Designing the Architecture of a Convolutional Neural Network Automatically for Diabetic Retinopathy Diagnosis
Abstract
1. Introduction
- We proposed a constructive data-dependent AutoML approach to design lightweight CNN models customized to DR screening under various clinical settings. It automatically determines the depth of the model, and the width of each COVN layer and initializes the learnable parameters using the fundus images dataset.
- To corroborate the usefulness of the proposed approach, we applied it to build an AutoML custom-designed lightweight CNN architecture for three datasets.
2. Previous Work
2.1. Data-Dependent and Auto-Deep Models
2.2. DR Screening Methods
3. Materials
3.1. KSU-DR
3.2. EyePACS
3.3. APTOS2019
4. Proposed Method
4.1. Problem Formulation
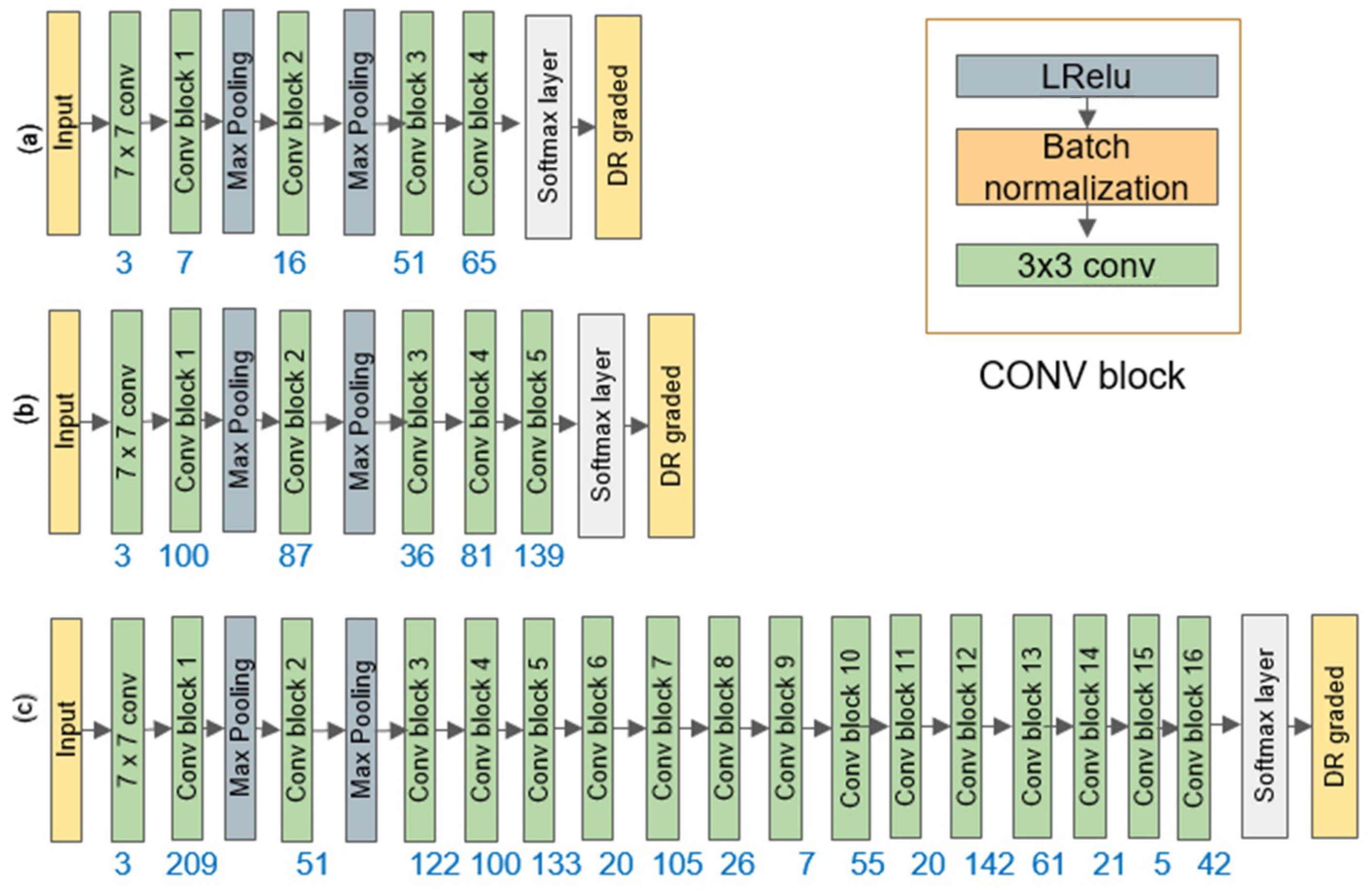
4.2. Custom-Designed CNN Model
4.2.1. Preprocessing
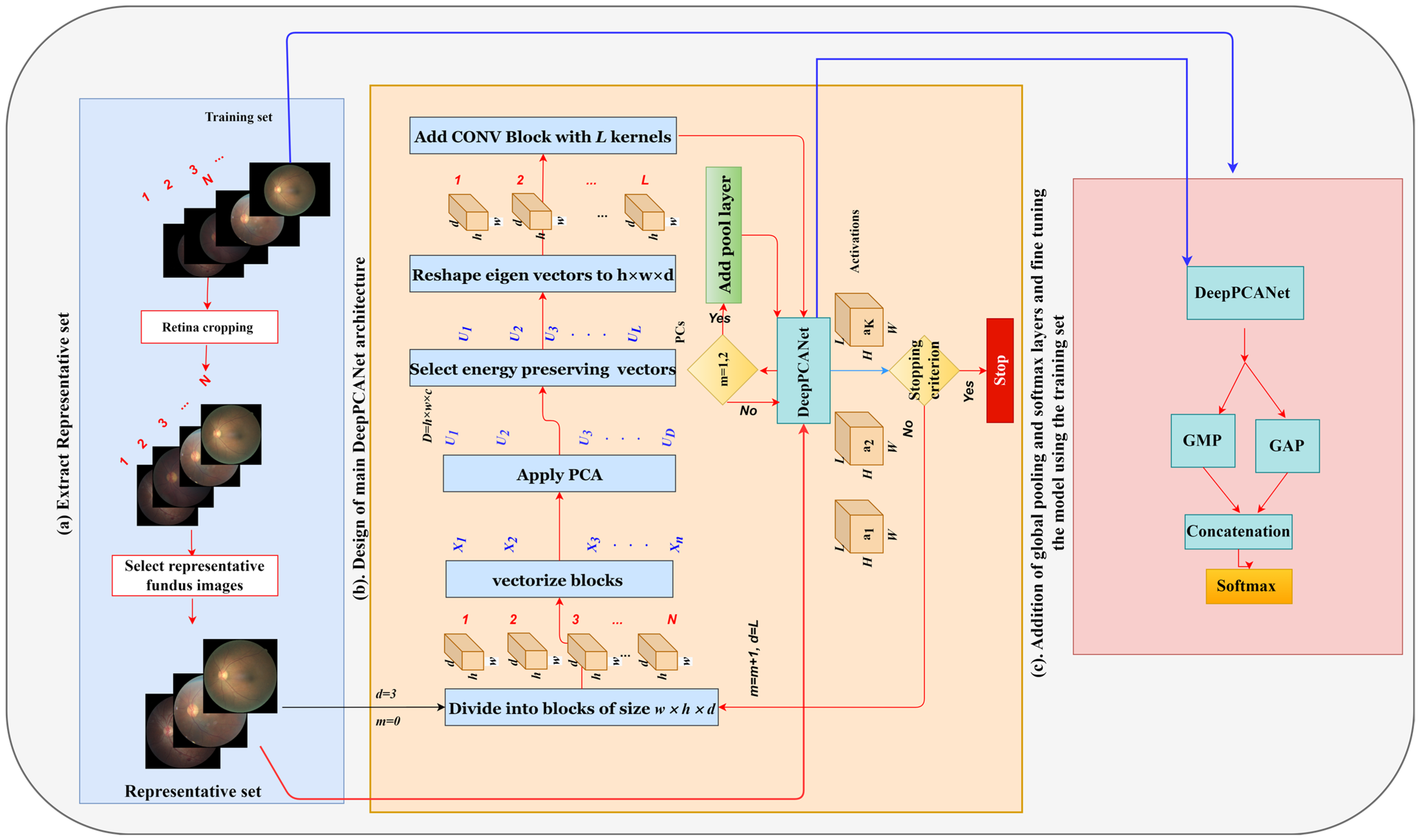
4.2.2. Selection of Representative Fundus Images
4.2.3. Designing the Main DeepPCANet Architecture
| Algorithm 1. To design the main DeepPCANet Architecture. |
| Input: Representative fundus images: RIj, j = 1, 2, …, K of size W × H and the class labels c = 1, 2, …, C; Energy threshold ε |
| Output: The main architecture of DeepPCANet Architecture |
| Processing |
| Step 1: Initialize DeepPCANet with an input layer and set w = 7, h = 7, d = 3, m = 0 (number of layers) |
| Step 2: Set aj = RIj, j = 1, 2, 3, …, K, and TRP (previous TR) = 0. |
| Step 3: Divide aj, j = 1, 2, 3, …, K, into blocks bij, i = 1, 2, 3, …, B, j = 1, 2, 3, …, K, of size w × h × d, where d is the number of channels (feature maps) in aj and B is the number of blocks created from each aj. |
| Step 4: Flatten bij into vectors , where M = K × B, and D = w × h × d. |
| Step 5: Compute zero-center vectors ϕi, such that , where . |
| Step 6: Compute the covariance matrix , where . Calculate the eigenvalues and eigenvectors of the covariance matrix C. |
| Step 7: Select L eigenvectors corresponding to the L largest eigenvalues such that , where ε determines the level of energy to be preserved (e.g., ε = 0.99, for 99% energy preservation). |
| Step 8: The eigenvectors corresponding to the are summed up to form a single eigenvector, and then stacked at the end of the L eigenvectors. |
| Step 9: Reshape to kernels of size W × H × D and add the CONV block to DeepPCANet; Update m = m + 1. |
| Step 10: If m = 1 or 2, add max pool layers with a pooling window of size 2 × 2 and stride 2 to DeepPCANet. |
| Step 11: Compute the activations aj, j = 1, 2, 3, …, K of representative fundus images RIj, j = 1, 2, 3, …, K such that aj = DeepPCANet (RIj). |
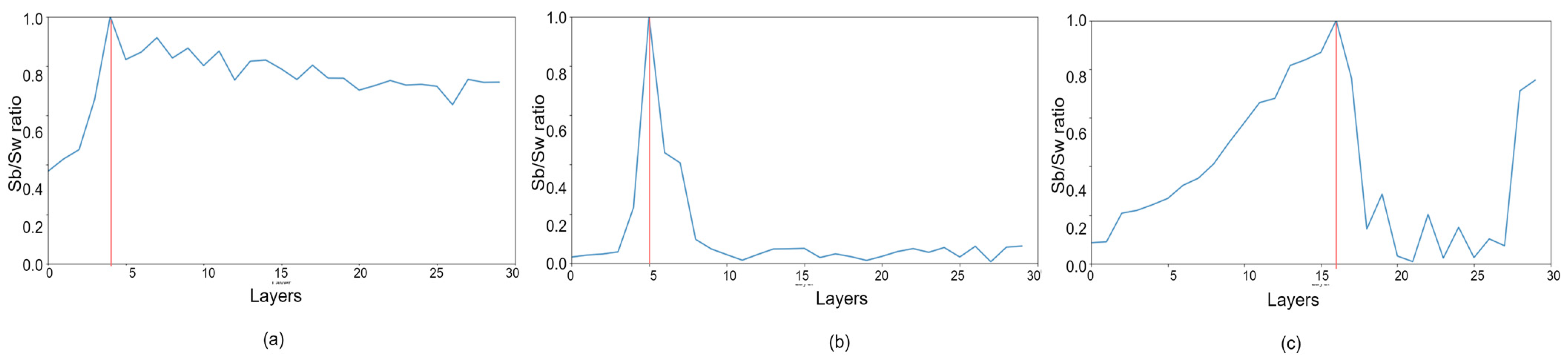
| Step 12: Compute the trace ratio between scatter between matrix (Sb) and within matrix (Sw) as where and . |
| Step 13: If , W = 3, H = 3, D = L, and go to Step 3, stop otherwise. |
4.2.4. Addition of Global Pool and Softmax Layers
