1. Introduction
Named Entity Recognition (NER) is one of the core objectives in natural language processing (NLP) [
1,
2], whose purpose is to determine the underlying entities and their categories from the unstructured text [
3]. As an essential component in many downstream NLP tasks, for instance, the correlation extraction [
4], information retrieval [
5], sarcasm detection [
6], and so on, NER is always a hot research direction and attracts much attention in the NLP community. In general, most of the previous works are devoted to the English NER task and achieve promising performances by integrating word-level features [
7]. Compared with English, the East Asian languages (e.g., Chinese) typically lack explicit word boundaries and have complex composition forms, which brings greater challenges for these languages for the development of a competitive NER model. For example, the property of present Chinese state-of-the-art (SOTA) models are much lower than the English SOTAs, with a gap of nearly 10% in terms of F1 metric [
8]. What is more, recent studies pay more attention to the domain-specific NER, e.g., medicine, which is much more complicated and requires external domain expertise [
9,
10].
In particular, in the current work, we pay attention to the research of Chinese Medical Named Entity Recognition (Chinese-MNER), which is considered as a character-level sequence labeling problem, while it is word level for English [
11]. Recently, deep learning methods have been extensively employed in Chinese-MNER [
10,
12,
13,
14,
15] due to their excellent ability in automatically extracting features from massive data. For instance, previous works attempt to leverage the Bi-directional Long Short-Term Memory (BiLSTM) network for acquiring sequence features and achieve comparable results [
16]. In addition, on account of the excellent ability of the pre-trained language models in extracting the contextual features, transformer-based models (e.g., BERT [
17]) are becoming a new paradigm for Chinese-MNER [
15,
18,
19,
20,
21].
Specially, in the medical domain, the external expertise is beneficial in understanding the technical terms and identifying the word boundary, which motivates recent research to incorporate the dictionary knowledge on the basis of traditional BiLSTM-CRF or BERT architecture [
9,
10]. However, the construction of high-quality dictionaries is typically time consuming and labor-intensive, which may also damage the generalization and robustness of NER models [
