Abstract
Medical images contain complex information, and the automated analysis of medical images can greatly assist doctors in clinical decision making. Therefore, the automatic segmentation of medical images has become a hot research topic in recent years. In this study, a novel architecture called a contoured convolutional transformer (CCTrans) network is proposed to solve the segmentation problem. A dual convolutional transformer block and a contoured detection module are designed, which integrate local and global contexts to establish reliable relational connections. Multi-scale features are effectively utilized to enhance semantic feature understanding. The dice similarity coefficient (DSC) is employed to evaluate experimental performance. Two public datasets with two different modalities are chosen as the experimental datasets. Our proposed method achieved an average DSC of 83.97% on a synapse dataset (abdominal multi-organ CT) and 92.15% on an ACDC dataset (cardiac MRI). Especially for the segmentation of small and complex organs, our proposed model achieves better segmentation results than other advanced approaches. Our experiments demonstrate the effectiveness and robustness of the novel method and its potential for real-world applications. The proposed CCTrans network offers a universal solution with which to achieve precise medical image segmentation.
Keywords:
transformer; medical image segmentation; visual attention mechanism; deep neural networks; machine learning MSC:
68T07; 68T01
1. Introduction
Computer vision (CV) [1] and machine learning (ML) [2] techniques have enabled the widespread use of deep neural networks in medical image analysis. The accurate, robust classification of medical images is essential for computer-aided and image diagnosis. The advancement of deep learning (DL) technology has greatly improved the accuracy and efficiency of medical image analysis. Deep neural networks (DNNs) can automatically recognize features from medical image data and handle complex non-linear relationships and large-scale data, thereby improving the robustness and accuracy of medical image analysis. In computer-aided diagnosis (CAD) and image diagnosis, the accurate and robust segmentation of medical images is crucial for improving the efficiency and diagnostic capabilities of doctors. By utilizing DNNs for medical image classification, doctors’ workloads can be reduced, and the accuracy and efficiency of disease diagnosis and treatment can be improved.
As U-Net [3,4] has achieved success in the field of medical image analysis, U-shaped frameworks, such as Res-Unet [5], Dense-Unet [6], etc., have become popular in modern medical image processing due to their effectiveness. The effectiveness of their methods can be attributed to the usage of skip connections, which merge feature maps with high-level abstractions generated by the decoder with the low-level feature maps generated by the encoder [7]. The application of this technique allows for the extraction of detailed information about target objects even in the presence of complex backgrounds, making it an essential tool for accurate medical image segmentation. Additionally, the incorporation of skip connections in U-shaped networks reduces the vanishing gradient problem and improves the flow of information across the network. As a result, models based on U-shaped frameworks have shown superior performance compared to regular neural network models in medical image segmentation tasks. Taking inspiration from the benefits of transformers in the field of natural language processing (NLP) [8,9,10], several medical image segmentation techniques have endeavored to employ transformers to tackle the problems of implicit assumptions. Combining the U-shaped architecture with a transformer can fully leverage the advantages of both methods. Such a combination of these two advanced techniques can better exploit local features and global contextual information, thereby improving segmentation accuracy and robustness.
Therefore, we developed the contoured convolutional transformer network (CCTrans), which is a kind of powerful medical-imaging segmentation technique. The proposed model adopts a U-shaped architecture, which consists of a dual convolutional (DC) transformer block and a contour detection module. The proposed method can integrate local and global contexts to establish reliable relational connections. In summary, our work contributes in the following ways:
- A novel segmentation model named the contoured convolutional transformer network (CCTrans) was designed for accurate medical image segmentation, which utilizes gated modules and skip connections. Both the dual convolutional (DC) transformer block and the contour detection module are designed to process important information contained in medical images.
- The DC transformer blocks utilize convolutional kernels with different sizes to capture multi-scale information. Short-distance and long-distance attention mechanisms are combined to extract local features and capture long-range dependencies, thereby enhancing the model’s interpretability.
- The contour detection module employs traditional CV techniques, which can help determine regions of interest and refine insignificant contoured segmentation information.
- Comprehensive experiments on two public datasets showed that the novel CCTrans model outperforms other state-of-the-art medical image segmentation methods. Diversified experimental results with illustrations are also presented in the paper.
