Optimal Model Averaging for Semiparametric Partially Linear Models with Censored Data
Abstract
1. Introduction
2. Model Setup and Parametric Estimation
3. Model-Averaging Estimator and Weight Choice Criterion
4. Asymptotic Optimality
- (Condition (C.1)) , where for any distribution function L.
- (Condition (C.2)) , where denotes the maximum singular value of a matrix, and is a constant.
- (Condition (C.3)) , a.s.
- (Condition (C.4)) and , a.s.
- (Condition (C.5)) , a.s.
- (Condition (C.6)) , a.s., where is a constant.
- (Condition (C.7)) The function g belongs to a class of functions , whose rth derivative exsits and is Lipschitz of order . That is,for some positive constant , where is the support of U, r is a nonnegative integer and such that .
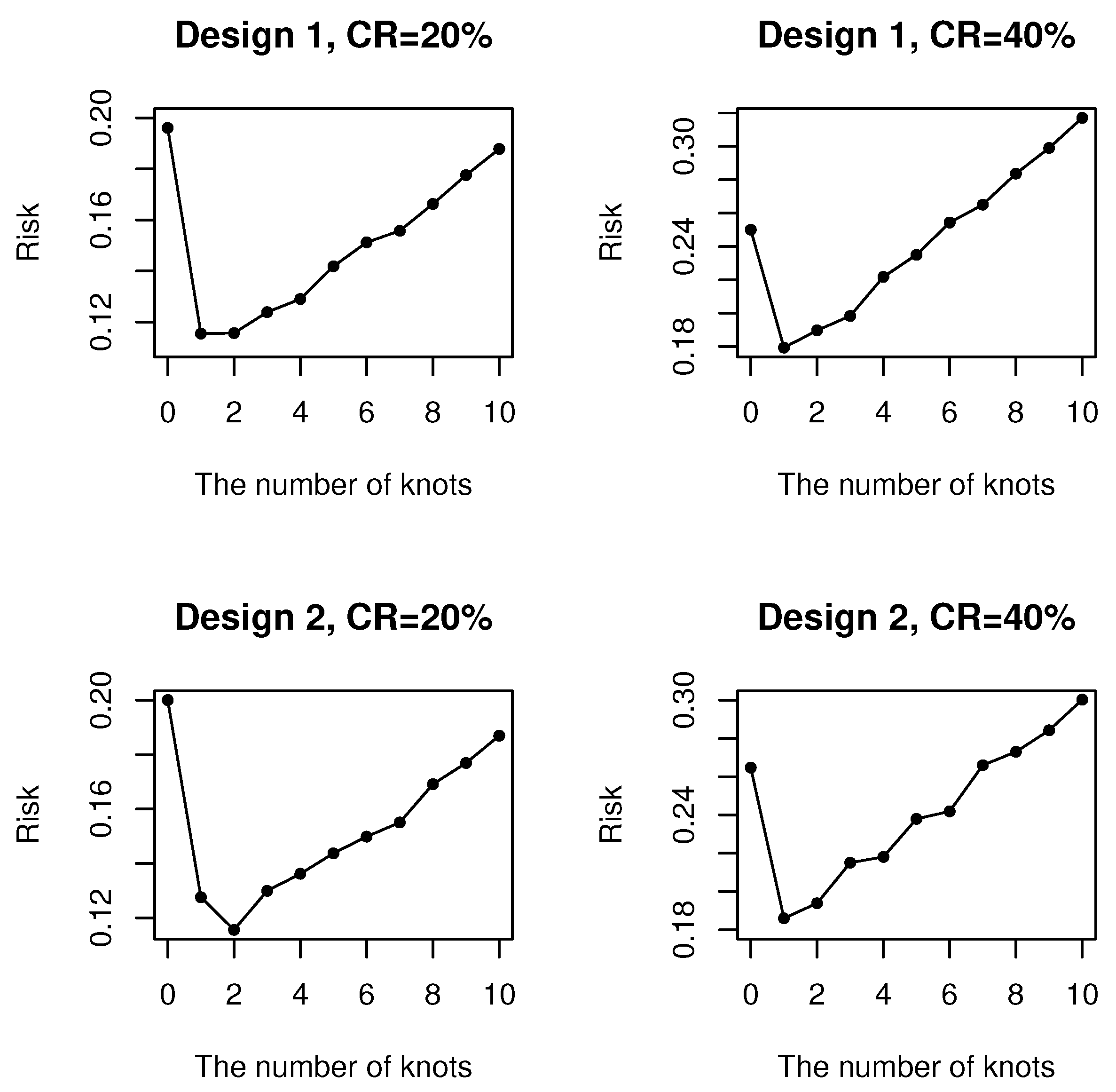
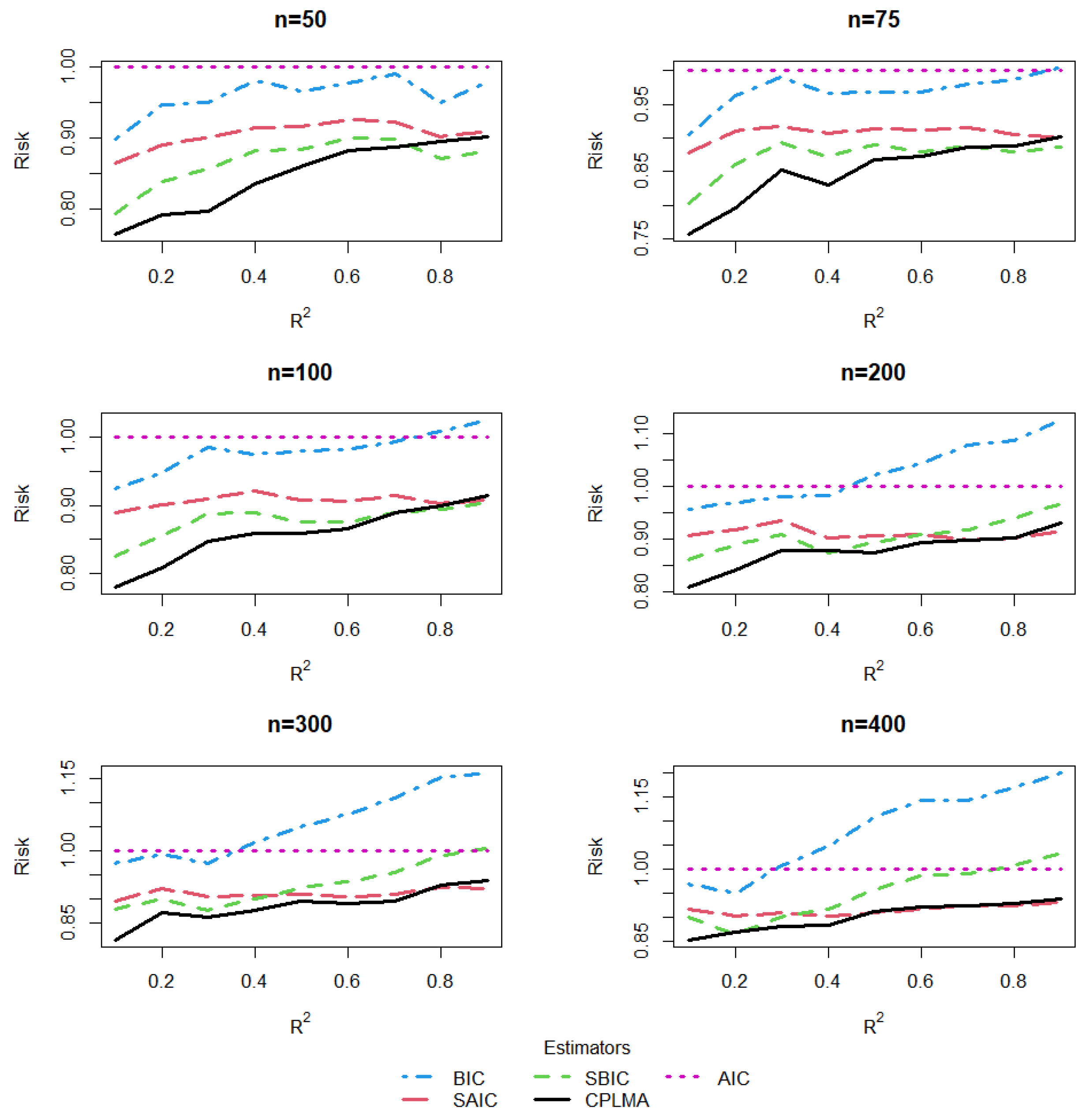
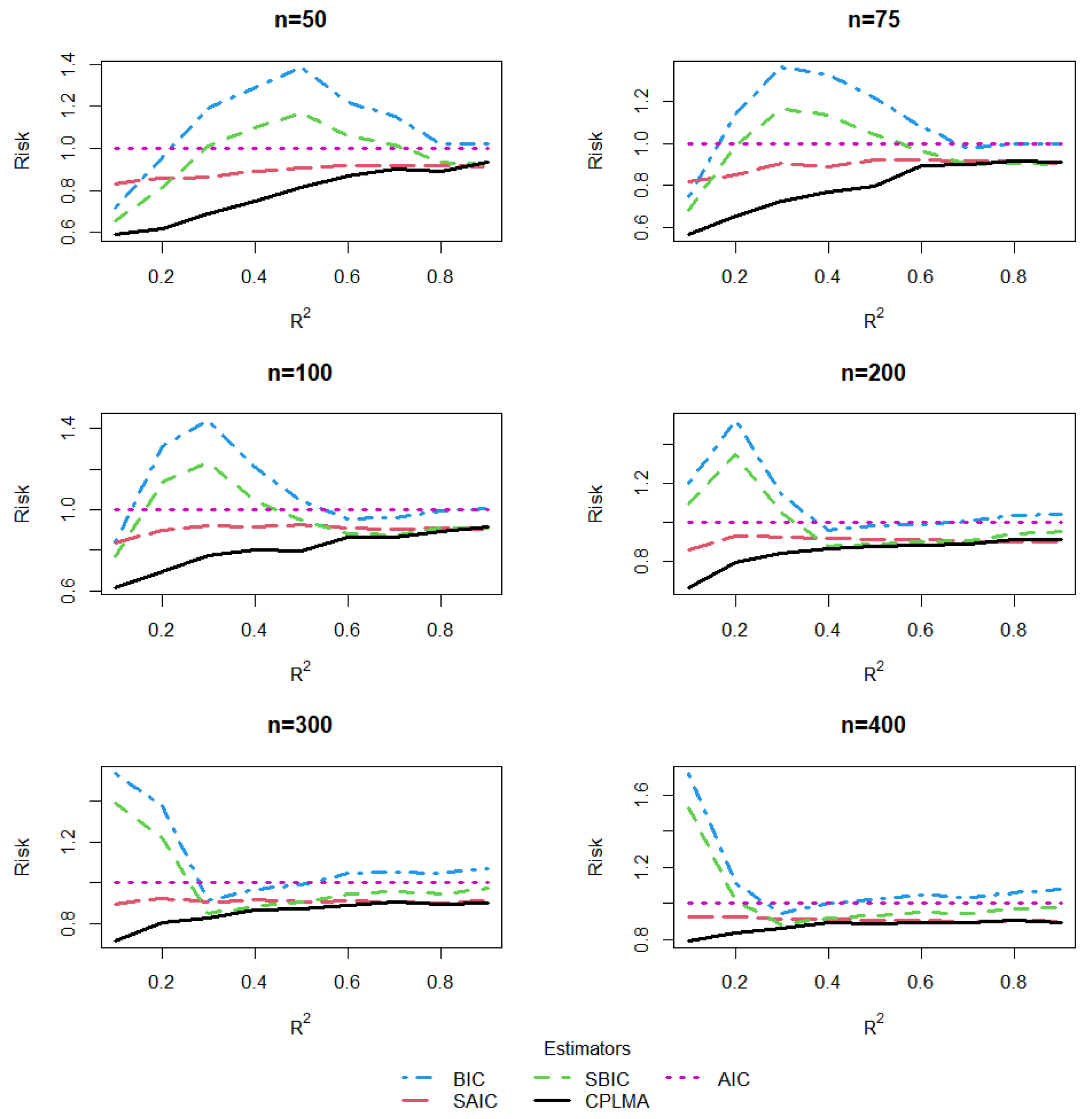
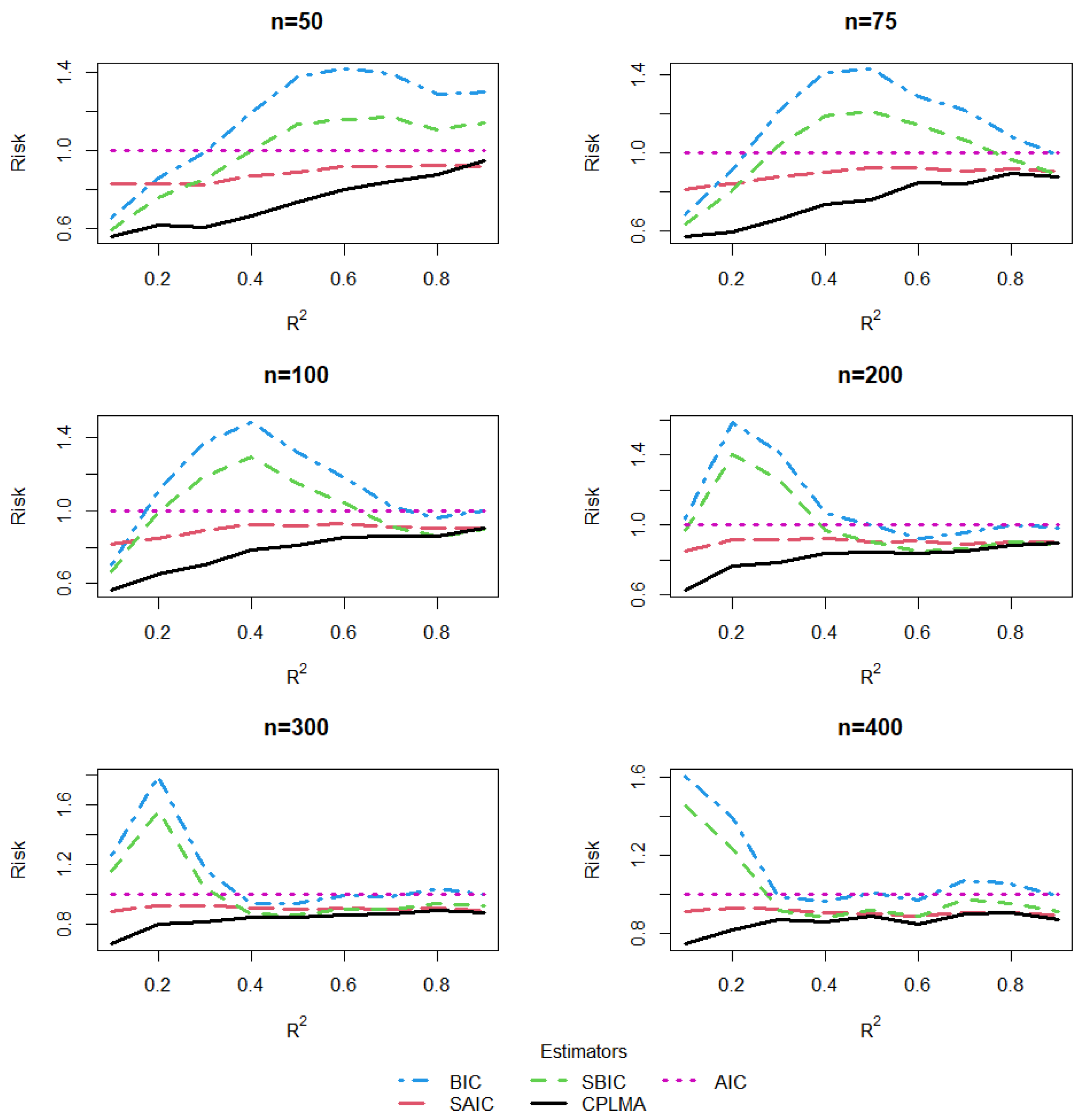
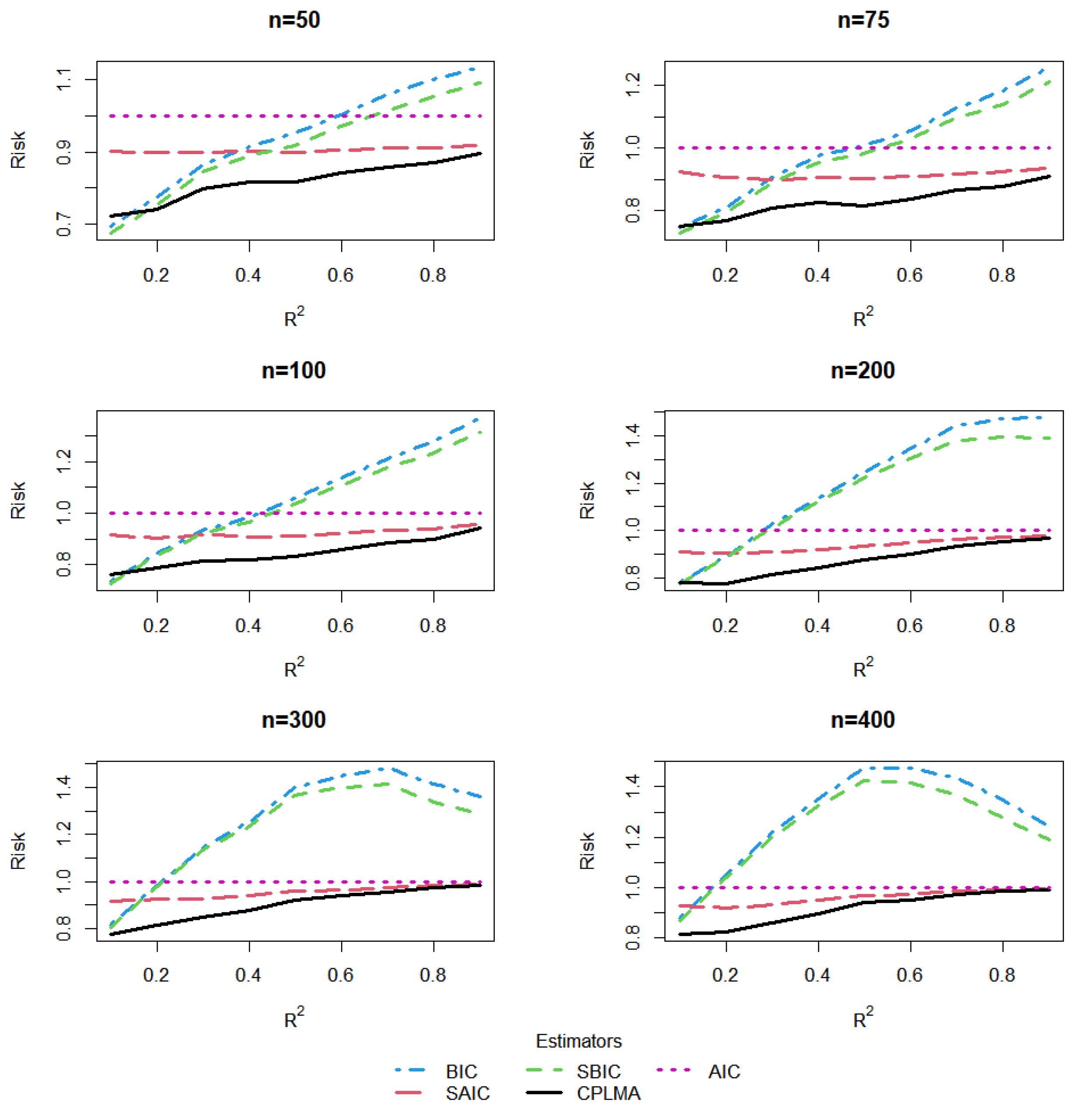
5. A Simulation Study
5.1. The Design of Simulation
5.2. Estimation and Comparison
5.3. Results
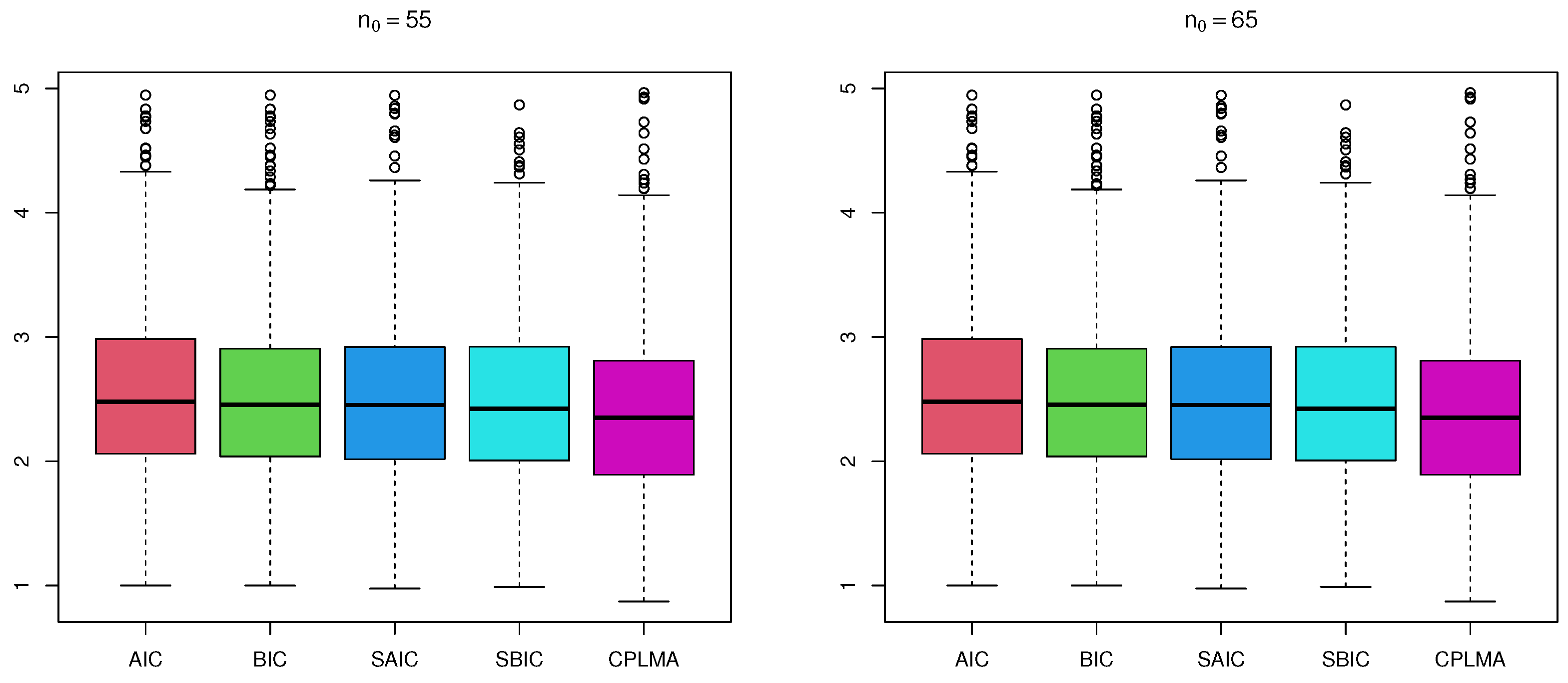
6. Real Data Analysis
6.1. Primary Biliary Cirrhosis Dataset Study
6.2. Mantle Cell Lymphoma Data Analysis
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Engle, R.F.; Granger, C.W.J.; Rice, J.; Weiss, A. Semiparametric estimates of the relation between weather and electricity sales. J. Am. Stat. Assoc. 1986, 81, 310–320. [Google Scholar] [CrossRef]