4.2.5. Finetuning the DeepPCANet Model
5. Experiments and Results
5.1. Evaluation Protocol
5.1.1. Five Class Problem (SC1)
5.1.2. Two Class Problem (SC2)
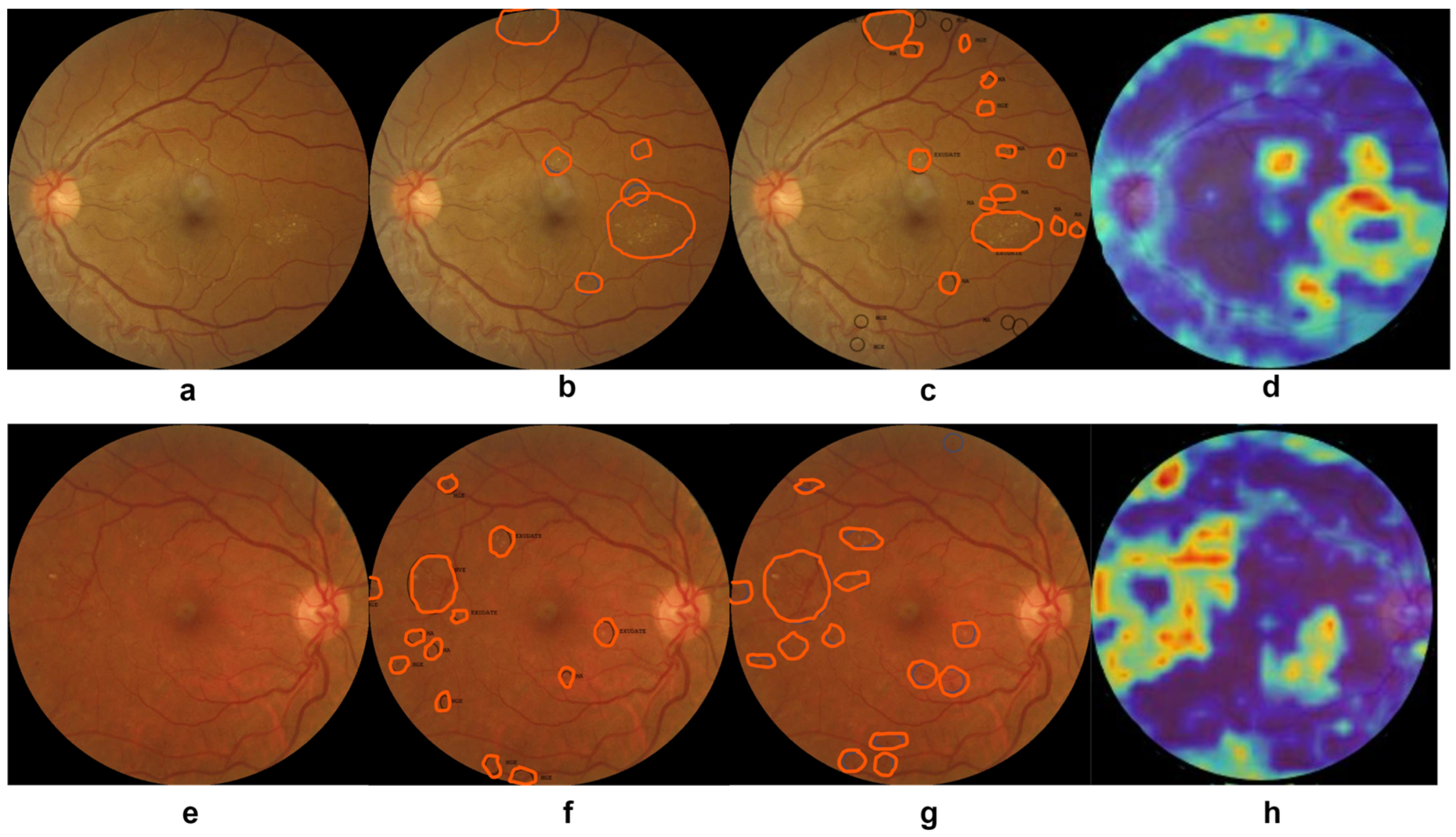
5.2. Visualization
6. Discussions
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yau, J.W.Y.; Rogers, S.L.; Kawasaki, R.; Lamoureux, E.L.; Kowalski, J.W.; Bek, T.; Chen, S.-J.; Dekker, J.M.; Fletcher, A.; Grauslund, J.; et al. Global Prevalence and Major Risk Factors of Diabetic Retinopathy. Diabetes Care 2012, 35, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Quellec, G.; Charrière, K.; Boudi, Y.; Cochener, B.; Lamard, M. Deep image mining for diabetic retinopathy screening. Med. Image Anal. 2017, 39, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Sreejini, K.; Govindan, V. Retrieval of pathological retina images using Bag of Visual Words and pLSA model. Eng. Sci. Technol. Int. J. 2019, 22, 777–785. [Google Scholar] [CrossRef]
- Hemanth, D.J.; Deperlioglu, O.; Kose, U. An enhanced diabetic retinopathy detection and classification approach using deep convolutional neural network. Neural Comput. Appl. 2020, 32, 707–721. [Google Scholar] [CrossRef]
- Stolte, S.; Fang, R. A Survey on Medical Image Analysis in Diabetic Retinopathy. Med. Image Anal. 2020, 64, 101742. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Grigorescu, S.; Trasnea, B.; Cocias, T.; Macesanu, G. A survey of deep learning techniques for autonomous driving. J. Field Robot. 2020, 37, 362–386. [Google Scholar] [CrossRef]
- Abou Arkoub, S.; Hajjam El Hassani, A.; Lauri, F.; Hajjar, M.; Daya, B.; Hecquet, S.; Aubry, S. Survey on Deep Learning Techniques for Medical Imaging Application Area. In Machine Learning Paradigms; Springer: Cham, Switzerland, 2020; pp. 149–189. [Google Scholar]
- Mohamadou, Y.; Halidou, A.; Kapen, P.T. A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19. Appl. Intell. 2020, 50, 3913–3925. [Google Scholar] [CrossRef]
- Zhu, H.; Shu, S.; Zhang, J. FAS-UNet: A Novel FAS-Driven UNet to Learn Variational Image Segmentation. Mathematics 2022, 10, 4055. [Google Scholar] [CrossRef]
- Lam, C.; Yu, C.; Huang, L.; Rubin, D. Retinal lesion detection with deep learning using image patches. Investig. Ophthalmol. Vis. Sci. 2018, 59, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Li, J.; Guo, J.; Chen, Y.; Yi, Z.; Zhong, J. Diagnosis of Diabetic Retinopathy Using Deep Neural Networks. IEEE Access 2019, 7, 3360–3370. [Google Scholar] [CrossRef]
- Shaik, N.S.; Cherukuri, T.K. Hinge attention network: A joint model for diabetic retinopathy severity grading. Appl. Intell. 2022, 52, 15105–15121. [Google Scholar] [CrossRef]
- Gao, Z.; Jin, K.; Yan, Y.; Liu, X.; Shi, Y.; Ge, Y.; Pan, X.; Lu, Y.; Wu, J.; Wang, Y.; et al. End-to-end diabetic retinopathy grading based on fundus fluorescein angiography images using deep learning. Graefe’s Arch. Clin. Exp. Ophthalmol. 2022, 260, 1663–1673. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G. Densely connected convolutional networks. CVPR 2017. arXiv 2016, arXiv:1608.06993. [Google Scholar]
