A Domain Adaptation-Based Method for Classification of Motor Imagery EEG
Abstract
:1. Introduction
2. Materials and Methods
2.1. CSP for Feature Extraction
2.2. LDA for Classification
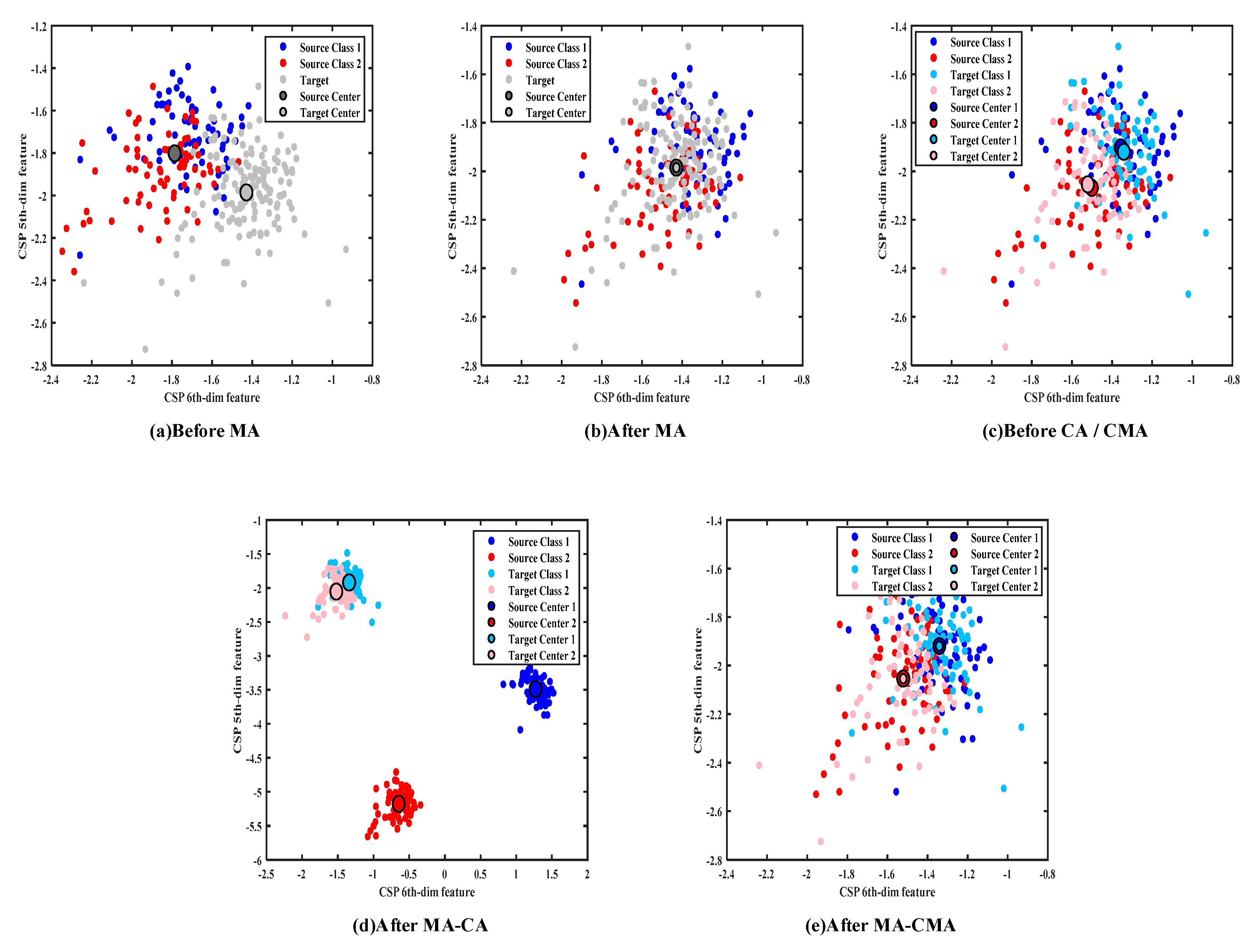
2.3. Overall Mean Alignment
2.4. Per-Class Covariance and Mean Alignment
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kubler, A.; Kotchoubey, B.; Kaiser, J.; Wolpaw, J.R.; Birbaumer, N. Brain-computer communication: Unlocking the locked in. Psychol. Bull. 2001, 127, 358–375. [Google Scholar] [CrossRef] [PubMed]
- Nijholt, A. BCI for Games: A ‘State of the Art’ Survey. In Proceedings of the 7th International Conference on Entertainment Computing (ICEC 2008), Pittsburgh, PA, USA, 25–27 September 2008; pp. 225–228. [Google Scholar]
- Wolpaw, J.R.; Birbaumer, N.; McFarland, D.J.; Pfurtscheller, G.; Vaughan, T.M. Brain-computer interfaces for communication and control. Clin. Neurophysiol. 2002, 113, 767–791. [Google Scholar] [CrossRef]
- Pfurtscheller, G.; Neuper, C. Motor imagery activates primary sensorimotor area in humans. Neurosci. Lett. 1997, 239, 65–68. [Google Scholar] [CrossRef]
- Wang, B.; Wong, C.M.; Kang, Z.; Liu, F.; Shui, C.; Wan, F.; Chen, C.L.P. Common Spatial Pattern Reformulated for Regularizations in Brain-Computer Interfaces. IEEE Trans. Cybern. 2021, 51, 5008–5020. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Wang, Q.; Zhao, Y.; Liu, X.; Bai, O.; Li, Y. Unsupervised domain adaptation techniques based on auto-encoder for non-stationary EEG-based emotion recognition. Comput. Biol. Med. 2016, 79, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Jayaram, V.; Alamgir, M.; Altun, Y.; Schoelkopf, B.; Grosse-Wentrup, M. Transfer Learning in Brain-Computer Interfaces. IEEE Comput. Intell. Mag. 2016, 11, 20–31. [Google Scholar] [CrossRef] [Green Version]
- Bamdadian, A.; Guan, C.T.; Ang, K.K.; Xu, J.X. Improving session-to-session transfer performance of motor imagery-based BCI using Adaptive Extreme Learning Machine. In Proceedings of the 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Osaka, Japan, 3–7 July 2013; pp. 2188–2191. [Google Scholar]
- Al-Saegh, A.; Dawwd, S.A.; Abdul-Jabbar, J.M. Deep learning for motor imagery EEG-based classification: A review. Biomed. Signal Processing Control 2021, 63, 102172. [Google Scholar] [CrossRef]
