Abstract
Automated segmentation of abdominal organs and tumors in medical images is a challenging yet essential task in medical image analysis. Deep learning has shown excellent performance in many medical image segmentation tasks, but most prior efforts were fragmented, addressing individual organ and tumor segmentation tasks with specialized networks. To tackle the challenges of abdominal organ and tumor segmentation using partially labeled datasets, we introduce Re-parameterizing Mixture-of-Diverse-Experts (RepMode) to abdominal organ and tumor segmentation. Within the RepMode framework, the Mixture-of-Diverse-Experts (MoDE) block forms the foundation, learning generalized parameters applicable across all tasks. We seamlessly integrate the MoDE block into a U-shaped network with dynamic heads, addressing multi-scale challenges by dynamically combining experts with varying receptive fields for each organ and tumor. Our framework incorporates task encoding in both the encoder–decoder section and the segmentation head, enabling the network to adapt throughout the entire system based on task-related information. We evaluate our approach on the multi-organ and tumor segmentation (MOTS) dataset. Experiments show that DoDRepNet outperforms previous methods, including multi-head networks and single-network approaches, giving a highly competitive performance compared with the original single network with dynamic heads. DoDRepNet offers a promising approach to address the complexities of abdominal organ and tumor segmentation using partially labeled datasets, enhancing segmentation accuracy and robustness.
MSC:
68T07
1. Introduction
Automated segmentation of abdominal organs and tumors in medical images is a key yet intricate task within medical image analysis [1,2,3]. This task holds significant importance in various computer-aided diagnosis applications, encompassing tasks such as lesion delineation, 3D reconstruction, and surgical planning. For abdominal multi-organ images, creating extensive fully annotated datasets for abdominal multi-organ images is a formidable undertaking, demanding both significant resources and time, especially in the case of 3D segmentation tasks. Currently, most benchmark datasets are typically limited in sample size, and most of them only annotate one or a few organs, rather than all abdominal organs, designating all task-irrelevant structures as background. For example, the pancreas-CT dataset exclusively provides labels for the pancreas and pancreas tumors, while the hepatic vessel dataset offers labels solely for hepatic vessels and tumors [4].
Recently, deep learning has made remarkable strides in medical image segmentation [5,6,7,8,9,10]. Previous abdominal organ and tumor segmentation tasks were typically addressed individually using specialized networks [11,12,13,14], leading to a dispersion of research efforts. Effectively acquiring representations of multiple organs and tumors by taking advantage of partially labeled datasets to enhance segmentation accuracy and robustness has attracted great interest. Broadly, prior approaches can be categorized into two main groups. One focuses on devising effective training strategies, which involve techniques such as knowledge distillation [15], cross-task consistency learning [16], and the design of jointly optimized loss functions [17]. The other strives to enhance a network’s structure [18,19]. They can be further grouped into multi-head networks [20] and a single network with dynamic heads [18,19]. A brief summary is provided in Table 1.
Multi-head networks consist of a partially shared architecture, incorporating a shared feature extractor and several task-dedicated decoders. Thanks to the common encoder, they can leverage the rich information from various partially annotated datasets. Nonetheless, multi-head networks face challenges in effectively co-training for multiple tasks, mainly due to the structural redundancy resulting from utilising individual decoders for each task. A single network with dynamic heads presents a versatile architecture designed to address various segmentation tasks by utilizing task encoding and a controller for generating segmentation heads equipped with dynamic convolutions. However, these models have a few limitations. Task-specific information, namely the dynamic convolution parameters, is set at the decoder’s end. This timing may pose challenges in making the model fully aware of the current task, especially when decoding sophisticated objectives. In addition, it is worth noting that abdominal organs and tumors naturally exhibit significant variations in size, giving rise to the multi-scale challenge. Currently, segmentation accuracy for small and highly deformable organs such as the pancreas, tubular structures, and tumors, is generally below 90%.
To address the above challenges inherent in abdominal organ and tumor segmentation using partially labeled datasets, we have adopted a unique approach known as Re-parameterizing Mixture-of-Diverse-Experts (RepMode) [21]. Its key feature is the dynamic organization of its parameters through the use of task-aware priors. Within the RepMode framework, the Mixture-of-Diverse-Experts (MoDE) block serves as a foundational component, designed to learn generalized parameters that can be applied across all the tasks. We have integrated RepMode into DoDNet, obtaining DoDNetRep, to suit the demands of abdominal organ and tumor segmentation, where each partially labeled dataset may encompass multiple labels, as illustrated in Figure 1.