22]. Compared with the dictionary knowledge, the part-of-speech (POS) tagging features [
23] are now readily available, which does not require additional manpower and material resources. The POS tagging features [
24] can be regarded as supervised signals to guide the model to explicitly identify the word boundary for the reason that it contains potential word segmentation information [
25]. Therefore, we argue that the POS tagging features are more suitable to be used for Chines-MNER than the dictionary knowledge. Last but not least, due to the restrictions of high specialization degree, ethics, and privacy, the annotated Chinese-MNER data are difficult to obtain and usually small in scale, which can result in the over-fitting problem easily when training the Chinese-MNER model [
26].
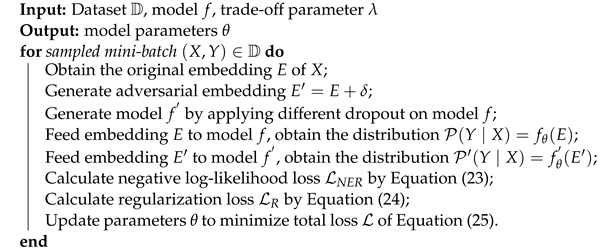
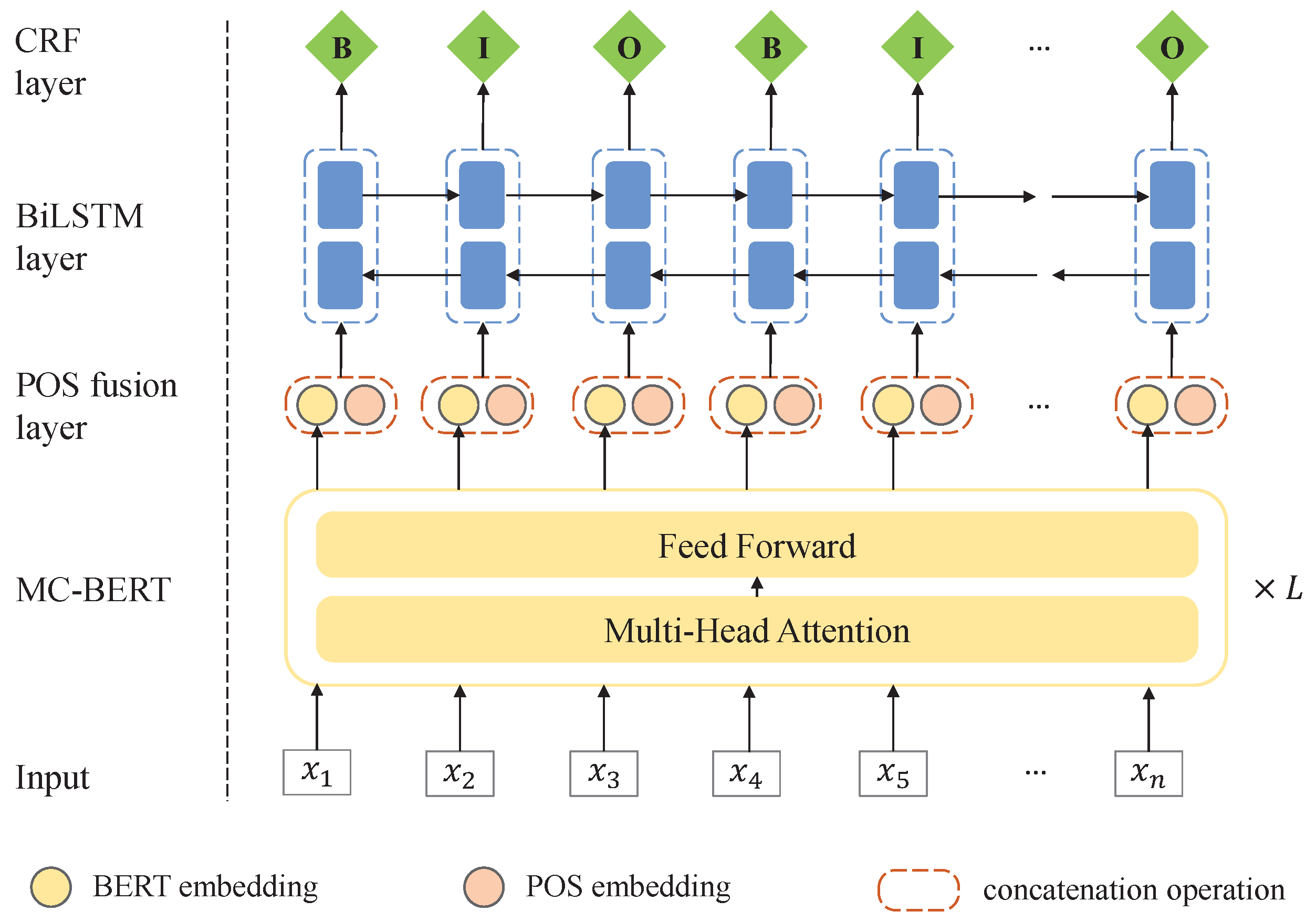
For the sake of alleviating the above issues, we present a
BERT-
BiLSTM-
CRF with
POS and
Regularization (BBCPR) model for Chines-MNER, which leverages a POS fusion layer to incorporate external syntax knowledge as well as introduces a novel
REgularization method with
Adversarial training and
Dropout (READ) to improve the model robustness. In general, our proposal is based on a combined MC-BERT [
27] and BiLSTM-CRF modeling framework. We first utilize the MC-BERT to generate the context representation of each token in the Chinese medical text. Then, we design a POS fusion layer to integrate the part of speech tagging features and send them into a BiLSTM module as inputs. Finally, a standard conditional random fields (CRF) [
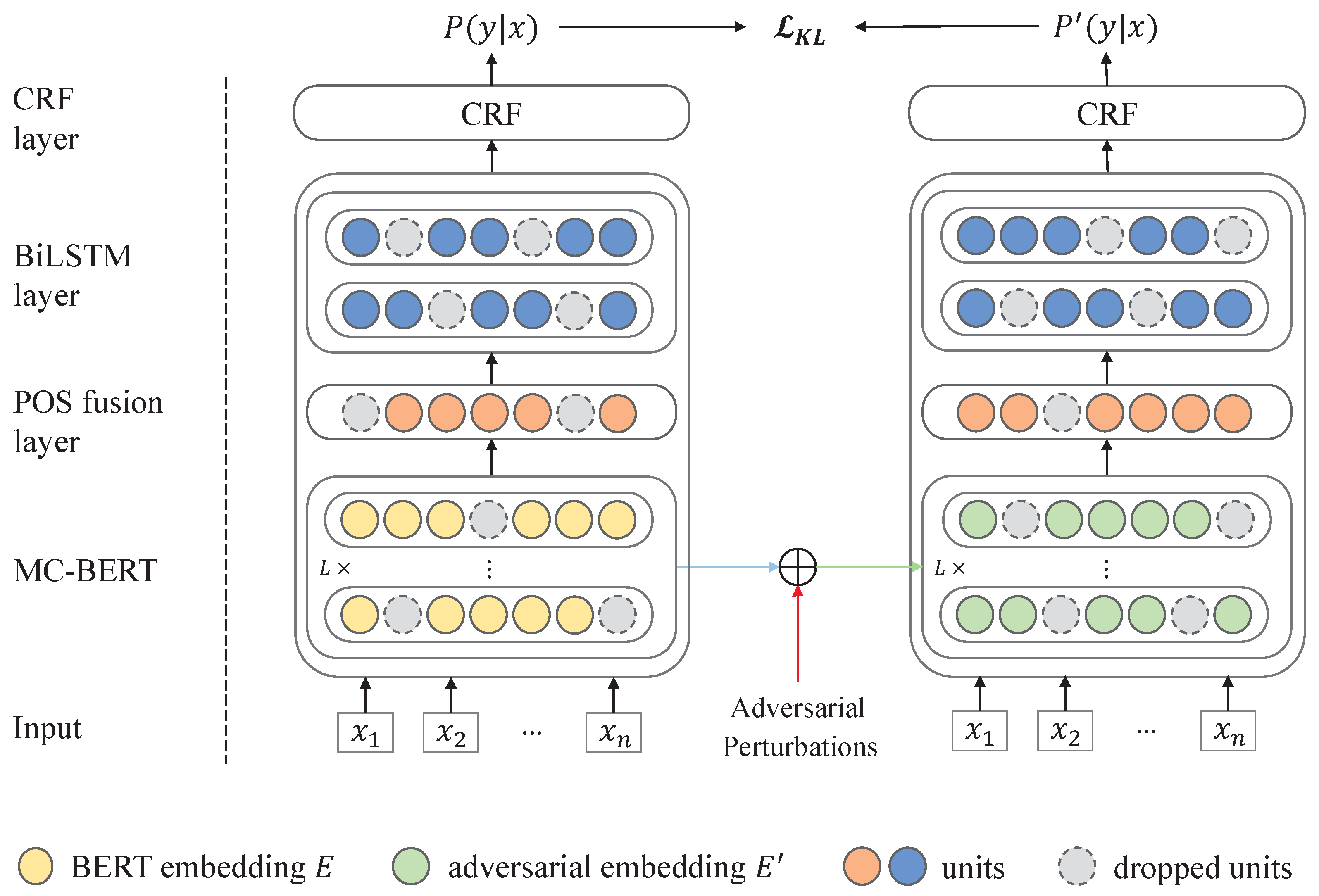
25] layer is employed for decoding the sequence labels. Particularly, besides the traditional learning objective, we introduce an external Kullback–Leibler (KL) divergence loss based on READ. In detail, READ can generate the adversarial word embeddings through a Fast Gradient Method (FGM) as well as a dropout mechanism, which are subsequently put into a Softmax layer for forecasting the label distributions. After that, we can regularize the model predictions through minimizing the bidirectional KL divergence between the adversarial output and original output distributions for the same sample [
28].
For proving the effectiveness of the proposal, we implement comprehensive experiments on two public data sets from the Chinese Biomedical Language Understanding Evaluation (ChineseBLUE) benchmark [
