Survival Risk Prediction of Esophageal Cancer Based on the Kohonen Network Clustering Algorithm and Kernel Extreme Learning Machine
Abstract
:1. Introduction
- The two risk levels of patients with esophageal cancer were divided by five-year survival [21,22]. In this study, the influences of different activation functions on the KELM model were studied, the results are verified theoretically and experimentally, the problem of activation function selection in the survival risk prediction model of esophageal cancer based on the KELM was solved.
2. Materials and Methods
2.1. Data Sources
2.2. Kohonen Clustering Network
- The network consists of two layers, namely an input layer and an output layer. The output layer is also called the competition layer and does not include the hidden layer.
- Each input node in the input layer is wholly connected to the output node.
- The output nodes are distributed in a two-dimensional structure, and there are lateral connections between the nodes.
- Step 1: Data normalization.
- Step 2: Determine the initial center point of the cluster.
- Step 3: Calculate the distance [29].
- Step 4: Adjust the position.
- Step 5: Determine whether the conditions for the end of the iteration are met.
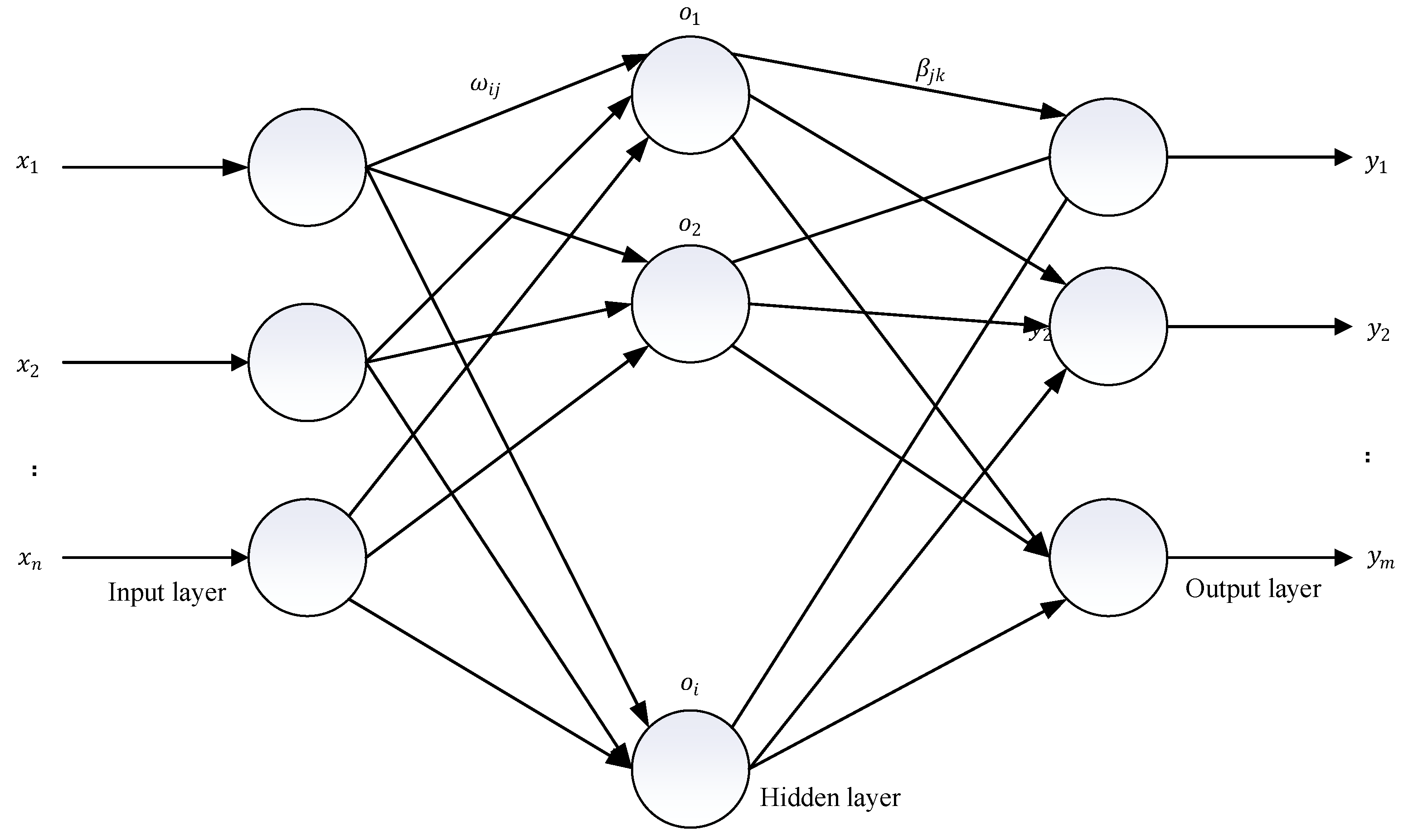
2.3. Kernel Extreme Learning Machine (KELM)
3. Optimized Kernel Extreme Learning Machine
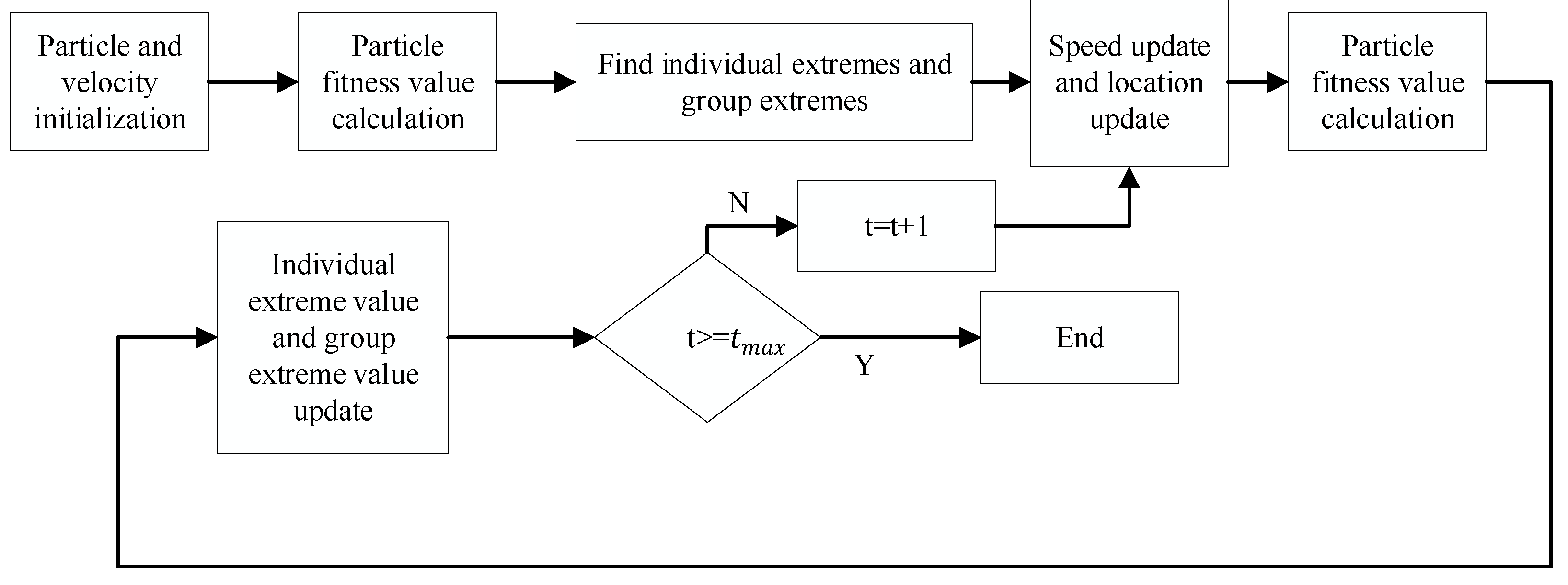