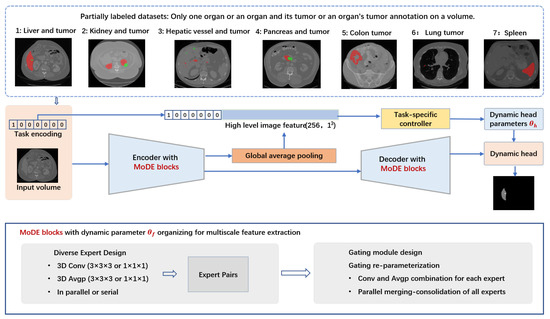
Figure 1.
In abdominal partially labeled datasets, only one organ or an organ and its tumors or an organ’s tumors are annotated on a volume. In the example images red denotes organ and green denotes tumor for tasks (1, 2, 3, 4); Red denotes tumor for tasks 5 and 6; Red denotes organ for task 7. Segmentation of targets in each partially labeled dataset is considered as a task, and designated a one-hot task embedding. Within the RepMode framework, the Mixture-of-Diverse-Experts (MoDE) block serves as a foundational component, designed to learn generalized parameters that can be applied across all tasks. We seamlessly integrate the MoDE block at each stage of both the encoder and decoder within the DoDNet. In our framework, we make use of task encoding in both the encoder–decoder section and the segmentation head. This strategy enables the network to adapt its behavior throughout the entire system, including the multi-stage encoder–decoder section () and the segmentation head (with dynamic head parameters ) based on task-related information. RepMode with MoDE blocks for organizing dynamic parameter , consists of diverse expert design, gating module design, and gating re-parameterization.
In our framework, we make use of task encoding in both the encoder–decoder section and the segmentation head. This strategy enables the network to adapt its behavior throughout the entire system, including the multi-stage encoder–decoder section and the segmentation head, based on task-related information. Notably, we have seamlessly integrated the MoDE block at each stage of both the encoder and decoder within the DoDNet. This adaptation effectively addresses the multi-scale challenges inherent in the segmentation of abdominal organs and tumors. The integration empowers our network to combine experts with varying receptive fields dynamically for each specific organ and tumor, facilitating the learning of multi-scale features in a task-oriented way. The dynamic kernels within the segmentation head are generated adaptively by a controller, with this process being conditioned on the assigned segmentation task. Task-specific priors guide the controller in generating dynamic head kernels for each segmentation task, ensuring that our network can effectively handle the intricacies of abdominal organ and tumor segmentation from a set of partially annotated datasets. We performed experiments on the multi-organ and tumor segmentation (MOTS) dataset [18]. Our model outperformed both multi-head networks and previous present single-network methods, and obtained highly competitive performance compared with the original single network with dynamic heads [18]. DoDRepNet offers a promising approach to address the complexities of abdominal organ and tumor segmentation using partially labeled datasets, enhancing segmentation accuracy and robustness. Our contributions can be summarized as follows:
(1) We introduced RepMode into DoDNet, obtaining DoDNetRep, for abdominal organ and tumor segmentation using partially labeled datasets. DoDNetRep takes advantage of task-related information in both the encoder–decoder section and the dynamic segmentation head, enabling our network to combine experts with varying receptive fields dynamically for each specific organ and tumor, thus facilitating the learning of multi-scale features in a task-oriented way.
(2) We demonstrated that our model outperformed both multi-head networks and previous present single-network methods, and obtained a highly competitive performance compared with the original single network with dynamic heads on MOTS.

Table 1.
Related work for medical image segmentation from partially labeled datasets.
Table 1.
Related work for medical image segmentation from partially labeled datasets.