27], i.e., cMedQANER and cEHRNER. The experimental results suggest that our presented BBCPR model is superior to the SOTA baseline, and the overall improvement for the F1 score on cEHRNER and cMedQANER datasets is
and
, respectively. Furthermore, the effectiveness of our designed modules is verified by the ablation studies.
In summary, the major contributions of this research can be concluded as below:
We design a POS fusion layer that can explicitly learn the word boundary feature for the task of Chinese-MNER by incorporating the POS tagging features.
We put forward a novel regularization approach READ to alleviate the over-fitting problem for Chinese-MNER and enhance the robustness of the model on small data.
We conduct comprehensive experiments on two public datasets. The performance comparisons over several competitive baselines indicate the superiority of our proposal.
5. Results and Discussion
In this section, we first discuss the comparison of overall performance between BBCPR and the competitive baseline models (see
Section 5.1). Next, we analyze the effectiveness of each component we propose in BBCPR (see
Section 5.2). Furthermore, we explore the influence brought by different pre-trained language models (see
Section 5.3) and different hyper-parameters (see
Section 5.4). Finally, we present a case study to clearly demonstrate the superiority of BBCPR (See
Section 5.5).
5.1. Overall Evaluation
To answer
RQ1, we check the overall entity recognition performance of the baselines and our proposal in terms of all evaluation metrics on the cMedQANER and cEHRNER datasets.
Table 6 shows the detailed outcomes between the models discussed.
First, we focus on the property of the baselines. In accordance with
Table 6, compared to the traditional statistical learning model, i.e., HMM, deep learning based-models report obvious improvements in terms of all metrics. For example, BiLSTM-CRF beats HMM by 23.95% and 44.50% for the score of F1 on the cMedQANER and cEHRNER datasets, respectively, which demonstrates that the capability of learning context features is essential for the Chinese-MNER task. Specially, we can obverse that those models using MC-BERT as an encoder show nearly 7.08–8.87% and 4.59–5.31% improvements for the metric F1 against BiLSTM-CRF on the cMedQANER and cEHRNER dataset, respectively, indicating an impressive superiority of BERT in representation learning. In addition, equipping the model with additional CRF layer yields better performance than the original ones, which may be attributed to the CRF being able to well model the dependencies between tag sequences.