2. Related Works
This section is divided into three parts, which address convolutional neural network methods, transformer-based architectures, and U-shaped architectures with transformers.
2.1. Convolutional Neural Network Methods for Medical Image Segmentation
In the past, medical image segmentation methods primarily relied on CV-based and ML-based algorithms [11]. However, with the emergence of convolutional neural networks (CNNs), Unet was designed for medical image segmentation [12]. Due to the superior performance and simplicity of the U-shaped structure, numerous convolutional networks have emerged, such as Unet++ [13], Att-Unet [14], and nnUnet [15]. In addition, it has also been applied to the domain of three-dimensional medical image segmentation, such as 3D-Unet [16] and V-Net [17]. CNNs have been highly successful in medical image segmentation owing to their strong representational abilities. However, modeling long-range dependencies and contextual relationships using CNNs is still a challenging task. Although some studies have attempted to address this issue by modeling long-range dependencies for convolution, the developed methods still faced significant limitations. In contrast, the transformer structure, which has been successful in the fields of NLP and CV, offers an effective solution for the modeling of long-range dependencies in medical image segmentation.
2.2. Transformer-Based Architectures for Medical Image Segmentation
Drawing inspiration from the success of transformers in the field of CV, several scholars have endeavored to incorporate transformer components to improve the performance of medical image segmentation. Vision transformers (ViTs) [18] have achieved performance comparable to convolutional neural networks (CNNs) in large-scale image classification tasks. A ViT uses two-dimensional image patches with positional embedding as an input sequence and applies transformers with global self-attention mechanisms to process full-sized images. ViTs constitute a pioneering effort that demonstrates the ability of a pure transformer-based structure to achieve exceptional performance in image recognition, particularly when pre-trained on large-scale datasets such as ImageNet-22K and JFT-300M.
2.3. U-Shaped Architectures with Transformers for Medical Image Segmentation
TransUnet [19] draws inspiration from ViTs and further enhances the performance of medical image segmentation by combining the strengths of transformers and Unet. It is the first model to integrate self-attention mechanisms through the combination of transformers and Unet, demonstrating the effectiveness of using transformers as robust encoders for medical image segmentation.
Based on TransUnet, TransUnet+ [20] was proposed, which redesigns the skip connection. The restructured skip connection incorporates a strengthening component that leverages the score matrix generated by the transformer block to effectively optimize skip features and refine global attention.
SwinUnet [21] is different from most previous transformer-based models as it has the flexibility to be used as an all-purpose backbone architecture, which is achieved by introducing a hierarchical architecture for dense prediction. The encoder in SwinUnet adopts the Swin transformer with shifted windows to extract context information, while a decoder based on a symmetric Swin transformer with a patch-expanding layer is proposed to carry out up-sampling operations and restore the spatial resolution of the feature maps.
C2Former [22] presents a fresh perspective that stems from SwinUnet. Wang et al. innovatively redesigned the cross-convolutional self-attention mechanism algorithm to model long- and short-distance dependencies, resulting in an improved understanding of semantic features.
Building upon these studies, the proposed network redesigns the basic transformer unit and integrates abundant contexts. The contour information contained in medical images is also introduced into the method as prior knowledge, which enhances the interpretability of the model and greatly improves the effectiveness of medical image segmentation.
3. Proposed Method
In this study, the proposed method is explained through the elaboration of three aspects. Firstly, the overall architecture is introduced. Secondly, the dual convolutional transformer is designed so that the component can handle the deeper features in greater detail. The third important component of our proposed model is the contour detection module, which is designed to improve the network’s ability to extract edge features and upgrade contoured segmentation information that may not be obvious.
3.1. Overall Architecture
The overall architecture proposed in this study is illustrated in Figure 1. On the basis of Swin-Unet [21], the architecture of the novel model utilizes a U-shaped structure, which consists of an encoder, a decoder, and skip connections. The U-shaped design helps to capture both local and global features, while the encoder and decoder components are responsible for feature extraction and reconstruction, respectively. The skip connections facilitate the integration of low-level and high-level features, which improves the model’s accuracy and robustness. The sequences with various resolutions are processed by different blocks and modules. Such operations allow the model to completely extract features and capture potential information. Overall, this design enables the model to effectively extract and utilize features from the input data to attain improved performance.