- Speckman, P. Kernel smoothing in partial linear models. J. R. Stat. Soc. Ser. B Stat. Methodol. 1988, 50, 413–436. [Google Scholar] [CrossRef]
- Heckman, N.E. Spline smoothing in a partly linear model. J. R. Stat. Soc. Ser. B Stat. Methodol. 1986, 48, 244–248. [Google Scholar] [CrossRef]
- Shi, J.; Lau, T. Empirical likelihood for partially linear models. J. Multivar. Anal. 2000, 72, 132–148. [Google Scholar] [CrossRef]
- Härdle, W.; Liang, H.; Gao, J. Partially Linear Models; Springer Science & Business Media: Berlin, Germany, 2000. [Google Scholar]
- Claeskens, G.; Hjort, N.L. Model Selection and Model Averaging; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Hansen, B.E.; Racine, J.S. Jackknife model averaging. J. Econom. 2012, 167, 38–46. [Google Scholar] [CrossRef]
- Racine, J.S.; Li, Q.; Yu, D.; Zheng, L. Optimal model averaging of mixed-data kernel-weighted spline regressions. J. Bus. Econ. Stat. 2022, in press. [CrossRef]
- Akaike, H. Statistical predictor identification. Ann. Inst. Statist. Math. 1970, 22, 203–217. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Statist. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Claeskens, G.; Hjort, N.L. The focused information criterion. J. Am. Stat. Assoc. 2003, 98, 900–916. [Google Scholar] [CrossRef]