Next, we focus on the comparison between the baselines and our proposal. As revealed in
Table 6, BBCPR achieves the best performance among all discussed models on both the cMedQANER and cEHRNER datasets. Specially, it can be found that our approach achieves the SOTA performance with the improvements of 2.44% and 2.87% in terms of F1 score against the best baseline MC-BERT-BiLSTM-CRF on two datasets, respectively. There is a similar phenomenon in terms of P and R. For the metric P, our proposal model beats the best baseline by 2.50% and 4.05% on the cMedQANER and cEHRNER dataset, respectively. For the metric R, our method shows 2.36% and 1.68% improvements over the best baseline on the cMedQANER and cEHRNER dataset, respectively. The improvements acquired from BBCPR can be explained by the fact that using POS tagging features, which imply potential word segmentation, can provide an extra supervision signal to distinguish the edges between ordinary words and the medical entities. In addition, the adversarial samples generated by FGM can enhance the model’s robustness.
5.2. Ablation Study
To answer
RQ2, we conduct comprehensive ablation studies on cMedQANER and cEHRNER datasets to verify the effectiveness of each key module of BBCPR. The detailed results of ablation studies are presented in
Table 7. It can be observed that when any certain module is taken out, the model performances decrease obviously in terms of almost all metrics, which verifies the effectiveness of our proposed modules in BBCPR. Specifically, for the metric F1 and R, removing the POS fusion layer and the DP mechanism (see Row 6,
Table 7) results in the biggest drop of model performance. Specifically, on both of the cMedQANER and cEHRNER datasets, model performances show a 1.54% and 2.11% decrease in terms of F1 and a 1.68% and 1.36% decrease in terms of R. This observation indicates that incorporating POS tagging features into the neural networks can evidently enhance the superiority of the Chinese-MNER model, for which POS tagging features add extra potential entities’ boundary information. At the same time, the DP mechanism can help alleviate the over-fitting problem of the model and thus reduces the prediction error. As for the metric P, the removal of the READ (see Row 2,
Table 7) declines the model performance most, with a 1.55% and 3.29% decrease on the cMedQANER and cEHRNER dataset, respectively. This can be due to the fact that the READ module amplifies the variability of the original samples, thus enhancing the learning ability of the model.
In addition, we can observe that removing AP (see Row 4,
Table 7) or removing DP (see Row 3,
Table 7) leads to a 0.70% and 0.82% decrease for the score of F1 on the cMedQANER dataset, respectively. Similar results can be found on the cEHRNER dataset, where the model reveals a 0.58% and 0.96% drop. The reason why DP performs better than AP may be that it is applied on the whole model, while AP only works on the BERT-embedding layer. In addition, it is worth mentioning that using either the AP or DP alone is not as effective as the combination of the two, i.e., the READ. For example, without the READ (see Row 2,
Table 7), the decreases in F1 score are 1.29% and 1.82% on two datasets, respectively. The reason may be that each of the perturbations is relatively simple; employing only one mechanism can only introduce a small perturbation. Meanwhile, diverse perturbations can increase the dissimilarity of the representation from the same sample.
Moreover, in the condition of using DP by default (See Row 7,
Table 7), adding AP (see Row 5,
Table 7) results in a 0.32% and 0.70% increase in terms of F1 on the cMedQANER and cEHRNER datasets, respectively. Similarly, when adding POS (see Row 4,
Table 7), the score of F1 increases by 0.41% and 0.90%, respectively. Compared with AP, POS has a greater impact on the neural network that has a dropout by default. We attribute this phenomenon to the fact that POS can directly increase the features of entities and thus bring useful information to the model. However, AP utilizes an indirect way to enhance the learning ability of the model by adding perturbations. Under the condition that uses DP by default, the F1 score drops by 1.10% and 1.48% on two datasets when removing POS and AP, which indicates that the diversity added by both POS and AP has a positive impact on the model performance.
5.3. Influence of Pre-Trained Language Models
To answer RQ3, we further conduct comparative experiments to analyze the effect brought by different pre-trained language models, such as BERT, BERT-WWM, RoBERTa, MacBERT, and MC-BERT. First, BERT is able to obtain more informative contextual representation through employing the masked language model and next sentence to forecast training targets. Furthermore, BERT-WWM employs a whole word masking strategy for the Chinese corpus. For the RoBERTa, it robustly optimizes the BERT pre-training method by four simple and effective modifications. MacBERT masks the words with their similar words in the Chinese corpus, while MC-BERT is further trained on the Chinese biomedical corpus based on BERT.
