Accuracy Analysis of the End-to-End Extraction of Related Named Entities from Russian Drug Review Texts by Modern Approaches Validated on English Biomedical Corpora
Abstract
1. Introduction
- An assessment of the end-to-end solution of NER and RE problems was carried out on the corpus of annotated Russian-language reviews of Internet users about medicines;
- The results of a comparison of sequential and joint approaches to the end-to-end problem were presented on different versions of the used corpus of reviews;
- On the material of English-language corpora, it was demonstrated that the use of modern-language models in combination with the hyperparameter selection procedure makes it possible to increase the accuracy of the joint solution to the state of the art.
2. Related Works
2.1. Named Entity Recognition
2.2. Relation Extraction
2.3. Joint Solution of the Tasks
3. Materials and Methods
3.1. Data
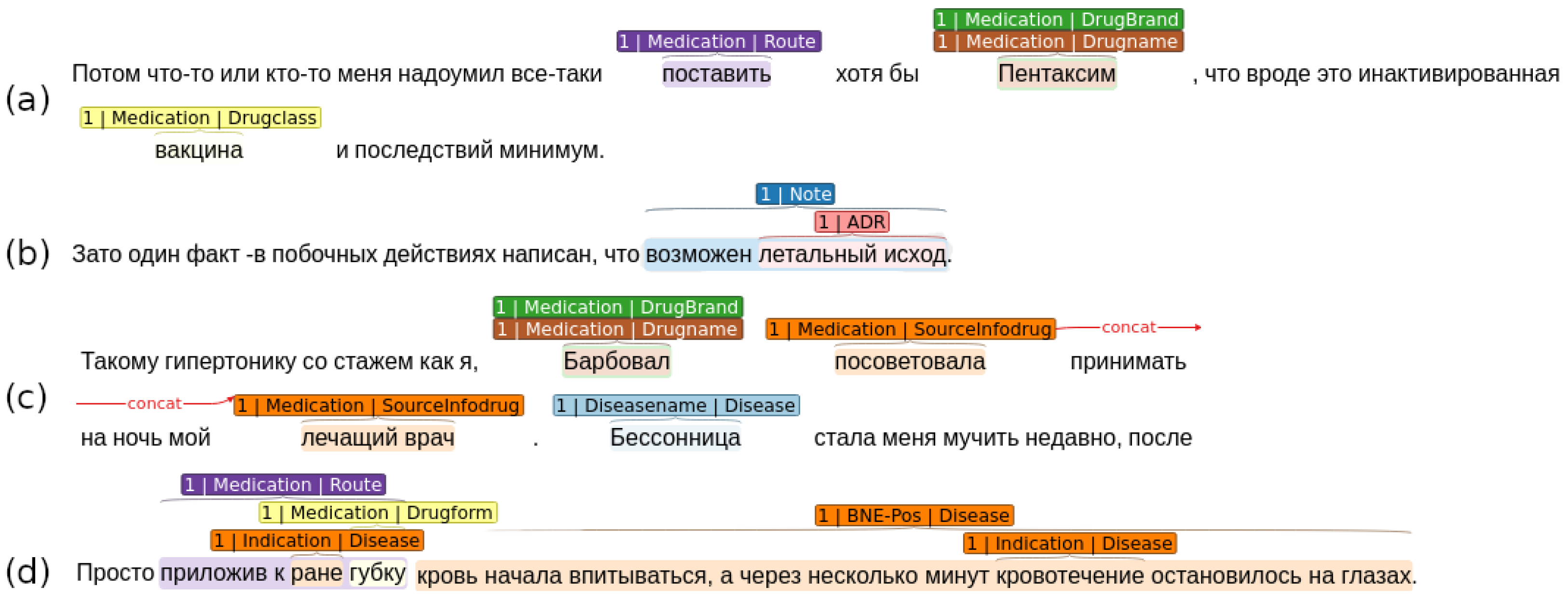
3.1.1. Russian Drug Review Corpus (RDRS)
- “Medication” describes drugs with the attributes: drug class, administration specifics, release form, etc.;
- “Disease” describes disease and related mentions: symptoms, course of the disease, etc.;
- “Adverse Drug Reaction” defines mentions of the adverse effects, described by the Internet users.
- ADR–Drugname—adverse effect of the particular medication;
- Drugname–SourceInfodrug—source of the information about medication (e.g., “my brother gave me advice”, “apothecary mentioned”);
- Drugname–Diseasename—a link between the disease and medication that user administrated against it;
- Diseasename–Indication—symptoms of the particular disease (e.g., “red rash”, “high temperature”).
3.1.2. English-Language Corpora
3.2. Sequential Approach
3.3. Joint Approach
3.4. Comparative Analysis of Both Approaches
3.5. Language Models
4. Experiments and Results
4.1. Description of the Experiments
- Estimation of joint-approach accuracy for known effective language models: RuBERT, RuDr-BERT, EnRuDr-BERT, RuBioRoBERTa, XLMR and XLMR_sag. The selection of hyperparameters was carried out using the GridSearch method. The criteria for the choice is a validation loss on each search, which is carried out on a separate dataset;
- Every approach accuracy was estimated on the XLMR and XLMR_sag language models, which showed the best results in the separate solution of the NER and RE problems in our previous studies, (see [69,83]). In the sequential use of the models, the following hyperparameters were used for both tasks: batch size 8, learning rate .
- Initial learning rate: 1, 5, , , , , , , , ;
- Epochs number: 10, 15, 20, 25, 30.