3.1. Particle Swarm Optimization to Optimize the KELM
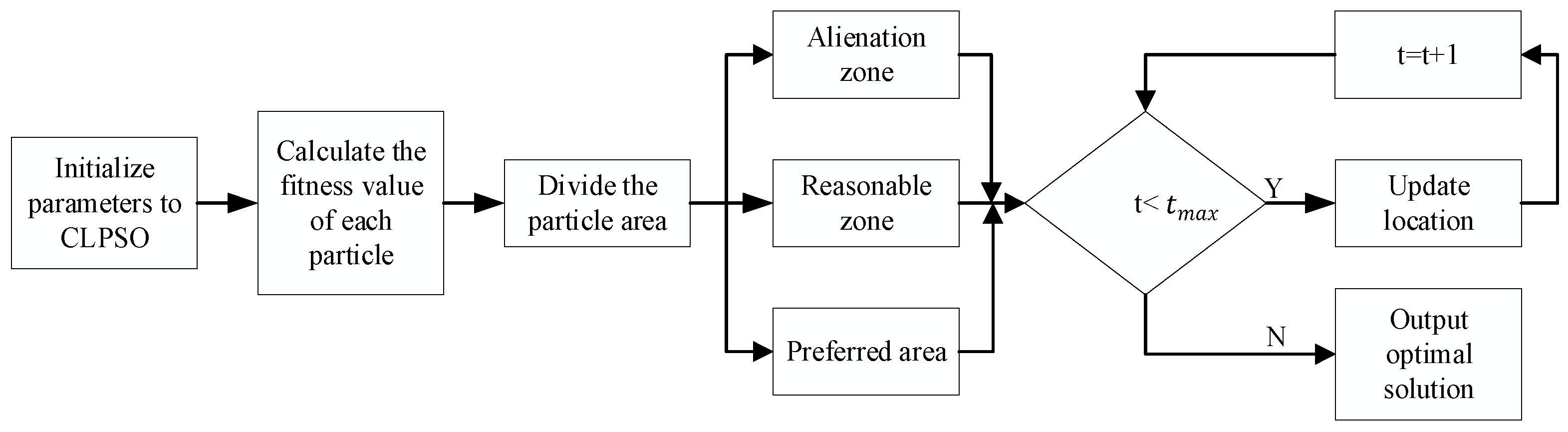
3.2. Particle Swarm Optimization Algorithm Based on Competitive Learning (CLPSP) to Optimize the KELM
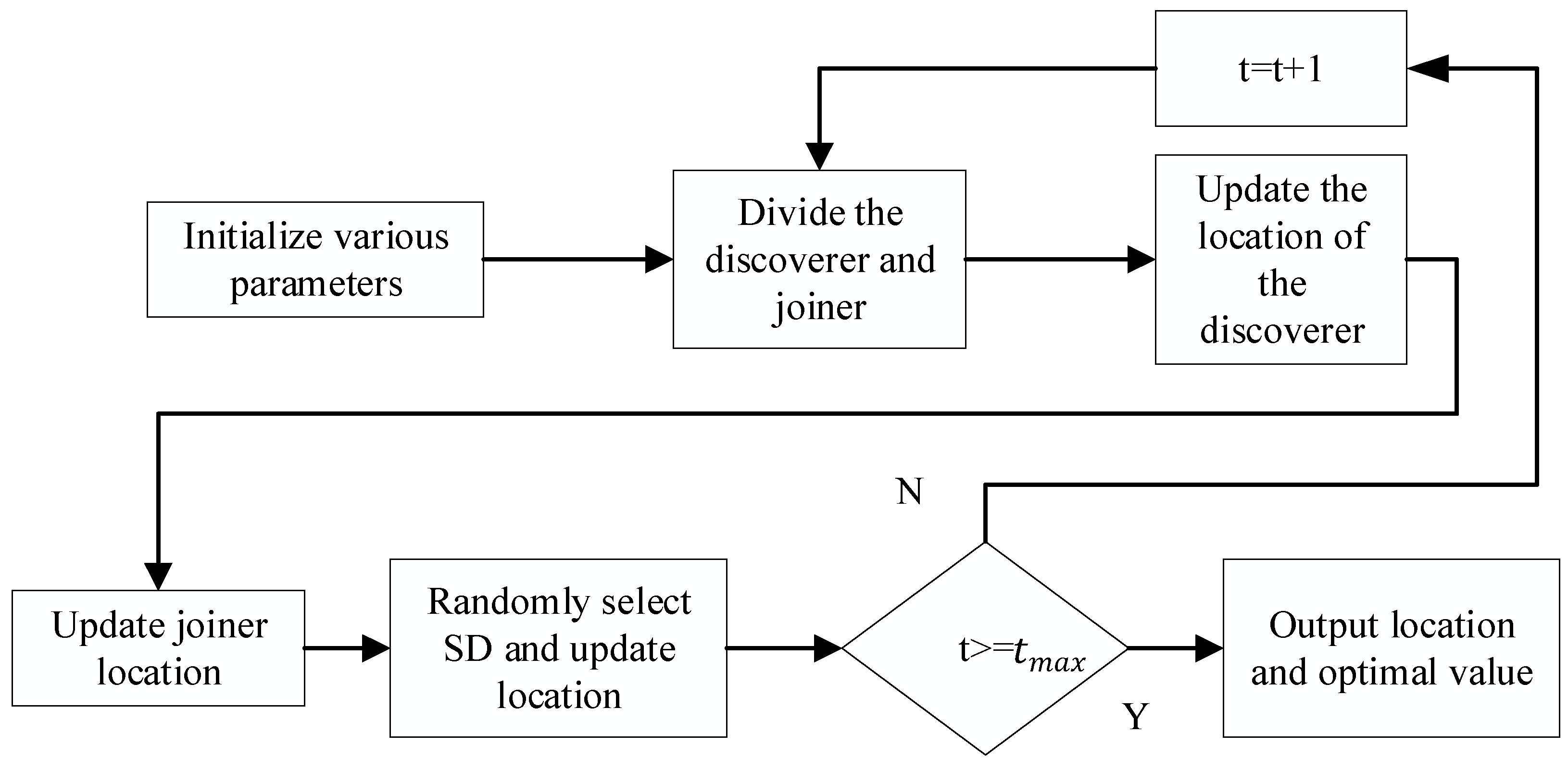
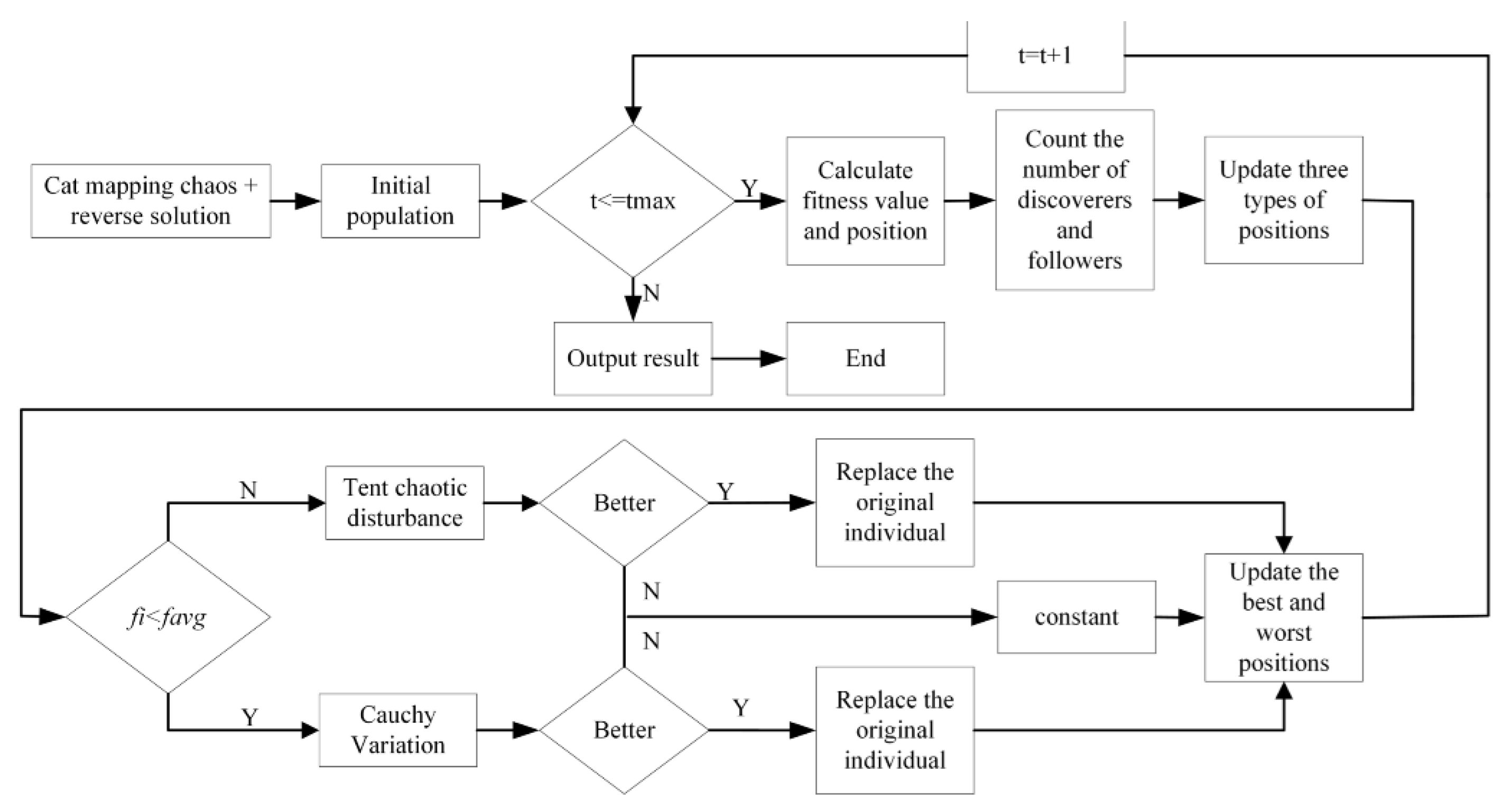
3.3. Sparrow Search Algorithm (SSA) to Optimize the KELM
- Under normal circumstances, the discoverer has a relatively high energy reserve, and the energy reserve corresponds to adaptability.