| Authors | Methods | Datasets |
|---|---|---|
| Zhang, L. et al. [15] | A multi-teacher knowledge distillation framework leveraging the soft labels | KiTS [22], MSD Spleen and Pancreas [23], TCIA [24], BTCV [25] |
| Shi, G. et al. [17] | Design of jointly optimized marginal loss and exclusion loss | BTCV [25], MSD Liver, MSD Spleen, MSD Pancreas [23], KiTS [22] |
| Chen., S. et al. [20] | Multi-head: transfer learning | LIDC [26], LiTS [27] |
| Fang, X. et al. [28] | Multi-head: pyramid input pyramid output feature abstraction network and a target adaptive loss | BTCV [25], LiTS [27], KiTS [22] and MSD Spleen [23] |
| Zhang, G. et al. [29] | Single network: conditional nnU-Net with a conditioning strategy for the decoder | LiTS [27], MSD Pancreas, MSD Spleen [23], KiTS [22], SLIVER07 [30], NIH pancreas [31], BTCV [25] |
| Zhang, J. et al. [18] | Single network: with dynamic heads leveraging one-hot task embedding | MOTS including LiTS [27], KiTS [22], and MSD Hepatic vessel and tumor, MSD Pancreas and tumor, MSD Colon tumor, MSD Lung tumor and MSD Spleen [23] |
| Liu, J. et al. [19] | Single network: with dynamic heads leveraging task embedding from Clip | MSD [23] and BTCV [25] |
2. Materials and Methods
2.1. Problem Definition
We consider each partially labeled dataset as a task (organ or tumor segmentation or organ and tumor segmentation) and assume there are P partially labeled dataset in total. Each dataset can have several labels. In this context, denotes the i-th partially labeled dataset, comprising labeled volumes. Each volume in is represented as , where indicates the dimensions of each slice, while D signifies the slice number. The ground truth segmentation corresponding to this is represented as , where the voxel labels belong to the set .
To address the challenges in abdominal organ and tumor segmentation using partially labeled datasets, one straightforward approach involves training P segmentation networks, individually on each of the P datasets. The same as in [18], we endeavor to tackle this issue by employing a single network denoted as f. This can be written as follows
Different from Multi-Net, maintaining an individual for each task, we adopt a single network, as in DoDNet [18], generating a dynamic head with parameters conditioned on the assigned task. What is more, we aim to utilize a shared across all tasks and dynamically structure to accommodate specific task requirements by using re-parameterize diverse experts in the encoder and decoder.
2.2. Network Architecture
Our network, DoDRepNet, which leverages the re-parameterization of diverse experts [21], is composed of several essential components similar to DoDNet [18]: a shared encoder–decoder featuring the MoDE block, a task-encoding module, a dynamic filter generation module, and a dynamic segmentation head (as depicted in Figure 1). In the following sections, we will briefly describe the shared encoder–decoder network. Other modules are the same as those in DoDNet; please reference [18] for details.
The input to DoDRepNet consists of randomly selected samples from the preprocessed dataset, including preprocessed images and annotated images, along with the task identifier corresponding to the subset dataset of that sample. The task identifier serves as input to the task-encoding module, generating a task code represented as a one-hot vector. The preprocessed images and task code are input to a shared encoder–decoder architecture. The multi-stage encoder produces feature maps at each stage, which serve as inputs to the respective stages of the decoder. The final stage of the encoder outputs feature maps that undergo global average pooling (GAP) to obtain high-level image features. These features are concatenated with the task code and serve as input to the dynamic filter generation module, producing a task-specific controller used to generate parameters for the dynamic segmentation head. , along with the output feature maps from the final stage of the decoder, are passed through the dynamic segmentation head to obtain the output of DoDRepNet, representing the segmented result of the preprocessed image predicted by the network. The loss function is employed to calculate the loss between the predicted result and the annotated image, and the network is trained using optimization algorithms. After training, given any sample (preprocessed image and annotated image) and the corresponding task identifier for that sample’s subset dataset, DoDRepNet can produce the segmentation result for the respective task.
The shared encoder–decoder relies on a 3D U-shaped design, primarily composed of downsampling and upsampling components, as seen in Figure 2. In detail, the downsampling path comprises two consecutive MoDE blocks, which are responsible for capturing task-specific feature maps and increasing the channel number. Following these blocks, there is a downsampling layer that uses a convolution with a kernel size and a stride of 2 to decrease the feature maps’ size by half. It is worth noting that we used group normalization and ReLU activation along with each MoDE block.
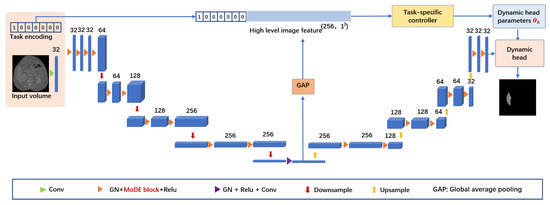
Figure 2.
Detailed structure of DoDRepNet with the shared encoder–decoder featuring the MoDE block.
Within each upsampling stage, feature maps are enlarged and their channel number is halved using an upsampling layer with a kernel and a stride of 2. Following that, the upsampled feature maps are combined with the corresponding feature maps transmitted from the encoder. These combined feature maps then go through additional enhancement via two successive MoDE blocks. Ultimately, a MoDE block, excluding batch normalization and ReLU, is used to decrease the number of channels to match the number of output classes, with the goal of producing the final predictions for each target class.