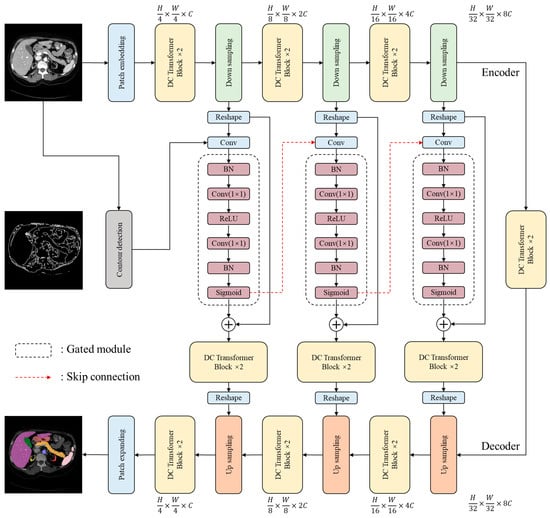
Figure 1.
The overall architecture of the proposed CCTrans network.
In the encoder stage, the input image is represented as , where the variable denotes height, the variable denotes width, and the variable represents the number of channels of the input image. equals 3 in this study. The common patch-embedding method involves splitting an input image into non-overlapping blocks at a size of [23,24,25]. However, models that use a single-size convolution kernel have weak generalization ability, are prone to overfitting, and cannot comprehensively capture the information contained in an image. To address these flaws, multi-scale sampling is performed by first splitting the image at different scales and then concatenating the sampled results to form a new patch. By adopting this approach, the loss of fine-grained information is minimized. The multiple-scale sampling process utilizes convolutional operations with four different kernel sizes (). Processed by the patch-embedding operation, the size of the processed patch is . This methodology was inspired by Wang et al. [26].
Detailed descriptions and formulaic expressions of the proposed dual convolutional (DC) transformer structure are provided in Section 3.2.
The gated module is composed of batch normalization (BN), convolution, ReLU, and Sigmoid layers, which was inspired by ResNet. It combines feature maps from the current and upper layers while filtering out irrelevant information to extract more comprehensive feature information. The encoder’s one-dimensional features are transformed into two-dimensional features and fused with edge information from contour detection in the gated module. The obtained fusion features are then transferred into the gated module to be further combined with various spatial features. The gated module takes intermediate data from the encoder and some information from the contour detection module as inputs. The output of the gated module will be used in the intermediate layers of the decoder. The calculation is expressed as a two-layer proposed block:
where represents the output part of the encoder; represents the input of the gated module after the reshaping operations; represents the output after the convolutional operations; represents the -th contoured features extracted by the contour detection module, whose detailed description is stated in Section 3.3; and represents the gate module, for which .
In the decoder stage, a linear extension is used to perform up-sampling. This involves linearly mapping sequences to a high-dimensional space, for which all the obtained features are utilized to generate the final results.
3.2. Dual Convolutional (DC) Transformer Block
The architecture of the DC transformer block is illustrated in Figure 2. Inspired by the Swin Transformer block [24], the dual convolutional (DC) transformer block is designed to capture features with multiple scales and expand the receptive field of the proposed technique. To improve the combination of the attention mechanism and the convolutional operation for medical image segmentation, a parallel convolutional attention mechanism was redesigned, which functions alongside self-attention. This approach allows for the simultaneous capture of information from the spatial and channel dimensions. Following the CBAM [27] approach, we start by generating average and maximum features over the spatial dimension for the input . These features are then passed through a fully connected network. The final output is obtained using the spatial and channel dimension attention as follows:
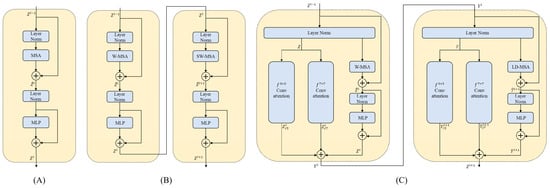
Figure 2.
The internal structure diagrams of different transformer architectures. (A) represents the basic transformer block. (B) represents the Swin transformer block [24]. (C) represents the proposed dual convolutional (DC) transformer block.
and refer to the operations of max- and average-pooling, respectively, and denotes the Sigmoid function.