- The identity between the discoverer and the joiner changes dynamically, and their ratio remains unchanged.
- The position of the joiner is proportional to the energy, the lower the power, the more likely it is to fly to other places for food.
- Joiners will always find discoverers who provide good food and fight with them.
- After the alarm value is greater than the safe value, the discoverer will lead the sparrow population into the safe area.
- When the entire population moves, the sparrows at the edge of the people will quickly move to a safe place. In contrast, the sparrows inside the population will randomly move to the surrounding sparrows. The location update of the discoverer
3.4. Adaptive Mutation Sparrow Search Algorithm (AMSSA) to Optimize the KELM
4. Result
4.1. Data Clustering
4.2. Choice of Kernel Function of the KELM
4.3. Optimization Algorithm Optimization Results
4.4. Results Predicted by Different Models
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anzolin, A.; Isenburg, K.; Toppi, A.; Yucel, M.; Ellingsen, D.; Gerber, J.; Ciaramidaro, A.; Astolfi, L.; Kaptchuk, T.; Napadow, V. Patient-Clinician Brain Response During Clinical Encounter and Pain Treatment. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; p. 19964246. [Google Scholar]
- Qiu, H.; Cao, S.; Xu, R. Cancer incidence, mortality, and burden in China: A time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun. 2021, 41, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Anji, R.; Soni, B.; Sudheer, R. Breast cancer detection by leveraging Machine Learning. ICT Express 2020, 6, 320–324. [Google Scholar]
- Chen, D.; Fan, N.; Jun, X.; Wang, W.; Qi, R.; Jia, Y. Multiple primary malignancies for squamous cell carcinoma and adenocarcinoma of the esophagus. J. Thorac. Dis. 2019, 11, 3292–3301. [Google Scholar] [CrossRef] [PubMed]
- Alberti, M.; Ruiz, J.; Fernandez, M.; Fassola, L.; Caro, F.; Roldan, I.; Paulin, F. Comparative survival analysis between idiopathic pulmonary fibrosis and chronic hypersensitivity pneumonitis. Pulmonology 2020, 26, 3–9. [Google Scholar] [CrossRef]
- Yu, X.; Wang, T.; Huang, S. How can gene-expression information improve prognostic prediction in TCGA cancers: An empirical comparison study on regularization and mixed Cox models. Front. Genet. 2020, 11, 920. [Google Scholar] [CrossRef]
- Emura, T.; Hsu, W.; Chou, W. A survival tree based on stabilized score tests for high-dimensional covariates. J. Appl. Stat. 2021, 1–27. [Google Scholar] [CrossRef]
- Gaudart, J.; Giusiano, B.; Huiart, L. Comparison of the performance of multi-layer perceptron and linear regression for epidemiological data. Comput. Stat. Data Anal. 2004, 44, 547–570. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Ding, X.; Wang, F. Determination of polynomial degree in the regression of drug combinations. IEEE/CAA J. Autom. Sin. 2017, 4, 41–47. [Google Scholar] [CrossRef]
- Witten, D.; Tibshirani, R. Survival analysis with high-dimensional covariates. Stat. Methods Med. Res. 2010, 19, 29–51. [Google Scholar] [CrossRef]
- Van Wieringen, W.; Kun, D.; Hampel, R.; Boulesteix, A. Survival prediction using gene expression data: A review and comparison. Comput. Stat. Data Anal. 2009, 53, 1590–1603. [Google Scholar] [CrossRef]
- Emura, T.; Matsui, S.; Chen, H. compound.Cox: Univariate feature selection and compound covariate for predicting survival. Comput. Methods Programs Biomed. 2019, 168, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.; Everson, M.; Haidry, R. Esophageal squamous dysplasia and cancer: Is artificial intelligence our best weapon? Baillière’s Best Pract. Res. Clin. Gastroenterol. 2020, 52, 101723. [Google Scholar] [CrossRef] [PubMed]