- Saini, M.; Susan, S. Diabetic retinopathy screening using deep learning for multi-class imbalanced datasets. Comput. Biol. Med. 2022, 149, 105989. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; pp. 249–256. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer, Santiago, Chile, 7–13 December 2015; Volume 22, pp. 1026–1034. [Google Scholar]
- Krähenbühl, P. Data-dependent initializations of convolutional neural networks. arXiv 2015, arXiv:1511.06856. [Google Scholar]
- Suau, X.; Apostoloff, N. Filter Distillation for Network Compression. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Piscataway, NJ, USA, 1–5 March 2020; pp. 3129–3138. [Google Scholar]
- Singh, V.K.; Joshi, K. Automated Machine Learning (AutoML): An overview of opportunities for application and research. J. Inf. Technol. Case Appl. Res. 2022, 24, 75–85. [Google Scholar] [CrossRef]
- Thornton, C. Auto-WEKA: Combined selection and hyperparameter optimization of classification algorithms. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 11–14 August 2013; pp. 847–855. [Google Scholar]
- Komer, B.; Bergstra, J.; Eliasmith, C. Hyperopt-sklearn: Automatic hyperparameter conTableuration for scikit-learn. In ICML Workshop on AutoML; Citeseer: Austin, TX, USA, 2014; Volume 9. [Google Scholar]
- Olson, R.S.; Bartley, N.; Urbanowicz, R.J.; Moore, J.H. Evaluation of a tree-based pipeline optimization tool for automating data science. In Proceedings of the Genetic and Evolutionary Computation Conference, Denver, CO, USA, 20–24 June 2016; pp. 485–492. [Google Scholar]
- Feurer, M.; Springenberg, J.; Hutter, F. Initializing bayesian hyperparameter optimization via meta-learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; Volume 29. No. 1. [Google Scholar]
- Jin, H.; Song, Q.; Hu, X. Auto-keras: An efficient neural architecture search system. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, New York, NY, USA, 4–8 August 2019; pp. 1946–1956. [Google Scholar]
- Doke, A.; Gaikwad, M. Survey on Automated Machine Learning (AutoML) and Meta learning. In Proceedings of the 2021 12th International Conference on Computing Communication and Networking Technologies (ICCCNT), Kharagpur, India, 6–8 July 2021; IEEE: Piscataway, NJ, USA, 2021. [Google Scholar]
- Stojadinovic, M.; Milicevic, B.; Jankovic, S. Improved predictive performance of prostate biopsy collaborative group risk calculator when based on automated machine learning. Comput. Biol. Med. 2021, 138, 104903. [Google Scholar] [CrossRef]
- Aloraini, T.; Aljouie, A.; Alniwaider, R.; Alharbi, W.; Alsubaie, L.; AlTuraif, W.; Qureshi, W.; Alswaid, A.; Eyiad, W.; Al Mutairi, F.; et al. The variant artificial intelligence easy scoring (VARIES) system. Comput. Biol. Med. 2022, 145, 105492. [Google Scholar] [CrossRef]