It is worth noting that MoDE blocks are used both in the encoder and decoder, effectively promoting the acquisition of task-specific features and thus contributing to superior performance. The detailed information such as the channel sizes of each convolution and output size of feature maps are presented in Table 2.

Table 2.
The detailed information such as the channel sizes of each convolution and output size of feature maps in key layers of DoDRepNet. Upsampling in each stage of the decoder is not listed in this table for simplicity.
2.3. Mixture-of-Diverse-Experts Block
In order to address the diverse prediction subtasks effectively and enhance the network’s representational capacity for robust generalization, RepMode with MoDE [21] for dynamic parameter organizing consists of diverse expert design, gating module design, and gating re-parameterization.
We use the MoDE block as a fundamental building block of the U-shaped encoder–decoder network as a potent alternative to the conventional convolutional layer. Within the MoDE block, an array of diverse experts is meticulously crafted, each responsible for exploring a distinctive convolutional configuration. Furthermore, a gating module is intricately devised to harness task-specific information, enabling the generation of gating weights for dynamic parameter organization.
For the diverse expert design, there are two distinct types of expert diversity to enhance its capabilities: shape diversity, to address the multi-scale challenges by using different experts to possess a variety of receptive fields, and kernel diversity, using combinations of different kernels. Following these principles, the concept of “expert pairs” is used to form the multiple branch structure. Each expert pair consists of two key components: 3D convolutions (Conv) and 3D average poolings (Avgp). In essence, the MoDE block comprises expert pairs with three distinct receptive fields, promoting shape diversity. For more details, please refer to [21].
As for the gating module design, we encode the task-aware prior associated with each volume, , by transforming its subtask indicator, , into a P-dimensional one-hot vector, , as in [18]. This encoding is expressed as follows:
where signifies the p-th element of . Next, the one-hot vector, , is introduced to the gating module, from which the gating weights, , are produced using a single-layer fully connected network, denoted as , as depicted below: , where . The subscript n in is omitted for brevity. Each represents the gating weights for the t-th expert, and these values are derived from , with representing the number of channels in the output feature maps. Lastly, undergoes further processing to yield , which is expressed as a set by applying the Softmax function. This operation ensures a balanced intensity among the various experts and can be defined as follows:
where (resp. ) denotes the i-th element of (resp. ). Utilizing the resulting gating weights, , RepMode can undertake the dynamic organization of parameters for these experts, which are task-agnostic and adapt their behavior based on the task-aware prior.
Then, for gating re-parameterization, GatRep is designed according to principles of homogeneity and additivity that are established in [32,33]. The initial phase of GatRep involves the amalgamation of Avgp and Conv operations into a unified kernel. To streamline the explanation, let us consider the example of an Avgp–Conv expert. Here, we use to represent the input feature maps. Then, the output feature maps, denoted as , can be articulated as follows:
where ⊛ means the convolution operation. By applying the associative property, we can achieve an equivalent transformation for the equation in Equation (4) by initially consolidating and . This process can be denoted as follows:
With this process, the kernels of Avgp and Conv can be combined into a unified kernel for use in the subsequent stage.
The second phase of GatRep involves the consolidation of all experts in a manner specific to the subtask at hand. To facilitate this, we introduce a mapping function denoted as . This function effectively transforms a kernel to the kernel space by employing zero-padding. Here, we set , which represents the largest receptive field size among these experts. We use to represent the ultimate task-specific feature maps. The transformation can be expressed as follows:
where ⊙ represents channel-wise multiplication, and represents the kernel of the t-th expert. It is important to note that is an integrated kernel for an Avgp–Conv expert, or simply a Conv kernel for a Conv expert. Ultimately, is the dynamically generated resulting task-specific kernel by GatRep.
3. Results
3.1. Datasets
We used the multi-organ and tumor segmentation (MOTS) dataset, as in [18]. MOTS was presented for abdominal organ and tumor segmentation using partially labeled datasets. It consists of datasets for Liver and tumor [27], Kidney and tumor [22], and five datasets of Hepatic vessel and tumor, Pancreas and tumor, Colon tumor, Lung tumor, and Spleen from the Medical Segmentation Decathlon [23]. The dataset comprises 1155 3D abdominal CT volumes, gathered from diverse clinical sites worldwide. Of these, 920 volumes are allocated for training purposes, while the remaining 235 are reserved for testing. Notably, all volumes have been uniformly re-sampled to a consistent voxel size of 1.5 × 0.8 × 0.8 mm3, following the data preprocessing in [18]. In our experiments, we found out there are two bad cases in the Liver and tumor training dataset and two bad cases in the Liver and tumor testing dataset after preprocessing. The resolution of the volumes and the corresponding labels are different in these four cases, so we removed them, resulting 918 training cases and 233 testing cases.