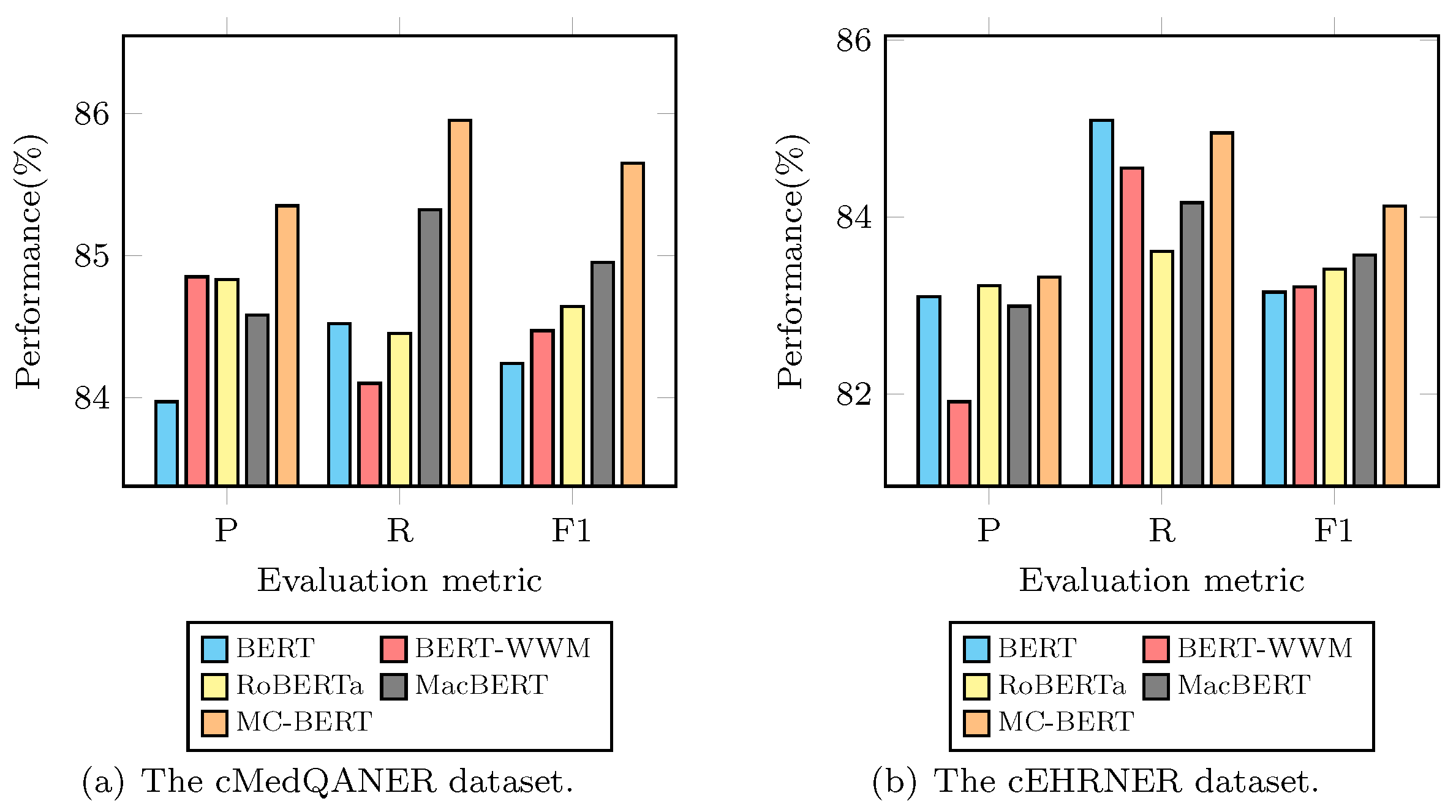
As shown in
Figure 3, MC-BERT significantly outperforms other pre-trained models in terms of all evaluation metrics. In particular, as reflected in
Figure 3a, it can be observed that MC-BERT shows nearly 0.83–1.67%, 0.59–1.65% and 0.74–2.21% improvements in terms of F1, P, and R score than other pre-trained language models on the cMedQANER dataset.
Figure 3b indicates that MC-BERT increases nearly 0.66–1.17%, 0.12–2.48%, and −0.16–1.60% of F1, P, and R scores compared to other pre-trained language models on the cEHRNER dataset. The possible reason may be due to the fact that MC-BERT adapts the whole entity masking strategy and the whole span masking strategy to inject medical domain knowledge for Chinese biomedical text, which can help generate better contextual representation for the Chinese-MNER task. Accordingly, we choose the MC-BERT model [
27] as the contextual embedding component in the following experiments.