- Zoph, B.; Le, Q.V. Neural architecture search with reinforcement learning. arXiv 2016, arXiv:1611.01578. [Google Scholar]
- Domhan, T.; Springenberg, J.T.; Hutter, F. Speeding up automatic hyperparameter optimization of deep neural networks by extrapolation of learning curves. In Proceedings of the Twenty-fourth International Joint Conference on Artificial Intelligence, Buenos Aires, Argentina, 25–31 July 2015. [Google Scholar]
- Tan, R.Z.; Chew, X.; Khaw, K.W. Neural Architecture Search for Lightweight Neural Network in Food Recognition. Mathematics 2021, 9, 1245. [Google Scholar] [CrossRef]
- Liu, H. Hierarchical representations for efficient architecture search. arXiv 2017, arXiv:1711.00436. [Google Scholar]
- Kaggle. Diabetic Retinopathy Detection (Kaggle). 2019. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection/data (accessed on 26 September 2021).
- Kaggle. APTOS Blindness Detection. 2019. Available online: https://www.kaggle.com/c/aptos2019-blindness-detection (accessed on 26 September 2021).
- Zhang, H. Resnest: Split-attention networks. arXiv 2020, arXiv:2004.08955. [Google Scholar]
- Mateen, M.; Wen, J.; Hassan, M.; Nasrullah, N.; Sun, S.; Hayat, S. Automatic Detection of Diabetic Retinopathy: A Review on Datasets, Methods and Evaluation Metrics. IEEE Access 2020, 8, 48784–48811. [Google Scholar] [CrossRef]
- Soomro, T.A.; Afifi, A.J.; Zheng, L.; Soomro, S.; Gao, J.; Hellwich, O.; Paul, M. Deep learning models for retinal blood vessels segmentation: A review. IEEE Access 2019, 7, 71696–71717. [Google Scholar] [CrossRef]
- Asiri, N.; Hussain, M.; Al Adel, F.; Alzaidi, N. Deep learning based computer-aided diagnosis systems for diabetic retinopathy: A survey. Artif. Intell. Med. 2019, 99, 101701. [Google Scholar] [CrossRef]
- Bhandari, S.; Pathak, S.; Jain, S.A.; Deshmukh, V. A Review on Swarm intelligence & Evolutionary Algorithms based Approaches for Diabetic Retinopathy Detection. In Proceedings of the 2022 IEEE World Conference on Applied Intelligence and Computing (AIC), Sonbhadra, India, 17–19 June 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar]
- Han, S.; Mao, H.; Dally, W.J. Deep compression: Compressing deep neural networks with pruning, trained quantization and huffman coding. arXiv 2015, arXiv:1510.00149. [Google Scholar]
- He, Y.; Zhang, X.; Sun, J. Channel pruning for accelerating very deep neural networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 1389–1397. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Jiang, C.; Li, G.; Qian, C.; Tang, K. Efficient DNN Neuron Pruning by Minimizing Layer-wise Nonlinear Reconstruction Error. In Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence (IJCAI-18), Stockholm, Sweden, 13–19 July 2018; Volume 2018, pp. 2298–2304. [Google Scholar]