3.2. Implementation Details
We performed experiments on a workstation equipped with two NVIDIA A100 GPUs. We adopted DoDNet from [18] as our baseline, then integrated the MoDE block with the GatRep module into DoDNet, obtaining our DoDRepNet. We maintained the remaining implementation settings consistent with those in [18] except with a batch size of 16. We reran the original DoDNet using their provided source code, and executed our DoDRepNet based on Re-parameterize Diverse Experts. To ensure fair comparisons, all models underwent the same training configurations, which included weight standardization, learning rate, optimizer, and other settings, as well as the inference strategy, except adding a few morphological operations for two cases for the spleen segmentation task in postprocessing.
3.3. Performance Metrics
This study employed the commonly used Dice similarity coefficient (Dice) and Hausdorff distance (HD) as performance metrics for evaluating the performance of segmentation methods. Dice is a measure of the spatial overlap between the predicted segmentation (output of an algorithm) and the ground truth segmentation (manual or reference segmentation). The Dice coefficient is calculated using the formula as follows:
where denotes the predicted segmentation and is the ground truth segmentation.
HD measures the maximum distance between any point in the predicted segmentation and its closest point in the ground truth, as well as the maximum distance between any point in the ground truth and its closest point in the predicted segmentation. The HD is defined as follows:
where and are points in and , and is the distance metric (e.g., Euclidean distance).
A lower HD indicates better agreement between the predicted and ground truth segmentations, as it represents the maximum distance between corresponding points. In the context of medical image segmentation, these metrics help quantify how well an algorithm delineates structures of interest (such as organs or tumors) compared with the manually annotated ground truth.
3.4. Comparisons with State-of-the-Art Approaches
We conduct a comparative analysis of our DoDRepNet against state-of-the-art methods designed for addressing the challenge of partially labeled data. This evaluation was carried out on seven partially labeled tasks using the MOTS test set, as in [18]. The competing methods include: two multi-head networks, namely Multi-Head [20] and TAL [28], a single-network method that operates without task conditioning, known as Cond-NO, and three single-network methods incorporating task conditioning, specifically Cond-Input [34], Cond-Dec [35], and DoDNet [18]. The results of the above methods are copied from [18]. We added our results of reran DoDNet and our DoDRepNet, as listed in Table 3. The best score of each metric in each task is denoted in red.

Table 3.
Performance (Dice, %, and Hausdorff distance (HD) ) of different approaches on MOTS test set, as in [18]. Please note that the term ’Average score’ serves as a composite metric, averaging the Dice or HD values across 11 organs and tumors.
Our DoDRepNet, designed to master the task-specific amalgamation of various task-agnostic experts, surpasses the performance of current methods on the liver (both on Dice and HD), kidney (on Dice), hepatic vessel (both on Dice and HD), and pancreas (on Dice), and achieves better overall performance than DoDNet. Figure 3 displays a sample of each task and the segmentations of Ground Truth (GT), DoDNet, DoDRepNet, and the 3D rendering of the three segmentations. From top to bottom, the tasks are Liver and Liver tumor, Kidney and Kidney tumor, Hepatic vessel and tumor, Pancreas and tumor, Colon tumor, Lung tumor, and Spleen. Specifically, (a) is a axial slice of the original resampled volume, (b,c) are its segmentations of Ground Truth (GT), DoDNet, and DoDRepNet, respectively, and (e,f) are the 3D rendering of the segmentations of Ground Truth (GT), DoDNet, and DoDRepNet, respectively. From Figure 3, we can see that the segmentations of DoDRepNet in most tasks are more similar to GT than those of DoDNet and both segmentations of DoDRepNet and DoDNet are more smooth than GT thanks to the dynamic parameter organizing based on Re-parameterize Diverse Experts.

Figure 3.