- Huang, X.; Xu, Y.; Hua, J.; Yi, W.; Yin, H.; Hu, R.; Wang, S. A Review on Signal Processing Approaches to Reduce Calibration Time in EEG-Based Brain-Computer Interface. Front. Neurosci. 2021, 15, 1066. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Azab, A.M.; Mihaylova, L.; Ang, K.K.; Arvaneh, M. Weighted Transfer Learning for Improving Motor Imagery-Based Brain-Computer Interface. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 1352–1359. [Google Scholar] [CrossRef]
- Zhang, D.; Yao, L.; Chen, K.; Wang, S. Ready for Use: Subject-Independent Movement Intention Recognition via a Convolutional Attention Model. In Proceedings of the 27th ACM International Conference on Information and Knowledge Management (CIKM), Torino, Italy, 22–26 October 2018; pp. 1763–1766. [Google Scholar]
- Zhang, R.; Zong, Q.; Dou, L.; Zhao, X.; Tang, Y.; Li, Z. Hybrid deep neural network using transfer learning for EEG motor imagery decoding. Biomed. Signal Processing Control 2021, 63, 102144. [Google Scholar] [CrossRef]
- Saha, S.; Baumert, M. Intra- and Inter-subject Variability in EEG-Based Sensorimotor Brain Computer Interface: A Review. Front. Comput. Neurosci. 2020, 13, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazli, S.; Daehne, S.; Samek, W.; Biessmann, F.; Mueller, K.-R. Learning From More Than One Data Source: Data Fusion Techniques for Sensorimotor Rhythm-Based Brain-Computer Interfaces. Proc. IEEE 2015, 103, 891–906. [Google Scholar] [CrossRef]
- Abdi, L.; Hashemi, S. Unsupervised Domain Adaptation Based on Correlation Maximization. IEEE Access 2021, 9, 127054–127067. [Google Scholar] [CrossRef]
- Li, P.; Ni, Z.; Zhu, X.; Song, J. Inter-class distribution alienation and inter-domain distribution alignment based on manifold embedding for domain adaptation. J. Intell. Fuzzy Syst. 2020, 39, 8149–8159. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, X.; Lan, L.; Luo, Z. Maximum Mean and Covariance Discrepancy for Unsupervised Domain Adaptation. Neural Processing Lett. 2020, 51, 347–366. [Google Scholar] [CrossRef]
- Lee, B.-H.; Jeong, J.-H.; Lee, S.-W. SessionNet: Feature Similarity-Based Weighted Ensemble Learning for Motor Imagery Classification. IEEE Access 2020, 8, 134524–134535. [Google Scholar] [CrossRef]
- Zheng, M.; Yang, B.; Xie, Y. EEG classification across sessions and across subjects through transfer learning in motor imagery-based brain-machine interface system. Med. Biol. Eng. Comput. 2020, 58, 1515–1528. [Google Scholar] [CrossRef]