- Choudhary, T.; Mishra, V.; Goswami, A.; Sarangapani, J. A transfer learning with structured filter pruning approach for improved breast cancer classification on point-of-care devices. Comput. Biol. Med. 2021, 134, 104432. [Google Scholar] [CrossRef]
- Chan, T.H.; Jia, K.; Gao, S.; Lu, J.; Zeng, Z.; Ma, Y. PCANet: A simple deep learning baseline for image classification? IEEE Trans. Image Process. 2015, 24, 5017–5032. [Google Scholar] [CrossRef] [PubMed]
- Seuret, M.; Alberti, M.; Liwicki, M.; Ingold, R. PCA-initialized deep neural networks applied to document image analysis. In Proceedings of the 2017 14th IAPR International Conference on Document Analysis and Recognition (ICDAR), Kyoto, Japan, 9–15 November 2017; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar]
- Zhong, Z.; Yang, Z.; Deng, B.; Yan, J.; Wu, W.; Shao, J.; Liu, C.L. Blockqnn: Efficient block-wise neural network architecture generation. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 2314–2328. [Google Scholar] [CrossRef] [PubMed]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA. In Automated Machine Learning; Springer: Cham, Switzerland, 2019; pp. 81–95. [Google Scholar]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.; Blum, M.; Hutter, F. Auto-sklearn: Efficient and robust automated machine learning. In Automated Machine Learning; Springer: Cham, Switzerland, 2019; pp. 113–134. [Google Scholar]
- Real, E.; Aggarwal, A.; Huang, Y.; Le, Q.V. Regularized evolution for image classifier architecture search. In Proceedings of the AAAI Conference on Artificial Intelligence, Palo Alto, CA, USA, 22 February–1 March 2022; No. 01. Volume 33. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Pham, H.; Guan, M.; Zoph, B.; Le, Q.; Dean, J. Efficient neural architecture search via parameters sharing. In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; PMLR 80. pp. 4095–4104. [Google Scholar]
- Liu, H.; Simonyan, K.; Yang, Y. Darts: Differentiable architecture search. arXiv 2018, arXiv:1806.09055. [Google Scholar]
- Bergstra, J.; Yamins, D.; Cox, D. Making a science of model search: Hyperparameter optimization in hundreds of dimensions for vision architectures. In Proceedings of the International Conference on Machine Learning, Atlanta, GA, USA, 17–19 June 2013; PMLR 28. pp. 115–123. [Google Scholar]
- Bisong, E. Google AutoML: Cloud vision. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Springer: Berkeley, CA, USA, 2019; pp. 581–598. [Google Scholar]
- Bisong, E. An Overview of Google Cloud Platform Services. Building Machine Learning and Deep Learning Models on Google Cloud Platform; Springer: Berkeley, CA, USA, 2019; pp. 7–10. [Google Scholar]
- Islam, S.M.S.; Hasan, M.M.; Abdullah, S. Deep Learning based Early Detection and Grading of Diabetic Retinopathy Using Retinal Fundus Images. arXiv 2018, arXiv:1812.10595. [Google Scholar]