The smaller convolutional kernels () are more suitable for the processing of local features such as texture and details, while the larger convolutional kernels () are more suitable for the processing of global features such as shape and contour. By using convolutional kernels of different sizes, multi-scale features can be better captured. The equations for computing the multi-scale convolution are as follows:
In addition, a window multi-head self-attention (W-MSA) mechanism and a long-distance multi-head self-attention (LD-MSA) mechanism are fully utilized. The difference between the two approaches is illustrated in Figure 3. The W-MSA mechanism can be used to develop relevant associations between local regions. It divides the input data into multiple windows with a size of for feature modelling. To incorporate fine-grained internal features and efficiently capture hidden details across various regions, tokens are employed. In the LD-MSA mechanism, represents the sampling interval. The LD-MSA mechanism was used to apply masking processing to unsampled image blocks. The feature graph is split into individual units with a length and width of , resulting in groups. Feature modeling is conducted within each group to obtain feature maps. On the other hand, the W-MSA mechanism samples adjacent image blocks to establish dependencies. The outputs of both mechanisms are combined to obtain the final feature representation. The calculations of the W-MSA and LD-MSA can be performed as follows:
where denotes the learnable weight matrix, represents the query, represents the key, represents the value, and represents the bias. The stacked blocks consist of W-MSA and LD-MSA.
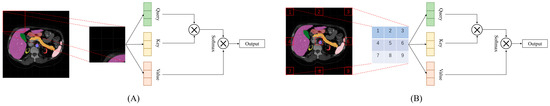
Figure 3.
The calculation processes of window multi-head self-attention (W-MSA) mechanism and long-distance multi-head self-attention (LD-MSA) mechanism. (A) represents the W-MSA approach and (B) represents the LD-MSA approach.
The standard operates through query, key, and value matrices. However, the self-attention mechanism has undeniable shortcomings with respect to modeling short-range dependencies. To address this issue, we incorporate the W-MSA to integrate the associations. By adopting this window-partitioning approach, the W-MSA can be calculated as follows:
where represents the output results of W-MSA of the -layer, while represents the output results of multilayer perceptron (MLP) of the -layer. To efficiently capture the interrelated features among different tokens, the design of the LD-MSA method referred to the transformer’s cross-scale attention mechanism [26]. The LD-MSA samples feature maps along a given length and width, allowing for self-attention within the obtained groups and improving the interaction between each window’s information element. The LD-MSA can be defined as follows:
where denotes the output results of the LD-MSA of the ()-th layer, while denotes the output results of the MLP of the ()-th layer.
Finally, represents the middle output of the first layer of the DC transformer block, while represents the output of the second layer of the DC transformer block.
3.3. Contour Detection Module
The contour detection module is an important component of our proposed CCTrans architecture. By utilizing the contour information contained in medical images, the module aims to optimize the interpretability of the model and improve the effectiveness of medical image segmentation. Specifically, the contour detection module is designed to preserve all contour information, including both internal and external contours, to provide the model with more reference information in order to acquire more accurate segmentation results. Compared to traditional segmentation methods that only focus on external contours, our approach can effectively capture and utilize more detailed contour information. This allows the model to better understand the shape and boundaries of the target objects in medical images, leading to more accurate segmentation results. The “findContours” function in the OpenCv package of Python is applied to detect and locate the contours of the object in a medical image, which is a variant of the Suzuki–Beck algorithm (Figure 4).
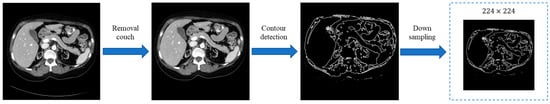
Figure 4.
The workflow of contour detection module.
As illustrated in Figure 4, the testing couch (or other interfering information) is first removed from the original image. If there is no such interference, this step can be omitted. Then, the contoured features are detected and extracted. The CV approaches are employed to finish contour detection, which can be used to accurately extract the contour information from images while preserving local features. Both the contoured and local features have high applicational value for tasks that require contour analysis and feature extraction in medical image processing. Finally, the contour detection module utilizes a down-sampling operation to match dimensions.