5.4. Analysis on Different Hyperparameters
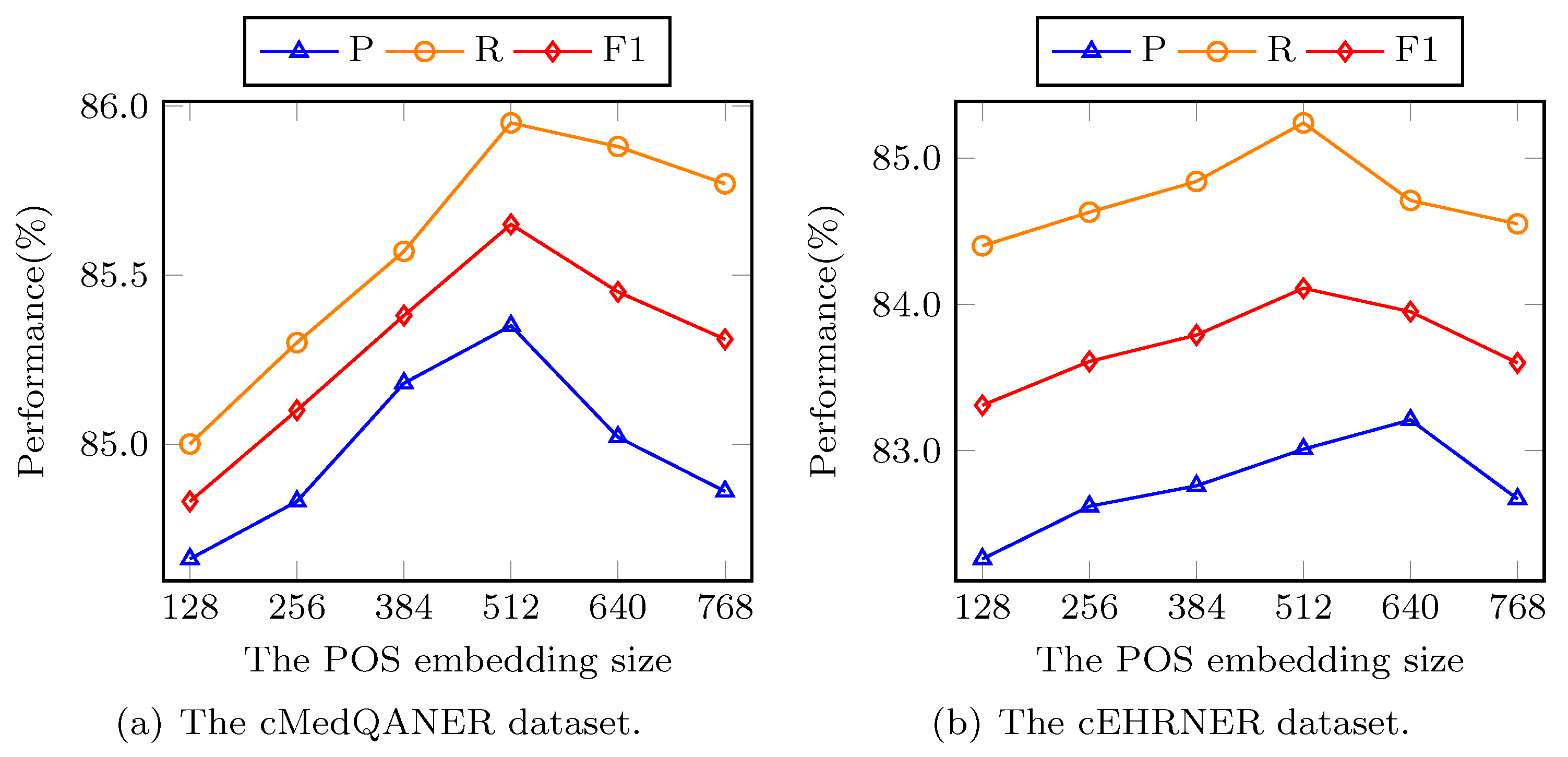
To answer
RQ4, we conduct the experiments on cMedQANER and cEHRNER datasets to explore the BBCPR property under different hyperparameters, i.e., POS embedding size, and trade-off parameter
. For the POS embedding, the size of it is set to 128, 256, 384, 512, 640, and 768 in our experiments, respectively. As displayed in
Figure 4a, the property of our model initially grows when the size of the POS embedding increases and the difference between the best and worst performance can be as high as 0.97%, 0.82%, and 1.12% in the terms of F1, P, and R, respectively on the cMedQANER dataset.
Figure 4b shows that there are similar outcomes on the cEHRNER dataset. This is probably explained through the fact that adding the neural network size can enhance the model complexity to obtain more powerful representation capability. However, when the size increases further, worse performance is achieved due to model over-fitting. As shown in
Figure 4, when the POS embedding size is 512, all the metrics achieve their best performance. Therefore, we choose 512 as the POS embedding size in our experiments.