4.2. Results on the RDRS Dataset
4.3. Results on the English-Language Datasets
5. Discussion
- There is a common prediction error with wrong borders of the entity resulting in formally incorrect relation extraction (for example, annotated entity: “oxolin”, predicted entity: “oxolin ointment”, “ointment” was originally annotated as Drugform, not Drugname; annotated entity of ADR class: “vomit is a side effect”, predicted entity: “vomit”. It could be concluded that described cases are not meaningful errors leading to false information extraction, in a sense;
- There are 240 unique texts for which the sequential approach predicted all relations correctly, and Joint did not; and 646 texts where situation is reverse (the joint approach did not misclassify relations). It shows that the approaches are comparable, and one is not directly better than the other.
- The joint model generates more possible relations than the sequential (false-positive relations number is 51% of all relations in the test set; in comparison, the sequential approach had a false positive rate equal to 42%).
6. Conclusions
- We first obtained estimates of the accuracy of end-to-end information extraction solutions for drug reviews from Internet users in Russian, including the recognition of named entities and the extraction of relations.
- We compared the sequential and joint approaches to the NER+RE task in the Russian Drug Reviews corpus and showed that although the sequential approach overcomes some of the shortcomings of the used implementation of the joint approach (inability to work with overlapping and discontinuous entities), both of them show very close accuracy rates to each other.
- We improved the accuracy of solving the end-to-end problem (NER+RE) for the ADE corpus. For a DDI corpus, our accuracy estimates are comparable to SoTA solutions. Thus, this confirms the reliability of the methods used in the work.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADE | Adverse Events Dataset |
| AdE | Side-effects entity type in the ADE corpus |
| ADR | Adverse reaction |
| BERT | Bidirectional encoder representations from transformers |
| biLSTM | Bidirectional LSTM |
| CADEC | CSIRO Adverse Drug Event Corpus |
| CDR | The BioCreative V Chemical-Disease Relation Corpus |
| CNN | Convolutional neural network |
| CRF | Conditional random field |
| DDI | Drug–Drug Interaction 2013 Dataset |
| GLoVe | Global vector representation |
| KECI | Knowledge-enhanced collective inference model |
| LSTM | Long short-term memory recurrent neural network |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MLP | Multilayer perceptron |
| n2c2 | National NLP clinical challenge |
| NER | Named-entity recognition |
| RDRS | Russian Drug Review Corpora |
| RE | Relation extraction |
| RNN | Recurrent neural network |
| SMM4H | Social Media Mining for Health Challenge |
| w2v | word2vec vector representation |
| XLMR | XLM-RoBERTa-large |
| XLMR_sag | XLM-RoBERTa-sag-large |
References
- Gydovskikh, D.V.; Moloshnikov, I.A.; Naumov, A.V.; Rybka, R.B.; Sboev, A.G.; Selivanov, A.A. A probabilistically entropic mechanism of topical clusterisation along with thematic annotation for evolution analysis of meaningful social information of internet sources. Lobachevskii J. Math. 2017, 38, 910–913. [Google Scholar] [CrossRef]
- Naumov, A.; Rybka, R.; Sboev, A.; Selivanov, A.; Gryaznov, A. Neural-network method for determining text author’s sentiment to an aspect specified by the named entity. In Proceedings of the Russian Advances in Artificial Intelligence, Moscow, Russia, 10–16 October 2020; Number 2648 in CEUR Workshop Proceedings. pp. 134–143. [Google Scholar]
- Fields, S.; Cole, C.L.; Oei, C.; Chen, A.T. Using named entity recognition and network analysis to distinguish personal networks from the social milieu in nineteenth-century Ottoman–Iraqi personal diaries. Digit. Scholarsh. Humanit. 2022, fqac047. [Google Scholar] [CrossRef]
- de Arruda, H.F.; Costa, L.d.F.; Amancio, D.R. Topic segmentation via community detection in complex networks. Chaos Interdiscip. J. Nonlinear Sci. 2016, 26, 063120. [Google Scholar] [CrossRef] [PubMed]
- Selivanov, A.A.; Moloshnikov, I.A.; Rybka, R.B.; Sboev, A.G. Keyword Extraction Approach Based on Probabilistic-Entropy, Graph, and Neural Network Methods. In Proceedings of the Russian Conference on Artificial Intelligence, Moscow, Russia, 10–16 October 2020; Springer International Publishing: Berlin/Heidelberg, Germany, 2020. Number 12412 in Lecture Notes in Computer Science. pp. 284–295. [Google Scholar] [CrossRef]