- Ni, X.; Zhang, H.; Zhang, D. Automatic model selection for partially linear models. J. Multivar. Anal. 2009, 100, 2100–2111. [Google Scholar] [CrossRef]
- Raheem, S.E.; Ahmed, S.E.; Doksum, K.A. Absolute penalty and shrinkage estimation in partially linear models. Comput. Stat. Data Anal. 2012, 56, 874–891. [Google Scholar] [CrossRef]
- Xie, H.; Huang, J. SCAD-penalized regression in high-dimensional partially linear models. Ann. Statist. 2009, 37, 673–696. [Google Scholar] [CrossRef]
- Peng, J.; Yang, Y. On improvability of model selection by model averaging. J. Econom. 2022, 229, 246–262. [Google Scholar] [CrossRef]
- Hoeting, J.A.; Madigan, D.; Raftery, A.E.; Volinsky, C.T. Bayesian model averaging: A tutorial. Statist. Sci. 1999, 14, 382–417. [Google Scholar]
- Hansen, B.E. Least squares model averaging. Econometrica 2007, 75, 1175–1189. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, G.; Carroll, R.J. Model averaging based on Kullback-Leibler distance. Stat. Sin. 2015, 25, 1583–1598. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Okui, R.; Yoshimura, A. Generalized least squares model averaging. Economet. Rev. 2016, 35, 1692–1752. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Wang, S.; Zou, G. Model averaging based on leave-subject-out cross-validation. J. Econom. 2016, 192, 139–151. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C. Model averaging prediction by K-fold cross-validation. J. Econom. 2022, in press.