A sample of each task and the segmentations of Ground Truth (GT), DoDNet, and DoDRepNet, and the 3D rendering of the three segmentations. From top to bottom, the tasks are Liver and Liver tumor, Kidney and Kidney tumor, Hepatic vessel and tumor, Pancreas and tumor, Colon tumor, Lung tumor, and Spleen. In the example images red denotes organ and green denotes tumor for the first four tasks; Red denotes colone tumor for Task 5 and lung tumor for Task 6; Red denotes spleen for Task 7. Specifically, (a) is a axial slice of the original resampled volume, (b–d) are its segmentations of Ground Truth (GT), DoDNet, and DoDRepNet, respectively, and (e–g) are the 3D rendering of the segmentations of Ground Truth (GT), DoDNet, and DoDRepNet, respectively.
In Figure 4, we compared the speed–accuracy trade-off of DoDRepNet with previous methods, as listed in [18]. From Figure 4, we can see that DoDRepNet has a few more parameters than DoDNet due to the MoDE blocks. It also has a little more inference time than DoDNet. However, DoDRepNet achieves the best accuracy.
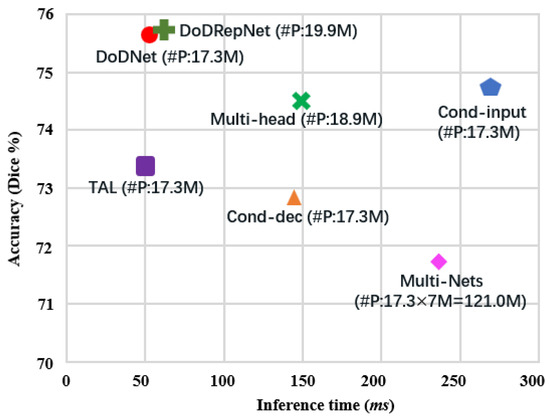
Figure 4.
Speed vs. accuracy. The accuracy refers to the overall Dice score on the MOTS test set. The inference time is computed based on a single input with 64 slices of spatial size , as in [18]. Part of the results are copied from [18]. ‘#P’: the number of parameters. ‘M’: Million.
4. Discussion
A single network with dynamic heads [18,19] offers a good way for leveraging partially annotated data for multi-organ abdominal and tumor segmentation tasks. However, substantial differences in the relative sizes of distinct target organs lead to imbalances in segmentation objectives. In the training process, the size differences between organs create substantial competition, which can be detrimental to smaller organs. In order to address the diverse prediction subtasks effectively and enhance the network’s representational capacity for robust generalization, we introduce the MoDE block as a fundamental building block of the U-shaped encoder–decoder network with dynamic segmentation heads. The MoDE block is put forth as a potent alternative to the conventional convolutional layer. Within the MoDE block, an array of diverse experts is meticulously crafted, each responsible for exploring a distinctive convolutional configuration. Furthermore, a gating module is intricately devised to harness task-specific information, enabling the generation of gating weights for dynamic parameter organization.
This design enables DoDRepNet to focus on learning dynamic compositions of experts with varying receptive fields specific to each organ and tumor. This capability make it able to acquire multi-scale features adaptively in a task-specific way, addressing the multi-scale challenges of abdominal organ and tumor segmentation. Compared with DoDNet, which only uses task-related information at the decoder end for the segmentation head, we utilize it in both the encoder–decoder section and the segmentation head. By incorporating task-dependent gating and Mixture-of-Diverse-Experts, DoDRepNet can learn the generalized parameters for all tasks by combine experts with diverse configurations, and generate the specialized parameters for each task with gating re-parameterization (GatRep) in the encoder–decoder section. This approach offers greater flexibility in modeling various scenarios, thus improving segmentation performance.
While DoDRepNet showed promising segmentation performance, there were still some instances of suboptimal predictions. Additionally, our best model did not surpass the performance of other methods on all the organs and tumors. One limitation of our model is that the task-related information is a simple one-hot vector, the same as in DoDNet, which cannot offer task-specific semantic information. In addition, the current method does not consider anatomical prior among organs or of organs, which makes it fail when the boundaries between organs are quite unclear, resulting in necessary postprocessing, as in DoDNet [18]. Future work includes task-specific semantic information and anatomical priors, which may improve MOTS results. In addition, ensemble deep learning [36], combining several individual deep neural network models to obtain better generalization performance, has shown potential in tumor detection [37] and classification [38], and its utilization in MOTS is also a direction worth exploring.