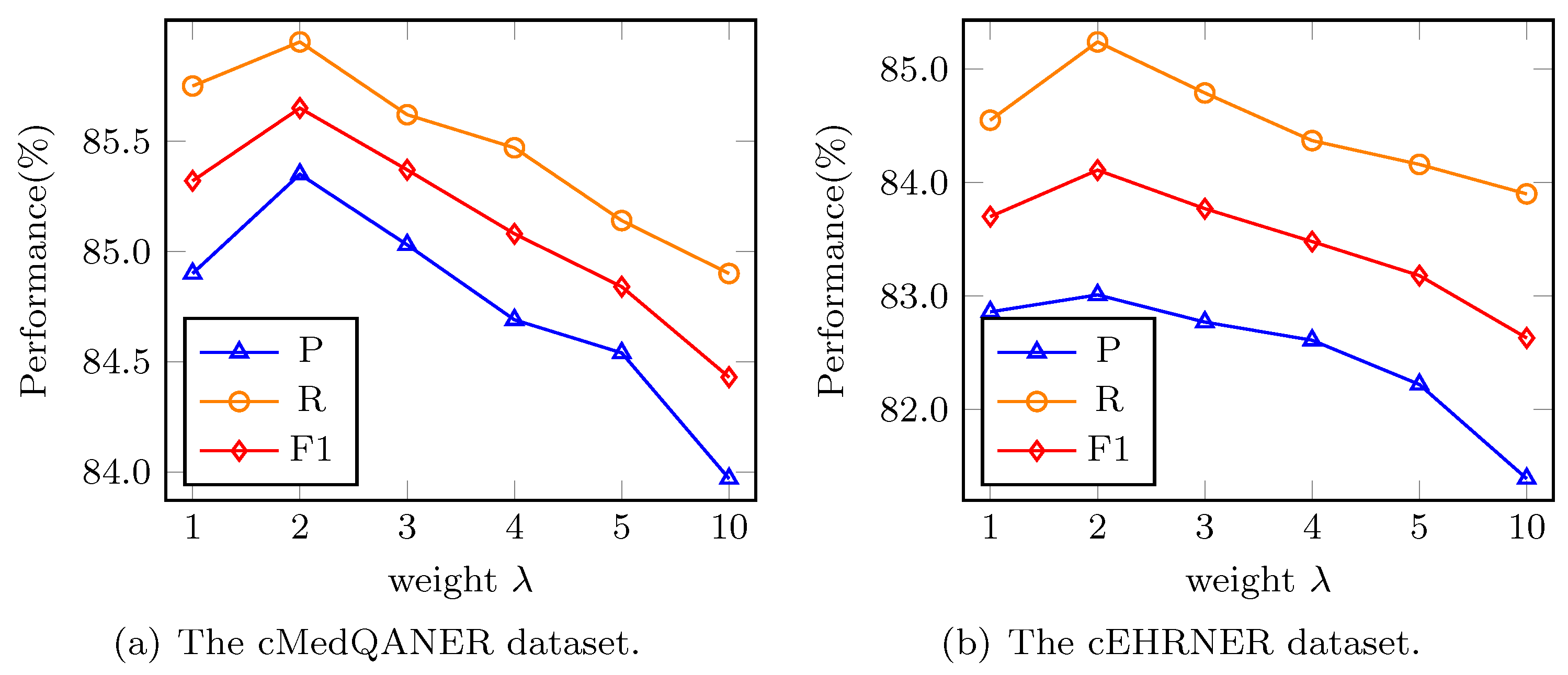
Further, we analyze the effect resulted from the trade-off parameter
. We vary the
in
and conduct extensive experiments. According to
Figure 5a,b, either too large or too small
will make our model perform poorly. When
, the model realizes the optimum property, and subsequently, our model performs worse and worse as
increases. Specifically, the performances of the model on the cMedQANER dataset have drops by 0.33–1.45%, as shown in
Figure 5a and 0.41–1.80% on the cEHRNER dataset, as shown in
Figure 5b. In terms of P and R, there are floats of 0.15–1.64% and 0.24–1.24% on the cMedQANER dataset, while they are 0.19–1.99% and 0.53–1.60% on the cEHRNER dataset. When the
is at 2, all the metrics achieve their best performance. Therefore, we select 2 as the regularization loss weight for our proposed BBCPR model.
5.5. Case Study
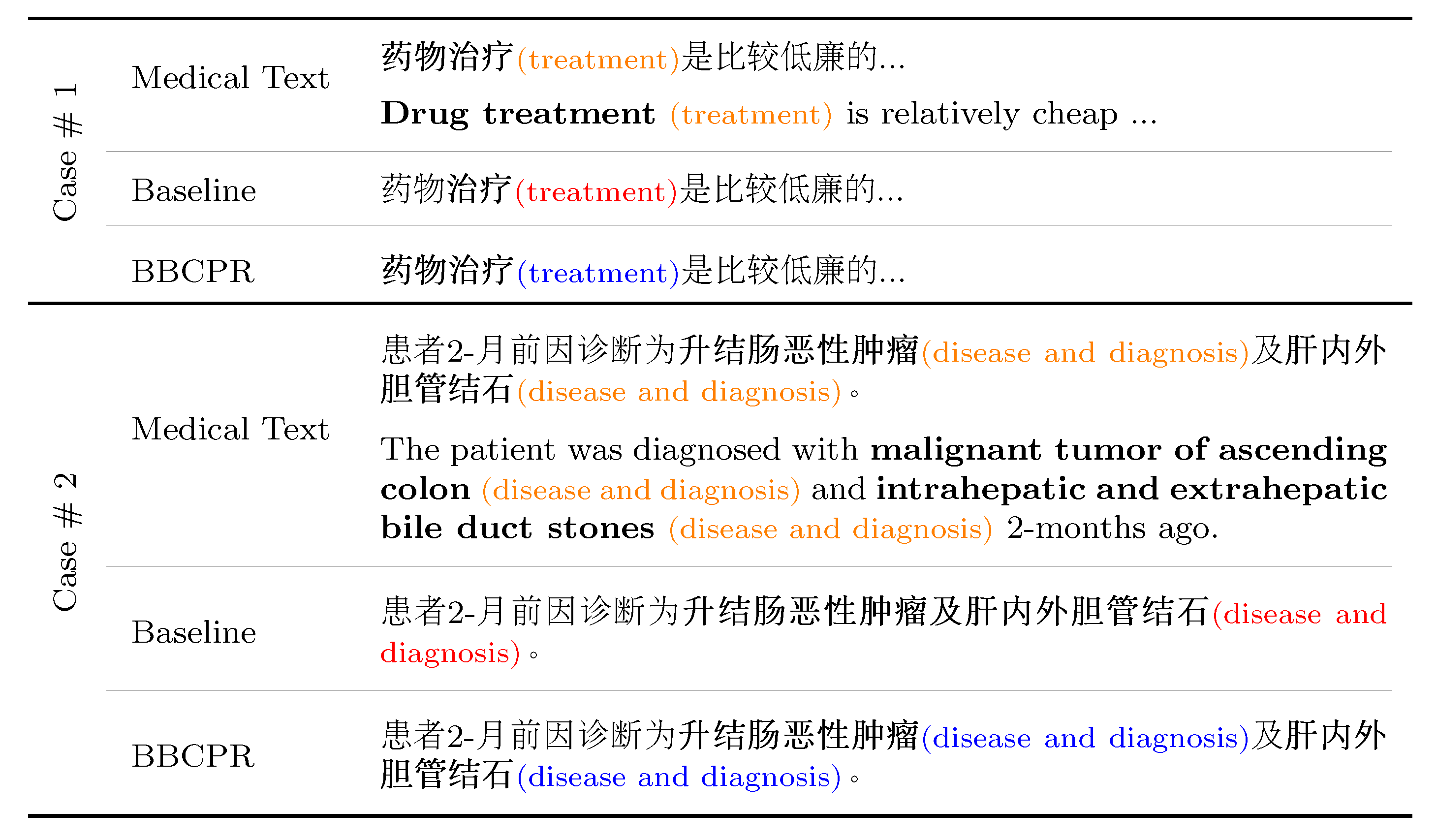
In the current section, we conduct a case study to demonstrate the superiority of our model against baselines. In particular, we compared the predictive results of our proposal and MC-BERT-BiLSTM-CRF on two cases from the cMedQANER and cEHRNER datasets, respectively. The detailed input medical text and the corresponding predictive results of the two cases are presented in
Figure 6.
UTF8gbsn As shown in
Figure 6, in Case #1, the baseline model fails to identify “药物治疗 (
drug treatment)” as a whole unit. However, our model could completely and correctly recognize the entity “药物治疗 (
drug treatment)” as the four tokens have the same part-of-speech (i.e., noun). Likewise, in Case #2, we can see that the baseline MC-BERT-BiLSTM-CRF identifies “升结肠恶性肿瘤及肝内外胆管结石 (
malignant tumor of ascending colon and intrahepatic and extrahepatic bile duct stones)” as an independent entity. However, the token “及 (
and)” is a conjunction, while “瘤 (