- Tjong Kim Sang, E.F.; De Meulder, F. Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition. In Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL 2003, Edmonton, AB, Canada, 31 May–1 June 2003; Association for Computational Linguistics: Stroudsburg, PA, USA, 2003; Volume 4, pp. 142–147. [Google Scholar]
- Liu, P.; Guo, Y.; Wang, F.; Li, G. Chinese named entity recognition: The state of the art. Neurocomputing 2022, 473, 37–53. [Google Scholar] [CrossRef]
- Eberts, M.; Ulges, A. Span-Based Joint Entity and Relation Extraction with Transformer Pre-Training. In Proceedings of the European Conference on Artificial Intelligence, Digital, 29 August–8 September 2020; IOS Press: Amsterdam, The Netherlands, 2020; pp. 2006–2013. [Google Scholar]
- Liu, X.; Chen, H. AZDrugMiner: An information extraction system for mining patient-reported adverse drug events in online patient forums. In Proceedings of the International Conference on Smart Health, Beijing, China, 3–4 August 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 134–150. [Google Scholar]
- Sarker, A.; Gonzalez, G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J. Biomed. Inform. 2015, 53, 196–207. [Google Scholar] [CrossRef]
- Kiritchenko, S.; Mohammad, S.M.; Morin, J.; de Bruijn, B. NRC-Canada at SMM4H shared task: Classifying Tweets mentioning adverse drug reactions and medication intake. arXiv 2018, arXiv:1805.04558. [Google Scholar]
- Rastegar-Mojarad, M.; Elayavilli, R.K.; Yu, Y.; Liu, H. Detecting signals in noisy data-can ensemble classifiers help identify adverse drug reaction in tweets. In Proceedings of the Social Media Mining & Shared Task Workshop at the Pacific Symposium on Biocomputing, Kohala, HI, USA, 4–8 January 2016. [Google Scholar]
- Rajapaksha, P.; Weerasinghe, R. Identifying adverse drug reactions by analyzing Twitter messages. In Proceedings of the 2015 Fifteenth International Conference on Advances in ICT for Emerging Regions (ICTer), Colombo, Sri Lanka, 24–26 August 2015; pp. 37–42. [Google Scholar]
- Miranda, D.S. Automated detection of adverse drug reactions in the biomedical literature using convolutional neural networks and biomedical word embeddings. arXiv 2018, arXiv:1804.09148. [Google Scholar]
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural Architectures for Named Entity Recognition. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, San Diego, CA, USA, 12–17 June 2016; pp. 260–270. [Google Scholar]
- Cocos, A.; Fiks, A.G.; Masino, A.J. Deep learning for pharmacovigilance: Recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J. Am. Med. Inform. Assoc. 2017, 24, 813–821. [Google Scholar] [CrossRef]
- Wen, X.; Zhou, C.; Tang, H.; Liang, L.; Jiang, Y.; Qi, H. Type-supervised sequence labeling based on the heterogeneous star graph for named entity recognition. arXiv 2022, arXiv:2210.10240. [Google Scholar]
- Ma, X.; Hovy, E. End-to-end Sequence Labeling via Bi-directional LSTM-CNNs-CRF. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 7–12 August 2016; pp. 1064–1074. [Google Scholar]
- Chowdhury, S.; Zhang, C.; Yu, P.S. Multi-task pharmacovigilance mining from social media posts. In Proceedings of the 2018 World Wide Web Conference, Lyon, France, 23–27 April 2018; pp. 117–126. [Google Scholar]
- Weissenbacher, D.; Gonzalez, G. Social Media Mining for Health Applications (# SMM4H) Workshop & Shared Task. In Proceedings of the Fourth Workshop, Florence, Italy, 2 August 2019; Association for Computational Linguistics: Stroudsburg, PA, USA, 2019. [Google Scholar]
- Chen, S.; Huang, Y.; Huang, X.; Qin, H.; Yan, J.; Tang, B. HITSZ-ICRC: A report for SMM4H shared task 2019-automatic classification and extraction of adverse effect mentions in tweets. In Proceedings of the Fourth Social Media Mining for Health Applications (# SMM4H) Workshop & Shared Task, Florence, Italy, 2 August 2019; pp. 47–51. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Miftahutdinov, Z.; Alimova, I.; Tutubalina, E. KFU NLP team at SMM4H 2019 tasks: Want to extract adverse drugs reactions from tweets? BERT to the rescue. In Proceedings of the Fourth Social Media Mining for Health Applications (# SMM4H) Workshop & Shared Task, Florence, Italy, 2 August 2019; pp. 52–57. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Aroyehun, S.T.; Gelbukh, A. Detection of adverse drug reaction in tweets using a combination of heterogeneous word embeddings. In Proceedings of the Fourth Social Media Mining for Health Applications (# SMM4H) Workshop & Shared Task, Florence, Italy, 2 August 2019; pp. 133–135. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Haq, H.U.; Kocaman, V.; Talby, D. Mining adverse drug reactions from unstructured mediums at scale. arXiv 2022, arXiv:2201.01405. [Google Scholar]
