysis. Int. J. Imaging Syst. Technol. 2023, 33, 445–464. [Google Scholar] [CrossRef]
- Bhalodia, R.; Elhabian, S.; Adams, J.; Tao, W.; Kavan, L.; Whitaker, R. DeepSSM: A blueprint for image-to-shape deep learning models. Med. Image Anal. 2024, 91, 103034. [Google Scholar] [CrossRef] [PubMed]








| Method | HD Similarity Computation | Iteration in Gradient-Based Optimization |
|---|---|---|
| SNE, t-SNE | ||
| BH t-SNE | ||
| Multi-scale t-SNE | ||
| Fast multi-scale t-SNE, FSS.t-SNE (ours) |
| TF Approach/Structure | Liver | Lungs | Kidneys | Bone |
|---|---|---|---|---|
| FSS.t-SNE-based TF (ours) | 0.8053 | 0.8899 | 0.6487 | 0.6692 |
| Intensity vs. Gradient Magnitude [25] | 0.6010 | 0.6075 | 0.2575 | 0.3497 |
| Intensity vs. Laplacian [7] | 0.4244 | 0.6671 | 0.5805 | 0.4055 |
| Statistical Properties [27] | 0.2932 | 0.8197 | 0.5307 | 0.6745 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serna-Serna, W.; Álvarez-Meza, A.M.; Orozco-Gutiérrez, Á. Fast Semi-Supervised t-SNE for Transfer Function Enhancement in Direct Volume Rendering-Based Medical Image Visualization. Mathematics 2024, 12, 1885. https://doi.org/10.3390/math12121885
Serna-Serna W, Álvarez-Meza AM, Orozco-Gutiérrez Á. Fast Semi-Supervised t-SNE for Transfer Function Enhancement in Direct Volume Rendering-Based Medical Image Visualization. Mathematics. 2024; 12(12):1885. https://doi.org/10.3390/math12121885
Chicago/Turabian StyleSerna-Serna, Walter, Andrés Marino Álvarez-Meza, and Álvaro Orozco-Gutiérrez. 2024. "Fast Semi-Supervised t-SNE for Transfer Function Enhancement in Direct Volume Rendering-Based Medical Image Visualization" Mathematics 12, no. 12: 1885. https://doi.org/10.3390/math12121885
APA StyleSerna-Serna, W., Álvarez-Meza, A. M., & Orozco-Gutiérrez, Á. (2024). Fast Semi-Supervised t-SNE for Transfer Function Enhancement in Direct Volume Rendering-Based Medical Image Visualization. Mathematics, 12(12), 1885. https://doi.org/10.3390/math12121885