4. Datasets and Experiments
This section is composed of three parts, including the introduction of the experimental datasets, the experimental settings, and the experimental results and analysis.
4.1. Experimental Datasets
There are two public datasets with two modalities (CT and MRI) utilized to evaluate the corresponding approaches in this study:
- The synapse abdominal multi-organ (Synapse) dataset: Synapse contains thirty CT volumes of eight kinds of abdominal organs (the aorta, gallbladder, spleen, left kidney, right kidney, liver, pancreas, spleen, and stomach), with a total of 3779 slices [28]. In the subsequent experiments, the training set consists of eighteen CT cases, while the testing set is composed of the remaining twelve cases.
- Automated cardiac diagnosis challenge (ACDC) dataset: The ACDC dataset contains one hundred MRI cases [29]. The MRI data consist of data concerning the right ventricle, myocardium, and left ventricle. In the subsequent experiments, seventy cases form the training set, ten cases form the validating set, and the remaining twenty cases form the testing set.
4.2. Experimental Settings
The proposed CCTrans architecture and experiments were developed using a universal Python package, called PyTorch [30] (see: https://pytorch.org/ (accessed on 30 March 2023)), which was run on the Linux operating system. All the experiments, including the model-training process, in this study were carried out on a high-powered workstation, which included an Intel Xeon E5-2620 CPU operating @2.4 GHz and three NVIDIA TITAN XP GPUs with 12 GB of RAM. The input image size was . The batch size was 24, and the number of training epochs was 200. We utilized basic data augmentation techniques for the subsequent experiments, such as random rotation and flipping. The SGD optimizer with an initial learning rate of 0.05 was used, which was via exponential decay. The momentum was equal to 0.9, while the weight decay was equal to 0.0001. The joint loss function was employed as the network-training strategy, which comprised dice loss and cross entropy loss:
Both and are two kinds of hyperparameters. is set to 0.6, while is set to 0.4.
The Dice Similarity Coefficient (DSC) [31] is a statistical measure employed to evaluate the similarity between two sets; it is commonly utilized to assess the performance of medical image segmentation methods. The DSC is applied to evaluate the segmentation accuracy in all experiments. The larger the value of DSC, the better the performance of the corresponding approach. The DSC can be defined as follows:
where represents the region segmented by the corresponding architecture, and represents the ground truth.
4.3. Experimental Results and Analysis
We conducted an evaluation of the proposed CCTrans architecture on the Synapse dataset. The experimental metrics, as listed in Table 1, indicate that our proposed CCTrans network outperforms other forefront methods with regard to the average DSC metric. Specifically, our proposed method achieves a 6.48% improvement over TransUnet and a 5.14% improvement over SwinUnet. The experimental metrics also demonstrate that all the models perform similarly for voluminous and normally shaped organs (including the stomach, spleen, and liver). However, for small-sized and complex organs (including the pancreas, gallbladder, and aorta), our proposed CCTrans network outperforms C2Former by 2.78%, 1.51%, and 1.11%, respectively. By incorporating the contour information as prior knowledge into the method, the proposed model not only enhances the model’s interpretability but also significantly improves the accuracy of medical image segmentation.

Table 1.
Segmentation performance of different methods with respect to the Synapse dataset.
To better understand the performance of our proposed approach, the visualization segmentation results of the Synapse dataset are illustrated in Figure 5. The poor segmentation performance of the TransUnet model may be attributed to the fact that its structural design is not suitable for the segmentation of complex and small organs. It can be observed that our proposed CCTrans network achieves outstanding performance in terms of segmenting small organs. To a certain extent, the network is still affected by the interference from surrounding tissues; thus, the segmented regions were slightly larger than the corresponding ground truth. These visualizations provide valuable insights into the strengths and weaknesses of our approach and can help guide future research directions.
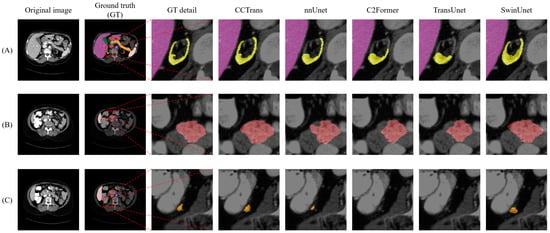
Figure 5.