5. Conclusions
We present DoDRepNet by integrating RepMode into DoDNet to suit the demands of abdominal organ and tumor segmentation. By dynamically organizing its parameters based on task-aware priors, we overcame the multi-scale challenge by combining experts with varying receptive fields, resulting in dynamic multi-scale feature learning. Task-dependent gating in both the encoder–decoder and segmentation head provides flexibility in modeling diverse scenarios. Our experiments on the MOTS dataset demonstrated that DoDRepNet outperformed other methods in several organ and tumor segmentation tasks, showcasing the effectiveness of DoDRepNet. However, some challenges remain, such as handling unclear organ boundaries and leveraging anatomical priors effectively.
Author Contributions
Conceptualization, G.C.; methodology, P.L. and C.G.; software C.G.; validation, P.L.; formal analysis, P.L. and Y.Q.; investigation, Y.Q.; resources, G.C.; data curation, C.G.; writing—original draft preparation, P.L.; writing—review and editing, B.W., Y.Q. and X.L.; visualization, Y.Q.; supervision, G.C. and B.W.; project administration, X.L. and Y.Q.; funding acquisition, P.L., Y.Q., X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key Research and Development Program of Guangzhou (No. 2023B01J0022), and The Regional Joint Fund of Guangdong under Grant 2021B1515130003 and 2021B1515120011, Guangdong Basic and Applied Basic Research Foundation (2020A1515111093), Natural Science Foundation of Guangdong Province (2021A1515011869), and Shenzhen Science and Technology Program (No. JCYJ20220818101401003).
Data Availability Statement
The data presented in this study are openly available in [22,23,27].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, Y.; Zhou, Y.; Shen, W.; Park, S.; Fishman, E.K.; Yuille, A.L. Abdominal multi-organ segmentation with organ-attention networks and statistical fusion. Med. Image Anal. 2019, 55, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, D.; Ge, R. Eye-Guided Dual-Path Network for Multi-organ Segmentation of Abdomen. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 23–32. [Google Scholar]
- Bilic, P.; Christ, P.; Li, H.B.; Vorontsov, E.; Ben-Cohen, A.; Kaissis, G.; Szeskin, A.; Jacobs, C.; Mamani, G.E.H.; Chartrand, G.; et al. The liver tumor segmentation benchmark (lits). Med. Image Anal. 2023, 84, 102680. [Google Scholar] [CrossRef]
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; et al. The medical segmentation decathlon. Nat. Commun. 2022, 13, 4128. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Taghanaki, S.A.; Abhishek, K.; Cohen, J.P.; Cohen-Adad, J.; Hamarneh, G. Deep semantic segmentation of natural and medical images: A review. Artif. Intell. Rev. 2020, 54, 137–178. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Jeyaseelan, L.; Li, Q.; Chiang, J.N.; Wu, Z.; Ding, X. Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Med. Image Anal. 2020, 63, 101693. [Google Scholar] [CrossRef] [PubMed]
- Suganyadevi, S.; Seethalakshmi, V.; Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retr. 2022, 11, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, I.; Yan, J.; Abbas, Q.; Shaheed, K.; Riaz, A.B.; Wahid, A.; Khan, M.W.J.; Szczuko, P. Medical image segmentation using deep semantic-based methods: A review of techniques, applications and emerging trends. Inf. Fusion 2023, 90, 316–352. [Google Scholar] [CrossRef]
- Heller, N.; Isensee, F.; Maier-Hein, K.H.; Hou, X.; Xie, C.; Li, F.; Nan, Y.; Mu, G.; Lin, Z.; Han, M.; et al. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge. Med. Image Anal. 2021, 67, 101821. [Google Scholar] [CrossRef]
- Gul, S.; Khan, M.S.; Bibi, A.; Khandakar, A.; Ayari, M.A.; Chowdhury, M.E. Deep learning techniques for liver and liver tumor segmentation: A review. Comput. Biol. Med. 2022, 147, 105620. [Google Scholar] [CrossRef]
- Dutande, P.; Baid, U.; Talbar, S. Deep residual separable convolutional neural network for lung tumor segmentation. Comput. Biol. Med. 2022, 141, 105161. [Google Scholar] [CrossRef]
- Ghorpade, H.; Jagtap, J.; Patil, S.; Kotecha, K.; Abraham, A.; Horvat, N.; Chakraborty, J. Automatic Segmentation of Pancreas and Pancreatic Tumor: A Review of a Decade of Research. IEEE Access 2023, 11, 108727–108745. [Google Scholar] [CrossRef]