- Lu, X.; Su, L. Jackknife model averaging for quantile regressions. J. Econom. 2015, 188, 40–58. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W. Optimal model averaging estimation for partially linear models. Stat. Sin. 2019, 29, 693–718. [Google Scholar] [CrossRef]
- Zhu, R.; Wan, A.T.K.; Zhang, X.; Zou, G. A Mallows-type model averaging estimator for the varying-coefficient partially linear model. J. Am. Stat. Assoc. 2019, 114, 882–892. [Google Scholar] [CrossRef]
- Xie, J.; Yan, X.; Tang, N. A model-averaging method for high-dimensional regression with missing responses at random. Stat. Sin. 2021, 31, 1005–1026. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, Q.; Liu, W. Model averaging for linear models with responses missing at random. Ann. Inst. Statist. Math. 2021, 73, 535–553. [Google Scholar] [CrossRef]
- Zhang, X.; Chiou, J.; Ma, Y. Functional prediction through averaging estimated functional linear regression models. Biometrika 2018, 105, 945–962. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, Y.; Carroll, R.J. MALMEM: Model averaging in linear measurement error models. J. R. Stat. Soc. Ser. B Stat. Methodol. 2019, 81, 763–779. [Google Scholar] [CrossRef]
- Ando, T.; Li, K.C. A model-averaging approach for high-dimensional regression. J. Am. Stat. Assoc. 2014, 109, 254–265. [Google Scholar] [CrossRef]
- Ando, T.; Li, K.C. A weight-relaxed model averaging approach for high-dimensional generalized linear models. Ann. Statist. 2017, 45, 2654–2679. [Google Scholar] [CrossRef]
- Zeng, D.; Lin, D. Efficient estimation for the accelerated failure time model. J. Am. Stat. Assoc. 2007, 102, 1387–1396. [Google Scholar] [CrossRef]
- Wang, H.J.; Wang, L. Locally weighted censored quantile regression. J. Am. Stat. Assoc. 2009, 104, 1117–1128. [Google Scholar] [CrossRef]
- Hjort, N.L.; Claeskens, G. Focused information criteria and model averaging for the Cox hazard regression model. J. Am. Stat. Assoc. 2006, 101, 1449–1464. [Google Scholar] [CrossRef]
- Du, J.; Zhang, Z.; Xie, T. Focused information criterion and model averaging in censored quantile regression. Metrika 2017, 80, 547–570. [Google Scholar] [CrossRef]
- Sun, Z.; Sun, L.; Lu, X.; Zhu, J.; Li, Y. Frequentist model averaging estimation for the censored partial linear quantile regression model. J. Statist. Plann. Inference 2017, 189, 1–15. [Google Scholar] [CrossRef]
- Yan, X.; Wang, H.; Wang, W.; Xie, J.; Ren, Y.; Wang, X. Optimal model averaging forecasting in high-dimensional survival analysis. Int. J. Forecast. 2021, 37, 1147–1155. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, X.; Zhou, Y. Mallows model averaging estimation for linear regression model with right censored data. Acta Math. Appl. Sin. E. 2022, 38, 5–23. [Google Scholar] [CrossRef]
- Koul, H.; Susarla, V.; Ryzin, J.V. Regression analysis with randomly right-censored data. Ann. Statist. 1981, 9, 1276–1288. [Google Scholar] [CrossRef]
- Xia, X. Model averaging prediction for nonparametric varying-coefficient models with B-spline smoothing. Stat. Pap. 2021, 62, 2885–2905. [Google Scholar] [CrossRef]
- De Boor, C. A Practical Guide to Splines; Springer: New York, NY, USA, 2001. [Google Scholar]
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Hu, G.; Cheng, W.; Zeng, J. Model averaging by jackknife criterion for varying-coefficient partially linear models. Comm. Statist. Theory Methods 2020, 49, 2671–2689. [Google Scholar] [CrossRef]
- Turlach, B.A.; Weingessel, A.; Moler, C. Quadprog: Functions to Solve Quadratic Programming Problems. R Package Version 1.5-8. 2019. Available online: https://CRAN.R-project.org/package=quadprog (accessed on 16 December 2022).
- Wei, Y.; Wang, Q. Cross-validation-based model averaging in linear models with response missing at random. Stat. Probab. Lett. 2021, 171, 108990. [Google Scholar] [CrossRef]
- Zhang, X.; Wan, A.T.K.; Zou, G. Model averaging by jackknife criterion in models with dependent data. J. Econom. 2013, 174, 82–94. [Google Scholar] [CrossRef]
- Wan, A.T.; Zhang, X.; Zou, G. Least squares model averaging by Mallows criterion. J. Econom. 2010, 156, 277–283. [Google Scholar] [CrossRef]
- Fan, J.; Ma, Y.; Dai, W. Nonparametric independence screening in sparse ultra-high-dimensional varying coefficient models. J. Am. Stat. Assoc. 2014, 109, 1270–1284. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.M.; Venables, W.N. Splines: Regression Spline Functions and Classes. R Package Version 3.6-1. 2019. Available online: https://CRAN.R-project.org/package=splines (accessed on 15 December 2022).
- Therneau, T.M.; Lumley, T.; Elizabeth, A.; Cynthia, C. Survival: Survival Analysis. R Package Version 3.4-0. 2022. Available online: https://CRAN.R-project.org/package=survival (accessed on 15 December 2022).