- Gurulingappa, H.; Rajput, A.M.; Roberts, A.; Fluck, J.; Hofmann-Apitius, M.; Toldo, L. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J. Biomed. Inform. 2012, 45, 885–892, Text Mining and Natural Language Processing in Pharmacogenomics. [Google Scholar] [CrossRef]
- Karimi, S.; Metke-Jimenez, A.; Kemp, M.; Wang, C. Cadec: A corpus of adverse drug event annotations. J. Biomed. Inform. 2015, 55, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Wu, F.; Wu, C.; Qi, T.; Huang, Y.; Xie, X. Fedner: Privacy-preserving medical named entity recognition with federated learning. arXiv 2020, arXiv:2003.09288. [Google Scholar]
- Stanovsky, G.; Gruhl, D.; Mendes, P. Recognizing mentions of adverse drug reaction in social media using knowledge-infused recurrent models. In Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics: Volume 1, Long Papers, Valencia, Spain, 3–7 April 2017; pp. 142–151. [Google Scholar]
- Lehmann, J.; Isele, R.; Jakob, M.; Jentzsch, A.; Kontokostas, D.; Mendes, P.N.; Hellmann, S.; Morsey, M.; Van Kleef, P.; Auer, S.; et al. Dbpedia—A large-scale, multilingual knowledge base extracted from wikipedia. Semant. Web 2015, 6, 167–195. [Google Scholar] [CrossRef]
- Bordes, A.; Weston, J.; Collobert, R.; Bengio, Y. Learning structured embeddings of knowledge bases. In Proceedings of the Twenty-Fifth AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 7–11 August 2011. [Google Scholar]
- Ding, J.; Berleant, D.; Nettleton, D.; Wurtele, E. Mining MEDLINE: Abstracts, sentences, or phrases? In Biocomputing 2002; World Scientific: Singapore, 2001; pp. 326–337. [Google Scholar]
- Jelier, R.; Jenster, G.; Dorssers, L.C.; van der Eijk, C.C.; van Mulligen, E.M.; Mons, B.; Kors, J.A. Co-occurrence based meta-analysis of scientific texts: Retrieving biological relationships between genes. Bioinformatics 2005, 21, 2049–2058. [Google Scholar] [CrossRef]
- Ono, T.; Hishigaki, H.; Tanigami, A.; Takagi, T. Automated extraction of information on protein–protein interactions from the biological literature. Bioinformatics 2001, 17, 155–161. [Google Scholar] [CrossRef]
- Divoli, A.; Attwood, T.K. BioIE: Extracting informative sentences from the biomedical literature. Bioinformatics 2005, 21, 2138–2139. [Google Scholar] [CrossRef]
- Zhou, G.; Su, J.; Zhang, J.; Zhang, M. Exploring various knowledge in relation extraction. In Proceedings of the 43rd Annual Meeting of the Association for Computational Linguistics (ACL’05), Ann Arbor, MI, USA, 25–30 June 2005; pp. 427–434. [Google Scholar]
- Airola, A.; Pyysalo, S.; Björne, J.; Pahikkala, T.; Ginter, F.; Salakoski, T. All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning. BMC Bioinform. 2008, 9, S2. [Google Scholar] [CrossRef]
- Xu, J.; Wu, Y.; Zhang, Y.; Wang, J.; Lee, H.J.; Xu, H. CD-REST: A system for extracting chemical-induced disease relation in literature. Database 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Muzaffar, A.W.; Azam, F.; Qamar, U. A relation extraction framework for biomedical text using hybrid feature set. Comput. Math. Methods Med. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Feldman, R.; Regev, Y.; Finkelstein-Landau, M.; Hurvitz, E.; Kogan, B. Mining biomedical literature using information extraction. Curr. Drug Discov. 2002, 2, 19–23. [Google Scholar]
- Skusa, A.; Rüegg, A.; Köhler, J. Extraction of biological interaction networks from scientific literature. Briefings Bioinform. 2005, 6, 263–276. [Google Scholar] [CrossRef]
- Rosario, B.; Hearst, M.A. Classifying semantic relations in bioscience texts. In Proceedings of the 42nd Annual Meeting of the Association for Computational Linguistics (ACL-04), Barcelona, Spain, 21–26 July 2004; pp. 430–437. [Google Scholar]
- Xu, Y.; Mou, L.; Li, G.; Chen, Y.; Peng, H.; Jin, Z. Classifying relations via long short term memory networks along shortest dependency paths. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 17–21 September 2015; pp. 1785–1794. [Google Scholar]
- Mehryary, F.; Björne, J.; Pyysalo, S.; Salakoski, T.; Ginter, F. Deep learning with minimal training data: TurkuNLP entry in the BioNLP shared task 2016. In Proceedings of the 4th BioNLP Shared Task Workshop, Berlin, Germany, 13 August 2016; pp. 73–81. [Google Scholar]
- Wang, L.; Cao, Z.; De Melo, G.; Liu, Z. Relation classification via multi-level attention cnns. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 7–12 August 2016; pp. 1298–1307. [Google Scholar]