- Zhang, L.; Feng, S.; Wang, Y.; Wang, Y.; Zhang, Y.; Chen, X.; Tian, Q. Unsupervised Ensemble Distillation for Multi-Organ Segmentation. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; pp. 1–5. [Google Scholar]
- Li, W.H.; Liu, X.; Bilen, H. Learning multiple dense prediction tasks from partially annotated data. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 18879–18889. [Google Scholar]
- Shi, G.; Xiao, L.; Chen, Y.; Zhou, S.K. Marginal loss and exclusion loss for partially supervised multi-organ segmentation. Med. Image Anal. 2021, 70, 101979. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Xia, Y.; Shen, C. DoDNet: Learning to segment multi-organ and tumors from multiple partially labeled datasets. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 1195–1204. [Google Scholar]
- Liu, J.; Zhang, Y.; Chen, J.N.; Xiao, J.; Lu, Y.; A Landman, B.; Yuan, Y.; Yuille, A.; Tang, Y.; Zhou, Z. Clip-driven universal model for organ segmentation and tumor detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Waikoloa, HI, USA, 3–7 January 2023; pp. 21152–21164. [Google Scholar]
- Chen, S.; Ma, K.; Zheng, Y. Med3d: Transfer learning for 3d medical image analysis. arXiv 2019, arXiv:1904.00625. [Google Scholar]
- Zhou, D.; Gu, C.; Xu, J.; Liu, F.; Wang, Q.; Chen, G.; Heng, P.A. RepMode: Learning to Re-parameterize Diverse Experts for Subcellular Structure Prediction. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 18–22 June 2023; pp. 3312–3322. [Google Scholar]
- Available online: https://kits19.grand-challenge.org/data/ (accessed on 18 June 2022).
- Available online: http://medicaldecathlon.com/ (accessed on 12 July 2021).
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Landman, B.; Xu, Z.; Igelsias, J.E.; Styner, M.; Langerak, T.R.; Klein, A. 2015 miccai multi-atlas labeling beyond the cranial vault workshop and challenge. In Proceedings of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Armato III, S.G.; McLennan, G.; McNitt-Gray, M.F.; Meyer, C.R.; Yankelevitz, D.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; Kazerooni, E.A.; MacMahon, H.; et al. Lung image database consortium: Developing a resource for the medical imaging research community. Radiology 2004, 232, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://competitions.codalab.org/competitions/17094 (accessed on 22 July 2019).
- Fang, X.; Yan, P. Multi-organ segmentation over partially labeled datasets with multi-scale feature abstraction. IEEE Trans. Med Imaging 2020, 39, 3619–3629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yang, Z.; Huo, B.; Chai, S.; Jiang, S. Multiorgan segmentation from partially labeled datasets with conditional nnU-Net. Comput. Biol. Med. 2021, 136, 104658. [Google Scholar] [CrossRef] [PubMed]
- Heimann, T.; van Ginneken, G.; Styner, M. Available online: http://www.sliver07.org (accessed on 20 June 2019).
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.C.; Liu, J.; Turkbey, E.B.; Summers, R.M. Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015. Proceedings, Part I 18. pp. 556–564. [Google Scholar]
- Ding, X.; Zhang, X.; Han, J.; Ding, G. Diverse branch block: Building a convolution as an inception-like unit. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 10886–10895. [Google Scholar]
- Ding, X.; Zhang, X.; Ma, N.; Han, J.; Ding, G.; Sun, J. Repvgg: Making vgg-style convnets great again. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13733–13742. [Google Scholar]
- Chen, Q.; Xu, J.; Koltun, V. Fast image processing with fully-convolutional networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2497–2506. [Google Scholar]
- Dmitriev, K.; Kaufman, A.E. Learning multi-class segmentations from single-class datasets. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 9501–9511. [Google Scholar]
- Ganaie, M.A.; Hu, M.; Malik, A.; Tanveer, M.; Suganthan, P. Ensemble deep learning: A review. Eng. Appl. Artif. Intell. 2022, 115, 105151. [Google Scholar] [CrossRef]
- Alsubai, S.; Khan, H.U.; Alqahtani, A.; Sha, M.; Abbas, S.; Mohammad, U.G. Ensemble deep learning for brain tumor detection. Front. Comput. Neurosci. 2022, 16, 1005617. [Google Scholar] [CrossRef]
- Tandel, G.S.; Tiwari, A.; Kakde, O.G.; Gupta, N.; Saba, L.; Suri, J.S. Role of Ensemble Deep Learning for Brain Tumor Classification in Multiple Magnetic Resonance Imaging Sequence Data. Diagnostics 2023, 13, 481. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).