- Liang, Y.; Ma, Y. A Cross-Session Feature Calibration Algorithm for Electroencephalogram-Based Motor Imagery Classification. J. Med. Imaging Health Inform. 2019, 9, 1534–1540. [Google Scholar] [CrossRef]
- Cheng, M.; Lu, Z.; Wang, H. Regularized common spatial patterns with subject-to-subject transfer of EEG signals. Cogn. Neurodyn. 2017, 11, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Wei, Q.; Zhang, H.; Hu, R.; Liu, J.; Hua, J.; Guo, F. Transfer Learning Based on Regularized Common Spatial Patterns Using Cosine Similarities of Spatial Filters for Motor-Imagery BCI. J. Circuits Syst. Comput. 2019, 28, 1950123. [Google Scholar] [CrossRef]
- Khalaf, A.; Akcakaya, M. A probabilistic approach for calibration time reduction in hybrid EEG-fTCD brain-computer interfaces. Biomed. Eng. Online 2020, 19, 295–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Q.; Zhu, F.; Qin, J.; Heng, P.-A. Multiclass support matrix machine for single trial EEG classification. Neurocomputing 2018, 275, 869–880. [Google Scholar] [CrossRef]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A review of classification algorithms for EEG-based brain-computer interfaces: A 10 year update. J. Neural Eng. 2018, 15, 031005. [Google Scholar] [CrossRef] [Green Version]
- Ramoser, H.; Muller-Gerking, J.; Pfurtscheller, G. Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE Trans. Rehabil. Eng. 2000, 8, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wen, D.; Li, C.; Zhu, R. Ensemble classifier based on optimized extreme learning machine for motor imagery classification. J. Neural Eng. 2020, 17, 026004. [Google Scholar] [CrossRef]
- Tao, D.; Li, X.; Wu, X.; Maybank, S.J. Geometric Mean for Subspace Selection. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 31, 260–274. [Google Scholar]
- Li, Y.; Wei, Q.; Chen, Y.; Zhou, X. Transfer Learning Based on Hybrid Riemannian and Euclidean Space Data Alignment and Subject Selection in Brain-Computer Interfaces. IEEE Access 2021, 9, 6201–6212. [Google Scholar] [CrossRef]
- Ma, L.; Crawford, M.M.; Zhu, L.; Liu, Y. Centroid and Covariance Alignment-Based Domain Adaptation for Unsupervised Classification of Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2019, 57, 2305–2323. [Google Scholar] [CrossRef]
- Dornhege, G.; Blankertz, B.; Curio, G.; Muller, K.R. Boosting bit rates in noninvasive EEG single-trial classifications by feature combination and multiclass paradigms. IEEE Trans. Bio-Med. Eng. 2004, 51, 993–1002. [Google Scholar] [CrossRef]