- Li, Y.-H.; Yeh, N.-N.; Chen, S.-J.; Chung, Y.-C. Computer-assisted diagnosis for diabetic retinopathy based on fundus images using deep convolutional neural network. Mob. Inf. Syst. 2019, 2019, 6142839. [Google Scholar] [CrossRef]
- Challa, U.K.; Yellamraju, P.; Bhatt, J.S. A Multi-class Deep All-CNN for Detection of Diabetic Retinopathy Using Retinal Fundus Images. In Proceedings of the International Conference on Pattern Recognition and Machine Intelligence, Tezpur, India, 17–20 December 2019; Springer: Cham, Switzerland, 2019; pp. 191–199. [Google Scholar]
- Tymchenko, B.; Marchenko, P.; Spodarets, D. Deep Learning Approach to Diabetic Retinopathy Detection. arXiv 2020, arXiv:2003.02261. [Google Scholar]
- Sikder, N.; Chowdhury, M.S.; Arif, A.S.M.; Nahid, A.A. Early Blindness Detection Based on Retinal Images Using Ensemble Learning. In Proceedings of the 2019 22nd International Conference on Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 18–20 December 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar]
- Sikder, N.; Masud, M.; Bairagi, A.K.; Arif, A.S.M.; Nahid, A.A.; Alhumyani, H.A. Severity classification of diabetic retinopathy using an ensemble learning algorithm through analyzing retinal images. Symmetry 2021, 13, 670. [Google Scholar] [CrossRef]
- Colas, E.; Besse, A.; Orgogozo, A.; Schmauch, B.; Meric, N.; Besse, E. Deep learning approach for diabetic retinopathy screening. Acta Ophthalmol. 2016, 94. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; IEEE: Piscataway, NJ, USA, 2009. [Google Scholar]
- Zhang, Q.; Couloigner, I. A new and efficient k-medoid algorithm for spatial clustering. In Proceedings of the International Conference on Computational Science and Its Applications, Singapore, 9–12 May 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 181–189. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Likas, A.; Vlassis, N.; Verbeek, J.J. The global k-means clustering algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef]
- Saeed, F.; Hussain, M.; Aboalsamh, H.A. Method for Fingerprint Classification. U.S. Patent 9530042, 13 June 2016. [Google Scholar]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Mika, S.; Ratsch, G.; Weston, J.; Scholkopf, B.; Mullers, K.R. Fisher discriminant analysis with kernels. In Neural Networks for Signal Processing IX: Proceedings of the 1999 IEEE Signal Processing Society Workshop (Cat. No. 98TH8468), Madison, WI, USA, 25 August 1999; IEEE: Piscataway, NJ, USA, 1999. [Google Scholar]
- Al Asbahi, A.A.M.H.; Gang, F.Z.; Iqbal, W.; Abass, Q.; Mohsin, M.; Iram, R. Novel approach of Principal Component Analysis method to assess the national energy performance via Energy Trilemma Index. Energy Rep. 2019, 5, 704–713. [Google Scholar] [CrossRef]
- Cook, A. Global Average Pooling Layers for Object Localization. 2017. Available online: https://alexisbcook.github.io/2017/global-average-pooling-layers-for-object-localization (accessed on 23 November 2022).