Visual comparison of segmentation results on the Synapse dataset reveals several issues. (A–C) represent three typical diagrams of Synapse dataset. The first row shows that nnUnet, C2Former, and TransUnet failed to recognize the smaller kidney structure (yellow region), while SwinUnet mistakenly identified more regions as the target area. In the second row, none of the other methods accurately identified the boundaries of the stomach (brown region). In the third row, both C2Former and TransUnet failed to diagnose the pancreas region (orange region). Only our proposed method achieved relatively satisfactory results in all three cases.
We also evaluated the proposed CCTrans architecture based on the ACDC dataset. The experimental results, as listed in Table 2, indicate that our proposed CCTrans network outperforms other advanced methods in relation to the average DSC metric. While this improvement may not appear substantial, it should be noted that the DSC metric is already at a high level, making this a notable achievement. To better understand the performance of our proposed approach, the visualization segmentation results of the ACDC dataset are shown in Figure 6.

Table 2.
Segmentation performance of comparative methods with respect to the ACDC dataset.
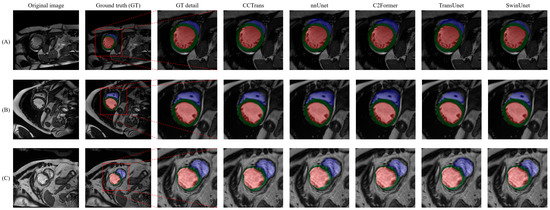
Figure 6.
Visual comparison of segmentation results with respect to the ACDC dataset reveals several issues. (A–C) represent three typical diagrams of ACDC dataset. Even under low-contrast conditions, our proposed method, C2Former, and nnUnet can achieve relatively accurate segmentation results with respect to the right ventricle (blue region) and left ventricle (green region), while TransUnet and SwinUnet are less accurate in identifying edges.
5. Discussion
Our proposed method has been validated through the aforementioned experiments. The gated module redesign enables the control of the relative weights between input and gating features, thus enhancing the model’s feature representation capacity. Additionally, the skip connection merges high-level feature maps generated by the decoder with low-level feature maps generated by the encoder, thereby enabling the recovery of fine-grained details of target organs, even in intricate backgrounds. The DC transformer block uses convolutional kernels of varying sizes to capture multi-scale information. It also combines short- and long-distance attention mechanisms to extract local features and capture long-range dependencies, thus improving interpretability. The contour detection module utilizes medical image contour information as prior knowledge, enhancing interpretability and greatly improving segmentation efficacy. Given that our input images are two-dimensional, our future research will explore the CCTrans network’s potential applications in three-dimensional medical image segmentation [32,33].
6. Conclusions
In this study, we proposed the CCTrans network, which incorporates a DC transformer block and a contour detection module. By utilizing gated modules and skip connections, the feature representation capacity of the model is enhanced. Our experiments, conducted on two public datasets, demonstrate that CCTrans outperforms the existing innovative methods. We aim to promote the use of the CCTrans network in clinics, where it will assist doctors in completing organ segmentation tasks quickly and improve diagnostic efficiency. The potential experimental results of this model when applied to other medical data, not just those concerning organ and cardiovascular segmentation, have also piqued our research interest. In future research, we will continue to improve the model’s performance and explore the use of more lightweight structures to further improve its computational speed and segmentation accuracy. Additionally, we will investigate the application of CCTrans in other data domains and further refine the model based on experimental results.
Author Contributions
Conceptualization, Z.Y. and H.Z.; methodology, J.W.; validation, H.Z. and J.W.; formal analysis, H.Z. and J.W.; investigation, J.W.; resources, J.W.; data curation, J.W.; writing—original draft preparation, J.W.; writing—review and editing, J.W. and H.Z.; visualization, J.W.; supervision, Z.Y.; project administration, Z.Y.; funding acquisition, Z.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Major Science and Technology Projects of China under Grant 2018AAA0100201, the Major Science and Technology Project from the Science & Technology Department of Sichuan Province: 2022ZDZX0023.