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Shows, J.H.; Lu, W.; Zhang, H.H. Sparse estimation and inference for censored median regression. J. Statist. Plann. Inference 2010, 140, 1903–1917. [Google Scholar] [CrossRef] [PubMed]
- Diebold, F.X.; Mariano, R.S. Comparing predictive accuracy. J. Bus. Econ. Stat. 1995, 13, 253–263. [Google Scholar]
- Rosenwald, A.; Wright, G.; Wiestner, A.; Chan, W.C.; Connors, J.M.; Campo, E.; Gascoyne, R.D.; Grogan, T.M.; Muller-Hermelink, H.K.; Smeland, E.B.; et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 2003, 3, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Du, P. Variable selection in partly linear regression model with diverging dimensions for right censored data. Stat. Sin. 2012, 22, 1003–1020. [Google Scholar] [CrossRef]




| Notations | Descriptions |
|---|---|
| The survival time of the ith subject | |
| The response variable, a transformation of | |
| The covariate vector of the ith subject | |
| The censoring indicator of the ith subject | |
| The last follow up time of the ith subject | |
| The observed time, equal to | |
| The cumulative distribution function of | |
| synthetic response vector | |
| conditional mean vector of the response | |
| B-spline basis matrix for the sth model | |
| linear regression coefficient vector for the sth model | |
| spline coefficient vector for the sth model | |
| The Kaplan–Meier estimator of | |
| The estimator of with | |
| The estimator of with | |
| The estimator of for the sth model with | |
| The model-averaging estimator of with | |
| or | the sth jackknife estimator of with or |
| or | the jackknife model-averaging estimator of with or |
| Method | BIC | SAIC | SBIC | CPLMA | |
|---|---|---|---|---|---|
| 140 | mean | 1.005 | 0.987 | 0.983 | 0.979 |
| median | 1.014 | 0.992 | 0.995 | 0.987 | |
| 160 | mean | 1.005 | 0.987 | 0.983 | 0.982 |
| median | 1.006 | 0.986 | 0.988 | 0.984 | |
| 180 | mean | 1.011 | 0.991 | 0.991 | 0.986 |
| median | 1.011 | 0.990 | 0.990 | 0.981 | |
| 200 | mean | 1.014 | 0.993 | 0.995 | 0.989 |
| median | 1.012 | 0.984 | 0.990 | 0.976 | |
| 220 | mean | 1.012 | 0.995 | 0.997 | 0.994 |
| median | 1.020 | 0.994 | 1.003 | 0.993 | |
| 240 | mean | 1.008 | 0.995 | 0.998 | 0.993 |
| median | 1.017 | 0.996 | 0.999 | 0.988 |
| Method | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 140 | DM | −3.013 | 15.130 | 12.335 | 14.858 | 16.743 | 25.341 | 16.633 | 4.973 | 6.361 | 2.834 |
| p-value | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.005 | |
| 160 | DM | −3.014 | 16.607 | 12.490 | 14.862 | 17.196 | 30.331 | 18.474 | 4.995 | 5.538 | 0.942 |
| p-value | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.347 | |
| 180 | DM | −8.082 | 12.355 | 7.874 | 11.238 | 22.554 | 34.561 | 21.914 | −0.348 | 4.439 | 5.679 |
| p-value | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.728 | 0.000 | 0.000 | |
| 200 | DM | −11.473 | 12.320 | 4.744 | 11.393 | 23.962 | 32.286 | 22.550 | −3.721 | 5.288 | 8.690 |
| p-value | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 220 | DM | −9.509 | 8.500 | 2.308 | 5.587 | 19.004 | 22.316 | 17.152 | −4.011 | 1.085 | 5.059 |
| p-value | 0.000 | 0.000 | 0.021 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.278 | 0.000 | |
| 240 | DM | −5.484 | 7.332 | 1.441 | 5.848 | 12.901 | 15.427 | 12.998 | −4.175 | 2.110 | 6.561 |
| p-value | 0.000 | 0.000 | 0.150 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.035 | 0.000 |
| Method | BIC | SAIC | SBIC | CPLMA | |
|---|---|---|---|---|---|
| 55 | mean | 1.011 | 0.982 | 0.988 | 0.923 |
| median | 0.951 | 0.965 | 0.937 | 0.918 | |
| 65 | mean | 0.992 | 0.976 | 0.987 | 0.947 |
| median | 0.982 | 0.970 | 0.973 | 0.939 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, G.; Cheng, W.; Zeng, J. Optimal Model Averaging for Semiparametric Partially Linear Models with Censored Data. Mathematics 2023, 11, 734. https://doi.org/10.3390/math11030734
Hu G, Cheng W, Zeng J. Optimal Model Averaging for Semiparametric Partially Linear Models with Censored Data. Mathematics. 2023; 11(3):734. https://doi.org/10.3390/math11030734
Chicago/Turabian StyleHu, Guozhi, Weihu Cheng, and Jie Zeng. 2023. "Optimal Model Averaging for Semiparametric Partially Linear Models with Censored Data" Mathematics 11, no. 3: 734. https://doi.org/10.3390/math11030734
APA StyleHu, G., Cheng, W., & Zeng, J. (2023). Optimal Model Averaging for Semiparametric Partially Linear Models with Censored Data. Mathematics, 11(3), 734. https://doi.org/10.3390/math11030734