- Fatourechi, M.; Bashashati, A.; Ward, R.K.; Birch, G.E. EMG and EOG artifacts in brain computer interface systems: A survey. Clin. Neurophysiol. 2007, 118, 480–494. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Ahn, M.; Ahn, S.; Kwon, M.; Jun, S.C. EEG datasets for motor imagery brain-computer interface. GigaScience 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Padfield, N.; Ren, J.; Qing, C.; Murray, P.; Zhao, H.; Zheng, J. Multi-segment Majority Voting Decision Fusion for MI EEG Brain-Computer Interfacing. Cogn. Comput. 2021, 13, 1484–1495. [Google Scholar] [CrossRef]
- Yu, Z.; Ma, T.; Fang, N.; Wang, H.; Li, Z.; Fan, H. Local temporal common spatial patterns modulated with phase locking value. Biomed. Signal Processing Control 2020, 59, 101882. [Google Scholar] [CrossRef]
- Hou, Y.; Chen, T.; Lun, X.; Wang, F. A novel method for classification of multi-class motor imagery tasks based on feature fusion. Neurosci. Res. 2021, 176, 40–48. [Google Scholar] [CrossRef]
- Gaur, P.; Gupta, H.; Chowdhury, A.; McCreadie, K.; Pachori, R.B.; Wang, H. A Sliding Window Common Spatial Pattern for Enhancing Motor Imagery Classification in EEG-BCI. IEEE Trans. Instrum. Meas. 2021, 70, 1–9. [Google Scholar] [CrossRef]
- Raza, H.; Rathee, D.; Zhou, S.-M.; Cecotti, H.; Prasad, G. Covariate shift estimation based adaptive ensemble learning for handling non-stationarity in motor imagery related EEG-based brain-computer interface. Neurocomputing 2019, 343, 154–166. [Google Scholar] [CrossRef]


| AA | AL | AV | AW | AY | Mean | |
|---|---|---|---|---|---|---|
| CSP | 72.14 | 93.57 | 66.43 | 92.14 | 91.43 | 83.14 |
| CSP-MA | 75 | 96.43 | 68.57 | 89.29 | 92.86 | 84.43 |
| CSP-MA-CA | 50 | 50 | 49.29 | 50 | 50 | 49.86 |
| CSP-MA-CMA | 75 | 96.43 | 70.71 | 92.14 | 92.86 | 85.43 |
| A01 | A02 | A03 | A04 | A05 | A06 | A07 | A08 | A09 | Mean | |
|---|---|---|---|---|---|---|---|---|---|---|
| CSP | 90.97 | 56.94 | 92.36 | 64.58 | 56.25 | 71.53 | 73.61 | 97.22 | 88.89 | 76.93 |
| CSP-MA | 90.97 | 58.33 | 99.31 | 79.17 | 58.33 | 71.53 | 77.08 | 97.22 | 88.19 | 80.02 |
| CSP-MA-CA | 50 | 50 | 50.69 | 50 | 50 | 50 | 50 | 50 | 50 | 50.08 |
| CSP-MA-CMA | 89.58 | 59.03 | 99.31 | 77.08 | 59.03 | 72.22 | 79.86 | 97.22 | 89.58 | 80.32 |
| Methods | Year | AA | AL | AV | AW | AY | Mean |
|---|---|---|---|---|---|---|---|