- Ting, W.; Chang, H.; Chang, C.; Lu, C. Developing a novel machine learning-based classification scheme for predicting SPCs in colorectal cancer survivors. Appl. Sci. 2020, 10, 1355. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.R.; Liu, Y.A. Breast cancer diagnosis and prediction model based on improved PSO-SVM based on gray relational analysis. In Proceedings of the International Symposium on Distributed Computing and Applications to Business Engineering & Science, Xuzhou, China, 16–19 October 2020; pp. 231–234. [Google Scholar]
- Li, N.; Luo, P.; Li, C.; Chen, Z. Analysis of related factors of radiation pneumonia caused by precise radiotherapy of esophageal cancer based on random forest algorithm. Math. Biosci. Eng. 2020, 18, 4477–4490. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, A.; Singh, A. eBreCaP: Extreme learning-based model for breast cancer survival prediction. IET Syst. Biol. 2020, 14, 160–169. [Google Scholar] [CrossRef]
- Kim, M.; Oh, I.; Ahn, J. An improved method for prediction of cancer prognosis by network learning. Genes 2018, 9, 478. [Google Scholar] [CrossRef] [Green Version]
- Hedjam, R.; Shaikh, A.; Luo, Z. Ensemble clustering using extended fuzzy k-means for cancer data analysis. Expert Syst. Appl. 2021, 172, 114622. [Google Scholar]
- Qadire, M. Symptom clusters predictive of quality of life among jordanian women with breast cancer. Semin. Oncol. Nurs. 2021, 37, 151144. [Google Scholar] [CrossRef]
- Hassen, H.; Teka, M.; Addissie, A. Survival status of esophageal cancer patients and its determinants in ethiopia: A facility based retrospective cohort study. Front. Oncol. 2021, 10, 3330. [Google Scholar] [CrossRef]
- Nguyen, T.; Bartscht, T.; Schild, S.E.; Rades, D. A scoring tool to estimate the survival of elderly patients with brain metastases from esophageal cancer receiving ehole-brain irradiation. Anticancer. Res. 2020, 40, 1661–1664. [Google Scholar] [CrossRef]
- Guo, H.; Liu, H.; Chen, J. Data mining and risk prediction based on apriori improved algorithm for lung cancer. J. Signal Process. Syst. 2021, 93, 795–809. [Google Scholar] [CrossRef]
- Raja, S.S.; Kunthavai, A. Hubness weighted svm ensemble for prediction of breast cancer subtypes. Technol. Health Care Off. J. Eur. Soc. Eng. Med. 2021, 1–14. [Google Scholar] [CrossRef]
- Barletta, V.; Caivano, D.; Nannavecchia, A.; Scalera, M. A kohonen SOM architecture for intrusion detection on in-vehicle communication networks. Appl. Sci. 2020, 10, 5062. [Google Scholar] [CrossRef]
- Barletta, V.; Caivano, D.; Nannavecchia, A.; Scalera, M. Intrusion detection for in-vehicle communication networks: An unsupervised Kohonen SOM approach. Future Internet 2020, 12, 119. [Google Scholar] [CrossRef]
- Chen, B.; Zhu, G.; Ji, M.; Yu, Y.; Zhao, J.; Liu, W. Air quality prediction based on Kohonen clustering and Relief feature selection. CMC-Comput. Mater. Cintinua 2020, 64, 1039–1049. [Google Scholar]
- Gupta, M.; Chandra, P. Effects of similarity/distance metrics on k-means algorithm with respect to its applications in IoT and multimedia: A review. Multimed. Tools Appl. 2021, 2021, 1–26. [Google Scholar] [CrossRef]
- Guo, A.; Jiang, A.; Lin, J.; Li, X. Data mining algorithms for bridge health monitoring: Kohonen clustering and LSTM prediction approaches. J. Supercomput. 2020, 76, 932–947. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Zhang, L.; Li, Z. Mining event logs for knowledge discovery based on adaptive efficient fuzzy Kohonen clustering network. Knowl. Based Syst. 2020, 209, 106482. [Google Scholar] [CrossRef]