- Li, H.; Zhang, J.; Wang, J.; Lin, H.; Yang, Z. DUTIR in BioNLP-ST 2016: Utilizing convolutional network and distributed representation to extract complicate relations. In Proceedings of the 4th BioNLP Shared Task Workshop, Berlin, Germany, 13 August 2016; pp. 93–100. [Google Scholar]
- Zhang, T.; Leng, J.; Liu, Y. Deep learning for drug–drug interaction extraction from the literature: A review. Briefings Bioinform. 2020, 21, 1609–1627. [Google Scholar] [CrossRef]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthc. HEALTH 2021, 3, 1–23. [Google Scholar] [CrossRef]
- Naseem, U.; Dunn, A.G.; Khushi, M.; Kim, J. Benchmarking for biomedical natural language processing tasks with a domain specific albert. BMC Bioinform. 2022, 23, 144. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Lai, P.T.; Wei, C.H.; Arighi, C.N.; Lu, Z. BioRED: A rich biomedical relation extraction dataset. Briefings Bioinform. 2022, 23, bbac282. [Google Scholar] [CrossRef]
- Milošević, N.; Thielemann, W. Comparison of biomedical relationship extraction methods and models for knowledge graph creation. J. Web Semant. 2023, 75, 100756. [Google Scholar] [CrossRef]
- Alvaro, N.; Miyao, Y.; Collier, N. TwiMed: Twitter and PubMed comparable corpus of drugs, diseases, symptoms, and their relations. JMIR Public Health Surveill. 2017, 3, e24. [Google Scholar] [CrossRef]
- Wührl, A.; Klinger, R. Recovering Patient Journeys: A Corpus of Biomedical Entities and Relations on Twitter (BEAR). arXiv 2022, arXiv:2204.09952. [Google Scholar]
- Zhang, T.; Lin, H.; Ren, Y.; Yang, L.; Xu, B.; Yang, Z.; Wang, J.; Zhang, Y. Adverse drug reaction detection via a multihop self-attention mechanism. BMC Bioinform. 2019, 20, 479. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.K.; Anand, A. Drug-drug interaction extraction from biomedical texts using long short-term memory network. J. Biomed. Inform. 2018, 86, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tang, B.; Chen, Q.; Wang, X. Drug-drug interaction extraction via convolutional neural networks. Comput. Math. Methods Med. 2016, 2016, 6918381. [Google Scholar] [CrossRef]
- Quan, C.; Hua, L.; Sun, X.; Bai, W. Multichannel convolutional neural network for biological relation extraction. BioMed Res. Int. 2016, 2016, 1850404. [Google Scholar] [CrossRef]
- Li, F.; Zhang, M.; Fu, G.; Ji, D. A neural joint model for entity and relation extraction from biomedical text. BMC Bioinform. 2017, 18, 198. [Google Scholar] [CrossRef]
- Henry, S.; Buchan, K.; Filannino, M.; Stubbs, A.; Uzuner, O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J. Am. Med. Inform. Assoc. 2019, 27, 3–12. [Google Scholar] [CrossRef]
- Fang, X.; Song, Y.; Maeda, A. Joint Extraction of Clinical Entities and Relations Using Multi-head Selection Method. In Proceedings of the 2021 International Conference on Asian Language Processing (IALP), Singapore, 11–13 December 2021; pp. 99–104. [Google Scholar]
- Santosh, T.; Chakraborty, P.; Dutta, S.; Sanyal, D.K.; Das, P.P. Joint Entity and Relation Extraction from Scientific Documents: Role of Linguistic Information and Entity Types. In Proceedings of the Workshop on Extraction and Evaluation of Knowledge Entities from Scientific Documents, Virtual, 30 September 2021. [Google Scholar]
- Zaikis, D.; Vlahavas, I. TP-DDI: Transformer-based pipeline for the extraction of Drug-Drug Interactions. Artif. Intell. Med. 2021, 119, 102153. [Google Scholar] [CrossRef]
- Fatehifar, M.; Karshenas, H. Drug-drug interaction extraction using a position and similarity fusion-based attention mechanism. J. Biomed. Inform. 2021, 115, 103707. [Google Scholar] [CrossRef]
- Wang, D.; Fan, H.; Liu, J. Drug-Drug Interaction Extraction via Attentive Capsule Network with an Improved Sliding-Margin Loss. In Proceedings of the International Conference on Database Systems for Advanced Applications, Taipei, Taiwan, 11–14 April 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 612–619. [Google Scholar]
- Xu, J.; Lee, H.J.; Ji, Z.; Wang, J.; Wei, Q.; Xu, H. UTH_CCB System for Adverse Drug Reaction Extraction from Drug Labels at TAC-ADR 2017. In Proceedings of the Text Analysis Conference (TAC), Gaithersburg, MA, USA, 13–14 November 2017. [Google Scholar]
- Li, F.; Zhang, Y.; Zhang, M.; Ji, D. Joint Models for Extracting Adverse Drug Events from Biomedical Text. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence (IJCAI), New York, NY, USA, 9–16 July 2016; Volume 2016, pp. 2838–2844. [Google Scholar]