Data Availability Statement
Data available in a publicly accessible repository.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Parker, J.R. Algorithms for Image Processing and Computer Vision; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.-W.; Heng, P.-A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. Unet 3+: A Full-Scale Connected Unet for Medical Image Segmentation. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1055–1059. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted Res-Unet for High-Quality Retina Vessel Segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 327–331. [Google Scholar]
- Cai, S.; Tian, Y.; Lui, H.; Zeng, H.; Wu, Y.; Chen, G. Dense-UNet: A novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant. Imaging Med. Surg. 2020, 10, 1275. [Google Scholar] [CrossRef]
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The Importance of Skip Connections in Biomedical Image Segmentation. In Proceedings of the International Workshop on Deep Learning in Medical Image Analysis, International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2016; pp. 179–187. [Google Scholar]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M. Transformers: State-of-the-Art Natural Language Processing. In Proceedings of the 2020 Conference on EMPIRICAL Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; Association for Computational Linguistics: Cedarville, OH, USA, 2020; pp. 38–45. [Google Scholar]
- Tetko, I.V.; Karpov, P.; Van Deursen, R.; Godin, G. State-of-the-art augmented NLP transformer models for direct and single-step retrosynthesis. Nat. Commun. 2020, 11, 5575. [Google Scholar] [CrossRef]
- Gillioz, A.; Casas, J.; Mugellini, E.; Abou Khaled, O. Overview of the Transformer-based Models for NLP Tasks. In Proceedings of the 2020 15th Conference on Computer Science and Information Systems (FedCSIS), Sofia, Bulgaria, 6–9 September 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 179–183. [Google Scholar]
- Sharma, N.; Aggarwal, L.M. Automated medical image segmentation techniques. J. Med. Phys. 2010, 35, 3–14. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Lian, S.; Luo, Z.; Zhong, Z.; Lin, X.; Su, S.; Li, S.; Representation, I. Attention guided U-Net for accurate iris segmentation. J. Vis. Commun. Image Represent. 2018, 56, 296–304. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Wang, C.; MacGillivray, T.; Macnaught, G.; Yang, G.; Newby, D. A two-stage 3D Unet framework for multi-class segmentation on full resolution image. arXiv 2018, arXiv:1804.04341. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 565–571. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Liu, Y.; Wang, H.; Chen, Z.; Huangliang, K.; Zhang, H. TransUNet+: Redesigning the skip connection to enhance features in medical image segmentation. Knowl. Based Syst. 2022, 256, 109859. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the Computer Vision–ECCV 2022 Workshops, Tel Aviv, Israel, 23–27 October 2022; Proceedings, Part III. Springer: Berlin/Heidelberg, Germany, 2023; pp. 205–218. [Google Scholar]
- Wang, J.; Zhao, H.; Liang, W.; Wang, S.; Zhang, Y. Biology, Cross-convolutional transformer for automated multi-organs segmentation in a variety of medical images. Phys. Med. Biol. 2023, 68, 035008. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Tian, Z.; Wang, Y.; Zhang, B.; Ren, H.; Wei, X.; Xia, H.; Shen, C. Twins: Revisiting the design of spatial attention in vision transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 9355–9366. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 568–578. [Google Scholar]
- Wang, W.; Yao, L.; Chen, L.; Lin, B.; Cai, D.; He, X.; Liu, W. CrossFormer: A versatile vision transformer hinging on cross-scale attention. arXiv 2021, arXiv:2108.00154. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Staffler, B.; Berning, M.; Boergens, K.M.; Gour, A.; Smagt, P.v.d.; Helmstaedter, M.J.E. SynEM, automated synapse detection for connectomics. Elife 2017, 6, e26414. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 8024–8035. Available online: https://proceedings.neurips.cc/paper_files/paper/2019/file/bdbca288fee7f92f2bfa9f7012727740-Paper.pdf (accessed on 11 April 2023).
- Thada, V.; Jaglan, V. Technology, Comparison of jaccard, dice, cosine similarity coefficient to find best fitness value for web retrieved documents using genetic algorithm. Int. J. Innov. Eng. Technol. 2013, 2, 202–205. [Google Scholar]
- Heimann, T.; Meinzer, H.-P. Statistical shape models for 3D medical image segmentation: A review. Med. Image Anal. 2009, 13, 543–563. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).