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A next-generation hyperparameter optimization framework. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, New York, NY, USA, 4–8 August 2019; pp. 2623–2631. [Google Scholar]
- Yu, M.; Wang, Y. Intelligent detection and applied research on diabetic retinopathy based on the residual attention network. Int. J. Imaging Syst. Technol. 2022, 32, 1789–1800. [Google Scholar] [CrossRef]
- Chowdhury, A.R.; Chatterjee, T.; Banerjee, S. A Random Forest classifier-based approach in the detection of abnormalities in the retina. Med. Biol. Eng. Comput. 2019, 57, 193–203. [Google Scholar] [CrossRef]
- Zhang, W.; Zhong, J.; Yang, S.; Gao, Z.; Hu, J.; Chen, Y.; Yi, Z. Automated identification and grading system of diabetic retinopathy using deep neural networks. Knowl.-Based Syst. 2019, 175, 12–25. [Google Scholar] [CrossRef]
- Zebin, T.; Rezvy, S. COVID-19 detection and disease progression visualization: Deep learning on chest X-rays for classification and coarse localization. Appl. Intell. 2021, 51, 1010–1021. [Google Scholar] [CrossRef]
- Ben-David, A. Comparison of classification accuracy using Cohen’s Weighted Kappa. Expert Syst. Appl. 2008, 34, 825–832. [Google Scholar] [CrossRef]
- Fernández-Fuentes, X.; Mera, D.; Gómez, A.; Vidal-Franco, I. Towards a fast and accurate eit inverse problem solver: A machine learning approach. Electronics 2018, 7, 422. [Google Scholar] [CrossRef]
- Shu, Y.; Wang, W.; Cai, S. Understanding architectures learnt by cell-based neural architecture search. arXiv 2019, arXiv:1909.09569. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-Cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Shorfuzzaman, M.; Hossain, M.S.; El Saddik, A. An Explainable Deep Learning Ensemble Model for Robust Diagnosis of Diabetic Retinopathy Grading. ACM Trans. Multimed. Comput. Commun. Appl. (TOMM) 2021, 17, 1–24. [Google Scholar] [CrossRef]
- Lahmar, C.; Idri, A. On the value of deep learning for diagnosing diabetic retinopathy. Health Technol. 2021, 12, 89–105. [Google Scholar] [CrossRef]
- Chetoui, M.; Akhloufi, M.A. Explainable end-to-end deep learning for diabetic retinopathy detection across multiple datasets. J. Med. Imaging 2020, 7, 044503. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.R.; Fletcher-Janzen, E. Handbook of Clinical Child Neuropsychology; Springer: Boston, MA, USA, 2013; ISBN 978-0-387-70708-22. [Google Scholar]


| Dataset | Model | ACC % | SE % |
|---|---|---|---|
| EyePACS | Random fundus images | 85.12 | 81.33 |
| K-means | 89.32 | 83.45 | |
| K-medoids | 94.22 | 86.56 |
| Dataset | Activation Function | Learning Rate | Patch’s Size | Optimizer | Dropout |
|---|---|---|---|---|---|
| KSU-DR | LReLU | 0.0001 | 10 | RMSprop | 0.50 |
| EyePACS | LReLU | 0.0055 | 10 | RMSprop | 0.38 |
| APTOS2019 | LReLU | 0.0007 | 5 | RMSprop | 0.40 |
| Dataset | Model | Performance (%) | |||
|---|---|---|---|---|---|
| ACC | SE | SP | Kappa | ||
| EyePACS (SC1) | PCANet model (mixed dataset) | 73 | 28 | 83 | 8 |
| APTOS2019 (SC1) | 88 | 36 | 91 | 51 | |
| KSU-DR (SC2) | 80 | 81 | 81 | 59 | |
| Dataset | Model | #FLOPs | # Parameters | ACC % | SE % | SP % | Kappa % |
|---|---|---|---|---|---|---|---|
| APTOS2019 | ResNet152 | 5.6 M | 60.19 M | 95.25 | 88.22 | 96.97 | 88.15 |
| DenseNet121 | 1.44 M | 7.98 M | 96.58 | 91.55 | 97.82 | 89.22 | |
| ResNeSt50 | 5.39 M | 27.5 M | 97.11 | 92.29 | 98.2 | 90.82 | |
| DeepPCANet-4 | 1.36 M | 63.7 K | 98.21 | 95.29 | 98.9 | 94.32 | |
| EyePACS | ResNet152 | 5.6 M | 60.19 M | 92.25 | 80.74 | 94.9 | 75.16 |
| DenseNet121 | 1.44 M | 7.98 M | 91.14 | 80.07 | 95 | 74.84 | |
| ResNeSt50 | 5.39 M | 27.5 M | 93.12 | 82.33 | 95.21 | 78 | |
| DeepPCANet-16 | 2.11 M | 557.68 K | 94.22 | 86.56 | 96.30 | 81.64 |
| Dataset | Model | #FLOPs | # Parameters | ACC % | SE % | SP % | Kappa % |
|---|---|---|---|---|---|---|---|
| KSU-DR dataset | ResNet152 | 5.6 M | 60.19 M | 97.98 | 97.83 | 97.83 | 95.75 |
| DenseNet121 | 1.44 M | 7.98 M | 98.51 | 96.06 | 98.86 | 96.4 | |
| ResNeSt50 | 5.39 M | 27.5 M | 99.47 | 99.46 | 99.46 | 98.93 | |
| DeepPCANet-5 | 1.375 M | 73.66 K | 99.5 | 99.5 | 99.5 | 98.99 | |
| APTOS2019 | ResNet152 | 5.6 M | 60.19 M | 95 | 94.44 | 94.44 | 89.80 |
| DenseNet121 | 1.44 M | 7.98 M | 99.32 | 98.8 | 98.8 | 98.73 | |
| ResNeSt50 | 5.39 M | 27.5 M | 98.33 | 96.54 | 96.53 | 94.22 | |