- Sboev, A.; Sboeva, S.; Moloshnikov, I.; Gryaznov, A.; Rybka, R.; Naumov, A.; Selivanov, A.; Rylkov, G.; Ilyin, V. Analysis of the Full-Size Russian Corpus of Internet Drug Reviews with Complex NER Labeling Using Deep Learning Neural Networks and Language Models. Appl. Sci. 2022, 12, 491. [Google Scholar] [CrossRef]
- Herrero-Zazo, M.; Segura-Bedmar, I.; Martínez, P.; Declerck, T. The DDI corpus: An annotated corpus with pharmacological substances and drug–drug interactions. J. Biomed. Inform. 2013, 46, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Sboev, A.; Selivanov, A.; Moloshnikov, I.; Rybka, R.; Gryaznov, A.; Sboeva, S.; Rylkov, G. Extraction of the Relations among Significant Pharmacological Entities in Russian-Language Reviews of Internet Users on Medications. Big Data Cogn. Comput. 2022, 6, 10. [Google Scholar] [CrossRef]
- Conneau, A.; Khandelwal, K.; Goyal, N.; Chaudhary, V.; Wenzek, G.; Guzmán, F.; Grave, E.; Ott, M.; Zettlemoyer, L.; Stoyanov, V. Unsupervised cross-lingual representation learning at scale. arXiv 2019, arXiv:1911.02116. [Google Scholar]
- Zhu, Y.; Kiros, R.; Zemel, R.; Salakhutdinov, R.; Urtasun, R.; Torralba, A.; Fidler, S. Aligning Books and Movies: Towards Story-Like Visual Explanations by Watching Movies and Reading Books. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015. [Google Scholar]
- Hamborg, F.; Meuschke, N.; Breitinger, C.; Gipp, B. news-please: A generic news crawler and extractor. In Proceedings of the 15th International Symposium of Information Science (ISI 2017), Berlin, Germany, 13–15 March 2017; pp. 218–223. [Google Scholar]
- Tutubalina, E.; Alimova, I.; Miftahutdinov, Z.; Sakhovskiy, A.; Malykh, V.; Nikolenko, S. The Russian Drug Reaction Corpus and neural models for drug reactions and effectiveness detection in user reviews. Bioinformatics 2020, 37, 243–249. [Google Scholar] [CrossRef]
- Kuratov, Y.; Arkhipov, M. Adaptation of deep bidirectional multilingual transformers for Russian language. In Proceedings of the Komp’juternaja Lingvistika i Intellektual’nye Tehnologii, Moscow, Russia, 29 May–1 June 2019; pp. 333–339. [Google Scholar]
- Yalunin, A.; Nesterov, A.; Umerenkov, D. RuBioRoBERTa: A pre-trained biomedical language model for Russian language biomedical text mining. arXiv 2022, arXiv:2204.03951. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. Roberta: A robustly optimized bert pretraining approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Yasunaga, M.; Leskovec, J.; Liang, P. LinkBERT: Pretraining Language Models with Document Links. In Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Dublin, Ireland, 22–27 May 2022; pp. 8003–8016. [Google Scholar]
- Lan, Z.; Chen, M.; Goodman, S.; Gimpel, K.; Sharma, P.; Soricut, R. ALBERT: A Lite BERT for Self-supervised Learning of Language Representations. In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Alrowili, S.; Shanker, V. BioM-Transformers: Building Large Biomedical Language Models with BERT, ALBERT and ELECTRA. In Proceedings of the 20th Workshop on Biomedical Language Processing, Online, 11 June 20221; Association for Computational Linguistics: Stroudsburg, PA, USA, 2021; pp. 221–227. [Google Scholar]
- Selivanov, A.; Gryaznov, A.; Rybka, R.; Sboev, A.; Sboeva, S.; Klyueva, Y. Relation Extraction from Texts Containing Pharmacologically Significant Information on base of Multilingual Language Models [in press]. In Proceedings of the 6th International Workshop on Deep Learning in Computational Physics (DLCP-2022), Dubna, Russia, 6–8 July 2022; Sissa Medialab Srl.: Trieste, Italy, 2022. [Google Scholar]
- Segura-Bedmar, I.; Martínez, P.; Herrero-Zazo, M. SemEval-2013 Task 9: Extraction of Drug-Drug Interactions from Biomedical Texts (DDIExtraction 2013). In Proceedings of the Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013), Atlanta, GA, USA, 14–15 June 2013; Association for Computational Linguistics: Stroudsburg, PA, USA, 2013; pp. 341–350. [Google Scholar]
- Lai, T.; Ji, H.; Zhai, C.X.; Tran, Q.H. Joint biomedical entity and relation extraction with knowledge-enhanced collective inference. In Proceedings of the Joint Conference of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing, ACL-IJCNLP 2021, Bangkok, Thailand, 1 August 2021; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2021; pp. 6248–6260. [Google Scholar]
- Luo, L.; Yang, Z.; Cao, M.; Wang, L.; Zhang, Y.; Lin, H. A neural network-based joint learning approach for biomedical entity and relation extraction from biomedical literature. J. Biomed. Inform. 2020, 103, 103384. [Google Scholar] [CrossRef]