tumor)” and “肝 (
liver)” are nouns. On the contrary, our model could accurately recognize two entities “升结肠恶性肿瘤 (
malignant tumor of ascending colon)” and “肝内外胆管结石 (
intrahepatic and extrahepatic bile duct stones)” through identifying obvious entity boundaries before and after “及 (
and)”. Overall, the above cases demonstrate that BBCPR can explicitly learn word boundary information by introducing the POS tagging features, which is conducive to enhance the entity recognition accuracy.
6. Conclusions and Future Work
In our work, we propose a model named BBCPR for improving the performance of the Chinese-MNER task, which leverages a POS fusion layer to explicitly learn word boundary information by incorporating external syntax knowledge. What is more, we also design a novel regularization method READ to deal with the over-fitting problem and improve the model robustness. In detail, READ regularizes the predictions of the two sub-models through minimizing the bidirectional KL-divergence between the adversarial output and original output distributions for the same sample. Comprehensive experiments conducted on two benchmark datasets confirm the advantage of our proposal for the Chinese-MNER task. In addition, an ablation study proves that the POS fusion layer and READ can effectively improve the model performance.
For future research, we want to explore how to obtain more features for entities by introducing the contrastive learning [
41], which can pull the same type of entities closer and push apart different types of entities [
52]. Furthermore, we have interests in verifying the effectiveness of our proposal in other domains, e.g., financial domain, legal domain. Finally, mining more potential supervisory signals from the unlabeled samples and then training the model in an unsupervised setting may also be a promising direction.