| MSMV [36] | 2021 | 79.64 | 94.64 | 75 | 78.57 | 94.64 | 84.51 |
| p-LTCSP [37] | 2020 | 77.68 | 100 | 71.94 | 92.41 | 74.21 | 83.25 |
| MFCSP [38] | 2021 | 77.68 | 100 | 73.98 | 84.82 | 88.1 | 84.91 |
| Proposed | 75 | 96.43 | 70.71 | 92.41 | 92.86 | 85.43 |
| Methods | Year | A01 | A02 | A03 | A04 | A05 | A06 | A07 | A08 | A09 | Mean |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SWCSP [39] | 2021 | 86.11 | 64.58 | 95.83 | 64.58 | 68.06 | 68.75 | 81.94 | 97.22 | 90.97 | 79.78 |
| CSCSP [40] | 2019 | 88.89 | 63.89 | 95.14 | 69.44 | 74.31 | 65.97 | 72.92 | 92.36 | 88.19 | 79.01 |
| DACSP [5] | 2021 | 91.67 | 53.47 | 95.84 | 72.92 | 64.58 | 73.61 | 78.47 | 95.83 | 92.37 | 79.48 |
| Proposed | 89.58 | 59.03 | 99.31 | 77.08 | 59.03 | 72.22 | 79.86 | 97.22 | 89.58 | 80.32 |
| Improvement of Accuracies (%) | Overall Mean Difference | Per-Class Covariance Difference | ||
|---|---|---|---|---|
| Dataset 1 | A04 | 12.5 | 0.66 | 0.13 |
| A03 | 6.94 | 0.64 | 0.17 | |
| A07 | 6.25 | 0.55 | 0.07 | |
| A05 | 2.78 | 0.45 | 0.06 | |
| A02 | 2.08 | 0.41 | 0.17 | |
| A06 | 0.69 | 0.40 | 0.22 | |
| A09 | 0.69 | 0.14 | 0.47 | |
| A08 | 0 | 0.26 | 0.09 | |
| A01 | −1.39 | 0.33 | 0.23 | |
| Dataset 2 | AV | 4.30 | 0.43 | 0.36 |
| AA | 2.86 | 0.22 | 0.31 | |
| AL | 2.86 | 0.29 | 0.46 | |
| AY | 1.43 | 0.17 | 0.10 | |
| AW | 0 | 0.14 | 0.29 | |
| Dataset 1 | Dataset 2 | |||
|---|---|---|---|---|
| Training Time (s) | Test Time (s) | Training Time (s) | Test Time (s) | |
| CSP | 0.4523 | 0.0016 | 0.4063 | 0.0016 |
| CSP-MA | 0.5754 | 0.0016 | 0.5111 | 0.0016 |
| CSP-MA-CA | 0.8981 | 0.0021 | 0.7841 | 0.0020 |
| CSP-MA-CMA | 1.038 | 0.0022 | 0.9710 | 0.0022 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Chen, M.; Zhang, L. A Domain Adaptation-Based Method for Classification of Motor Imagery EEG. Mathematics 2022, 10, 1588. https://doi.org/10.3390/math10091588
Li C, Chen M, Zhang L. A Domain Adaptation-Based Method for Classification of Motor Imagery EEG. Mathematics. 2022; 10(9):1588. https://doi.org/10.3390/math10091588
Chicago/Turabian StyleLi, Changsheng, Minyou Chen, and Li Zhang. 2022. "A Domain Adaptation-Based Method for Classification of Motor Imagery EEG" Mathematics 10, no. 9: 1588. https://doi.org/10.3390/math10091588
APA StyleLi, C., Chen, M., & Zhang, L. (2022). A Domain Adaptation-Based Method for Classification of Motor Imagery EEG. Mathematics, 10(9), 1588. https://doi.org/10.3390/math10091588