| Named Entity (Contexts) | Named Entity (Contexts) | Relation Type |
|---|---|---|
| Alcon Torbex (1,3) | doctor (1) | Drugname-SourceInfodrug_1 |
| Torbex (1,3) | doctor (1) | Drugname-SourceInfodrug_1 |
| Torbex (1,3) | doctor (1) | Drugname-SourceInfodrug_1 |
| Alcon Torbex (1,3) | wrote out (1) | Drugname-SourceInfodrug_1 |
| Torbex (1,3) | wrote out (1) | Drugname-SourceInfodrug_1 |
| Torbex (1,3) | wrote out (1) | Drugname-SourceInfodrug_1 |
| Alcon Torbex (1,3) | maternity hospital (3) | Drugname-SourceInfodrug_1 |
| Torbex (1,3) | maternity hospital (3) | Drugname-SourceInfodrug_1 |
| Torbex (1,3) | maternity hospital (3) | Drugname-SourceInfodrug_1 |
| conjuctivitis (1,2) | my eyes hurt (1,2) | Diseasename-Indications_1 |
| conjuctivitis (1,2) | couldn’t open them (1,2) | Diseasename-Indications_1 |
| Alcon Torbex (1,3) | conjuctivitis (1,2) | Drugname-Diseasename_1 |
| Torbex (1,3) | conjuctivitis (1,2) | Drugname-Diseasename_1 |
| Torbex (1,3) | conjuctivitis (1,2) | Drugname-Diseasename_1 |
| red spots appeared under my eyes (2) | Alcon Torbex (1,3) | ADR-Drugname_0 |
| red spots appeared under my eyes (2) | Torbex (1,3) | ADR-Drugname_0 |
| red spots appeared under my eyes (2) | Torbex (1,3) | ADR-Drugname_0 |
| Alcon Torbex (1,3) | ophtalmologist (2) | Drugname-SourceInfodrug_0 |
| Torbex (1,3) | ophtalmologist (2) | Drugname-SourceInfodrug_0 |
| Torbex (1,3) | ophtalmologist (2) | Drugname-SourceInfodrug_0 |
| Alcon Torbex (1,3) | wrote out (2) | Drugname-SourceInfodrug_0 |
| Torbex (1,3) | wrote out (2) | Drugname-SourceInfodrug_0 |
| Torbex (1,3) | wrote out (2) | Drugname-SourceInfodrug_0 |
| conjuctivitis (1,2) | eyes start to turn sour (3) | Diseasename-Indication_0 |
| Value | Total in Main | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | Extension |
|---|---|---|---|---|---|---|---|
| text number | 2798 | 559 | 559 | 560 | 560 | 560 | 1023 |
| avg text length (in words) | 157 | 157 | 159 | 156 | 156 | 156 | 176 |
| ADR number | 1809 | 380 | 390 | 329 | 382 | 328 | 3241 |
| Drugname number | 8467 | 1695 | 1664 | 1736 | 1686 | 1686 | 3345 |
| SourceInfodrug number | 2595 | 530 | 523 | 508 | 519 | 515 | 1167 |
| Diseasename number | 4026 | 801 | 769 | 760 | 842 | 854 | 908 |
| Indication number | 4843 | 1014 | 982 | 965 | 936 | 946 | 2613 |
| ADR_Drugname relations number | 4274 (910) | 844 (166) | 832 (168) | 812 (210) | 1004 (212) | 782 (154) | 8443 (2288) |
| Drugname_Disease relations number | 11135 (2108) | 2168 (383) | 2107 (361) | 2138 (429) | 2311 (541) | 2411 (394) | 3012 (510) |
| Drugname_Info relations number | 7056 (1279) | 1481 (297) | 1404 (229) | 1381 (316) | 1382 (225) | 1408 (212) | 3251 (927) |
| Disease_Indication relations number | 7093 (742) | 1469 (144) | 1223 (177) | 1443 (126) | 1446 (136) | 1512 (159) | 2117 (572) |
| Number of | Total | Folds | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| sentences | 4272 | 427 | 427 | 427 | 427 | 427 | 427 | 427 | 427 | 427 | 429 |
| entities | 10,839 | 1070 | 1126 | 1091 | 1068 | 1054 | 1126 | 1071 | 1085 | 1079 | 1069 |
| AdE | 5776 | 576 | 616 | 574 | 564 | 561 | 607 | 552 | 577 | 582 | 567 |
| Drug | 5063 | 494 | 510 | 517 | 504 | 493 | 519 | 519 | 508 | 497 | 502 |
| Drug-AdE | 6821 | 666 | 724 | 688 | 657 | 648 | 732 | 666 | 704 | 688 | 648 |
| Value | Total | Train | Valid | Test |
|---|---|---|---|---|
| sentence number | 8920 | 6970 | 642 | 1308 |
| avg text length (in words) | 20 | 22 | 24 | 14 |
| Total entity number | 19,179 | 14,677 | 1438 | 3064 |
| Drug entity number | 12,123 | 9364 | 883 | 1876 |
| Group entity number | 4373 | 3375 | 330 | 668 |
| Brand entity number | 1906 | 1435 | 102 | 369 |
| Drug_n entity number | 777 | 503 | 123 | 151 |
| Total relation number | 11,811 | 3941 | 2109 | 5761 |
| Mechanism relations number | 1755 | 1319 | 134 | 302 |
| Effect relations number | 2141 | 1608 | 173 | 360 |
| Int relations number | 295 | 188 | 11 | 96 |
| Advise relations number | 1134 | 826 | 87 | 221 |
| Generated_false relations number | 28,568 | 22,082 | 1704 | 4782 |
| Difference Feature | Sequential Approach | Joint Approach |
|---|---|---|
| NER solver | Token classification | Span classification |
| Overlapping entites | Yes | No |
| Discontionous entities | Yes | No |
| Number of models | Model for each step | One model |
| Loss function | NER loss, RE Loss | Joint NER-RE loss |
| Training process | One for each step | One for joint stage |
| Language Model | H # | HLS | ILS | AH # | Vocab | Params |
|---|---|---|---|---|---|---|