| DeepPCANet-4 | 1.36 M | 63.7 K | 99.7 | 99.44 | 99.44 | 99.3 | |
| EyePACS | ResNet152 | 5.6 M | 60.19 M | 91.36 | 90.94 | 92.25 | 82.53 |
| DenseNet121 | 1.44 M | 7.98 M | 91.51 | 91.75 | 91.75 | 82.72 | |
| ResNeSt50 | 5.39 M | 27.5 M | 90.53 | 90.92 | 90.92 | 79.04 | |
| DeepPCANet-16 | 2.11 M | 557.68 K | 94.44 | 94.28 | 94.28 | 88.71 |
| Dataset | Model | #FLOPs | # Parameters | ACC % | SE % | PR % |
|---|---|---|---|---|---|---|
| EyePACS | Auto-Keras | 0.31 M | 15 M | 73 | 73 | 53 |
| Google-AutoML | Hidden | Hidden | -- | 71.43 | 79.1 | |
| DeepPCANet-16 | 2.11 M | 557.68 K | 94.44 | 94.28 | 96.12 |
| Paper | Method | Dataset | Performance (%) | |||
|---|---|---|---|---|---|---|
| ACC | SE | SP | Kappa | |||
| Five classes (SC1) | ||||||
| Sikder et al., 2019 [65] | Colored features extraction using ensemble | APTOS2019 | 91 | 89.54 | - | - |
| Tymchenko et al., 2020 [64] | An ensembled models with 3 CNN architectures fficientNet-B4, EfficientNet-B5, and SE-ResNeXt50 | APTOS2019 | 91.9 | 84 | 98 | 96.9 |
| Shorfuzzaman et al., 2021 [89] | CNN-based transfer learning ensemble | aptos2019 | 96.2 | 94.00 | - | - |
| Sikder et al. (2021) | Histogram and GLCM features tuning using XGBoostand genetic algorithm. | aptos2019 | 94.20 | - | - | - |
| DeepPCANet-4 | DeepPCANet model customized for APTOS2019 dataset | APTOS2019 (test: 40% of public dataset) | 98.21 | 95.28 | 98.85 | 94.32 |
| Lahmar et al., 2021 [90] | Transfer learning (MobileNet V2) | APTOS2019 (SC1) | 93.09 | 89.27 | 92.69 | - |
| Islam et al., 2018 [61] | A CNN model consisting of 18 layers with 3 × 3 and 4 × 4 kernels | EyePACS (test: 4% of dataset) | 94.5 | 90.2 | ||
| Li et al., 2019 [62] | Deep learning model based on DCNN | EyePACS (test: 10% of dataset) | 86.17 | - | - | - |
| DeepPCANet-16 | DeepPCANet model customized for EyePACS dataset | EyePACS (test: 10% of public dataset) | 94.22 | 89.56 | 96.30 | 81.64 |
| Normal vs. DR (all DR levels) (two classes) (SC2) | ||||||
| Tymchenko et al., 2020 [64] | Ensembled models with 3 CNN architectures EfficientNet-B4, EfficientNet-B5, and SE-ResNeXt50 | APTOS2019 | 99.3 | 99.3 | 99.3 | 98.6 |
| DeepPCANet-4 | DeepPCANet model customized for APTOS2019 dataset | APTOS2019 (test: 10% of public dataset) | 99.7 | 99.44 | 99.44 | 99.3 |
| Islam et al., 2018 [61] | A CNN model consisting of 18 layers with 3 × 3 and 4 × 4 kernels | EyePACS (test: 4% of public dataset) | 94.5 | 90.2 | - | |
| Li et al., 2019 [62] | Features extraction using deep learning model based on DCNN and SVM classification | EyePACS | 91.05 | 89.30 | 90.89 | - |
| Chetoui et al., 2020 [87] | Pretrained Inception-Resnet-v2 DCNN | EyePACS (SC2) | 97.9 | 95.8 | 97.1 | 98.6 |
| DeepPCANet-16 | DeepPCANet model customized for EyePACS dataset | EyePACS (test: 10% of public dataset) | 94.44 | 94.28 | 94.28 | 88.71 |
| Non-referral (Normal and DR grad 1) vs. referral (DR grade 2 to highest grade) (two classes) (SC3) | ||||||
| Colas et al., 2016 [67] | A CNN model. End to end training. | EyePACS (train: 89%, test: 11%) | 96.2 | 66.6 | 94.6 | |
| Islam et al., 2018 [61] | A CNN model | EyePACS (train: 96%, test: 4%) | 98 | 94 | ||
| DeepPCANet-16 | DeepPCANet model derived from EyePACS dataset | EyePACS (test: 10% of public dataset) | 94.59 | 94.86 | 94.87 | 89.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeed, F.; Hussain, M.; Aboalsamh, H.A.; Al Adel, F.; Al Owaifeer, A.M. Designing the Architecture of a Convolutional Neural Network Automatically for Diabetic Retinopathy Diagnosis. Mathematics 2023, 11, 307. https://doi.org/10.3390/math11020307
Saeed F, Hussain M, Aboalsamh HA, Al Adel F, Al Owaifeer AM. Designing the Architecture of a Convolutional Neural Network Automatically for Diabetic Retinopathy Diagnosis. Mathematics. 2023; 11(2):307. https://doi.org/10.3390/math11020307
Chicago/Turabian StyleSaeed, Fahman, Muhammad Hussain, Hatim A. Aboalsamh, Fadwa Al Adel, and Adi Mohammed Al Owaifeer. 2023. "Designing the Architecture of a Convolutional Neural Network Automatically for Diabetic Retinopathy Diagnosis" Mathematics 11, no. 2: 307. https://doi.org/10.3390/math11020307
APA StyleSaeed, F., Hussain, M., Aboalsamh, H. A., Al Adel, F., & Al Owaifeer, A. M. (2023). Designing the Architecture of a Convolutional Neural Network Automatically for Diabetic Retinopathy Diagnosis. Mathematics, 11(2), 307. https://doi.org/10.3390/math11020307