| RuBERT | 12 | 768 | 3072 | 12 | 119,547 | 178 M |
| RuDr-BERT | 12 | 768 | 3072 | 12 | 119,547 | 178 M |
| EnRuDr-BERT | 12 | 768 | 3072 | 12 | 119,547 | 178 M |
| BioALBERT * | 12 | 4096 | 16,384 | 64 | 30,000 | 222 M * |
| BioLink BERT | 24 | 1024 | 4096 | 16 | 28,895 | 333 M |
| RuBioRoBERTa | 24 | 1024 | 4096 | 16 | 50,265 | 355 M |
| XLMR | 24 | 1024 | 4096 | 16 | 250,002 | 559 M |
| XLMR-sag | 24 | 1024 | 4096 | 16 | 250,002 | 559 M |
| Corpora | Model | ADR-Drug | Drug-Dis | Drug-Info | Dis-Ind | F1-macro |
|---|---|---|---|---|---|---|
| RDRS- 2800 | RuBERT | 41.6 | 63.1 | 45.3 | 34.2 | 46.0 |
| RuDr BERT | 42.2 | 64.0 | 44.5 | 33.9 | 46.1 | |
| EnRuDr BERT | 41.7 | 62.7 | 44.1 | 33.0 | 45.4 | |
| RuBioRoBERTa | 41.9 | 63.1 | 43.5 | 33.5 | 45.5 | |
| XLMR | 51.2 | 69.3 | 49.2 | 38.6 | 52.1 | |
| XLMR-sag | 51.0 | 68.3 | 49.1 | 38.9 | 51.8 | |
| RDRS- 3800 | RuBERT | 44.0 | 64.7 | 44.7 | 34.0 | 46.8 |
| RuDr BERT | 45.2 | 64.2 | 44.6 | 34.0 | 47.0 | |
| EnRuDr BERT | 46.2 | 64.1 | 45.9 | 35.1 | 47.8 | |
| RuBioRoBERTa | 41.2 | 62.1 | 43.3 | 33.8 | 45.1 | |
| XLMR | 52.5 | 69.6 | 49.3 | 39.0 | 52.6 | |
| XLMR-sag | 55.6 | 69.8 | 49.6 | 39.1 | 53.5 |
| Corpus | A. | Model | ADR-Drug | Drug-Dis | Drug-Info | Dis-Ind | F1-macro |
|---|---|---|---|---|---|---|---|
| RDRS- 2800 | J. | XLMR | 51.2 | 69.4 | 49.2 | 38.6 | 52.1 |
| XLMR-sag | 51.1 | 68.3 | 49 | 38.9 | 51.8 | ||
| S. | XLMR | 46.1 | 69.2 | 45.1 | 32.2 | 48.1 | |
| XLMR-sag | 49.4 | 70.4 | 48.3 | 36.7 | 51.2 | ||
| RDRS- 3800 | J. | XLMR | 52.5 | 69.6 | 49.3 | 39.0 | 52.6 |
| XLMR-sag | 55.5 | 69.8 | 49.6 | 39.1 | 53.5 | ||
| S. | XLMR | 54.7 | 71.1 | 49.8 | 34.6 | 52.6 | |
| XLMR-sag | 55.7 | 71.4 | 49.7 | 35.8 | 53.2 |
| Corpus | A. | Model | ADR | Drug | Disease | Info | Indication | F1-macro |
|---|---|---|---|---|---|---|---|---|
| RDRS-2800 | J. | XLMR | 64.8 | 95.7 | 89.4 | 62.5 | 72.9 | 77.1 |
| XLMR-sag | 63.8 | 96.0 | 89.7 | 63.3 | 73.2 | 77.2 | ||
| S. | XLMR | 49.6 | 95.1 | 87.7 | 55.6 | 64.7 | 70.5 | |
| XLMR-sag | 54.7 | 95.3 | 88.3 | 60.0 | 67.2 | 73.1 | ||
| RDRS-3800 | J. | XLMR | 64.4 | 96.0 | 89.5 | 63.1 | 72.5 | 77.1 |
| XLMR-sag | 65.4 | 96.3 | 89.9 | 63.5 | 72.8 | 77.6 | ||
| S. | XLMR | 60.1 | 95.7 | 89.2 | 60.8 | 68.9 | 74.9 | |
| XLMR-sag | 60.1 | 95.7 | 89.0 | 60.3 | 69.0 | 74.8 |
| Source | Language Model | NER + RE F1-macro | NER F1-macro |
|---|---|---|---|
| ours | Joint (BioLinkBERT) | 80.5 | 90.4 |
| ours | Joint (XLMR-sag) | 80.6 | 90.4 |
| ours | Joint (XLMR) | 81.9 | 91.0 |
| ours | Joint (BioALBERT) | 84.2 | 92.1 |
| [63] | SpERT (BioBERT) | 82.0 | 91.2 |
| [85] | KECI | 81.7 | 90.7 |
| [8] | SpERT (BERT) | 78.9 | 89.3 |
| [60] | Stacked biLSTM | 71.4 | 84.6 |
| Source | Language Model | NER + RE F1-micro | NER + RE F1-macro | NER F1-macro |
|---|---|---|---|---|
| ours | Joint (BioLinkBERT) | 74.2 | 59.3 | 75.9 |
| ours | Joint (XLMR) | 73.4 | 63.5 | 78.0 |
| ours | Joint (XLMR-sag) | 77.3 | 68.4 | 78.7 |
| ours | Joint (BioALBERT) | 79.9 | 73.7 | 81.8 |
| [64] | TP-DDI | 82.4 | - | - |
| [86] | Att-BiLSTM-CRF | 75.1 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sboev, A.; Rybka, R.; Selivanov, A.; Moloshnikov, I.; Gryaznov, A.; Naumov, A.; Sboeva, S.; Rylkov, G.; Zakirova, S. Accuracy Analysis of the End-to-End Extraction of Related Named Entities from Russian Drug Review Texts by Modern Approaches Validated on English Biomedical Corpora. Mathematics 2023, 11, 354. https://doi.org/10.3390/math11020354
Sboev A, Rybka R, Selivanov A, Moloshnikov I, Gryaznov A, Naumov A, Sboeva S, Rylkov G, Zakirova S. Accuracy Analysis of the End-to-End Extraction of Related Named Entities from Russian Drug Review Texts by Modern Approaches Validated on English Biomedical Corpora. Mathematics. 2023; 11(2):354. https://doi.org/10.3390/math11020354
Chicago/Turabian StyleSboev, Alexander, Roman Rybka, Anton Selivanov, Ivan Moloshnikov, Artem Gryaznov, Alexander Naumov, Sanna Sboeva, Gleb Rylkov, and Soyora Zakirova. 2023. "Accuracy Analysis of the End-to-End Extraction of Related Named Entities from Russian Drug Review Texts by Modern Approaches Validated on English Biomedical Corpora" Mathematics 11, no. 2: 354. https://doi.org/10.3390/math11020354
APA StyleSboev, A., Rybka, R., Selivanov, A., Moloshnikov, I., Gryaznov, A., Naumov, A., Sboeva, S., Rylkov, G., & Zakirova, S. (2023). Accuracy Analysis of the End-to-End Extraction of Related Named Entities from Russian Drug Review Texts by Modern Approaches Validated on English Biomedical Corpora. Mathematics, 11(2), 354. https://doi.org/10.3390/math11020354