- Sun, J.; Han, J.; Wang, Y.; Liu, P. Memristor-based neural network circuit of emotion congruent memory with mental fatigue and emotion inhibition. IEEE Trans. Biomed. Circuits Syst. 2021, 15, 606–616. [Google Scholar] [CrossRef]
- Fan, J.; Sun, H.; Su, Y.; Huang, J. MuSpel-Fi: Multipath subspace projection and ELM-based fingerprint localization. IEEE Signal Processing Lett. 2022, 29, 329–333. [Google Scholar] [CrossRef]
- Yahia, S.; Said, S.; Zaied, M. A novel classification approach based on extreme learning machine and wavelet neural networks. Multimed. Tools Appl. 2020, 79, 13869–13890. [Google Scholar] [CrossRef]
- Sun, J.; Han, J.; Liu, P.; Wang, Y. Memristor-based neural network circuit of pavlov associative memory with dual mode switching. AEU-Int. J. Electron. Commun. 2021, 129, 153552. [Google Scholar] [CrossRef]
- Zhang, H.; Nguyen, H.; Bui, X.; Pradhan, B.; Mai, N.; Vu, D. Proposing two novel hybrid intelligence models for forecasting copper price based on extreme learning machine and meta-heuristic algorithms. Resour. Policy 2021, 73, 102195. [Google Scholar] [CrossRef]
- Lahoura, V.; Singh, H.; Aggarwal, A.; Sharma, B.; Cengiz, K. Cloud computing-based framework for breast cancer diagnosis using extreme learning machine. Diagnostics 2021, 11, 241. [Google Scholar] [CrossRef]
- Dereli, S. A new modified grey wolf optimization algorithm proposal for a fundamental engineering problem in robotics. Neural Comput. Appl. 2021, 33, 14119–14131. [Google Scholar] [CrossRef]
- Mohanty, F.; Rup, S.; Dash, B. Automated diagnosis of breast cancer using parameter optimized kernel extreme learning machine. Biomed. Signal Process. Control. 2020, 62, 102108. [Google Scholar] [CrossRef]
- Lu, H.; Du, B.; Liu, J.; Xia, H.; Yeap, W. A kernel extreme learning machine algorithm based on improved particle swam optimization. Memetic Comput. 2017, 9, 121–128. [Google Scholar] [CrossRef]
- Nguyen, T.; Nguyen, P.; Tran, Q.; Vo, N. Cancer classification from microarray data for genomic disorder research using optimal discriminant independent component analysis and kernel extreme learning machine. Int. J. Numer. Methods Biomed. Eng. 2020, 36, e3372. [Google Scholar] [CrossRef]
- Wang, W.; Yang, S.; Chen, G. Blood glucose concentration prediction based on VMD-KELM-AdaBoost. Med. Bioligical Eng. Comput. 2021, 59, 2219–2235. [Google Scholar]
- Liang, R.; Chen, Y.; Zhu, R. A novel fault diagnosis method based on the KELM optimized by whale optimization algorithm. Machines 2022, 10, 93. [Google Scholar] [CrossRef]
- Parida, N.; Mishra, D.; Das, K.; Rout, N. Development and performance evaluation of hybrid KELM models for forecasting of agro-commodity price. Evol. Intell. 2021, 14, 529–544. [Google Scholar] [CrossRef]
- Chen, P.; Zhao, X.; Zhu, Q. A novel classification method based on ICGOA-KELM for fault diagnosis of rolling bearing. Appl. Intell. 2020, 50, 2833–2847. [Google Scholar] [CrossRef]
- Hamza, M.; Yap, H.; Choudhury, I. Recent advances on the use of meta-heuristic optimization algorithms to optimize the type-2 fuzzy logic systems in intelligent control. Neural Comput. Appl. 2017, 28, 979–999. [Google Scholar] [CrossRef]
- Yu, V.; Jewpanya, P.; Redi, A.; Tsao, Y. Adaptive neighborhood simulated annealing for the heterogeneous fleet vehicle routing problem with multiple cross-docks. Comput. Oper. Res. 2021, 129, 105205. [Google Scholar] [CrossRef]
- Glover, F.; Lu, Z. Focal distance tabu search. Sci. China Inf. Sci. 2021, 64, 150101. [Google Scholar] [CrossRef]
- Misevicius, A.; Verene, D. A hybrid genetic-hierarchical algorithm for the quadratic assignment problem. Entropy 2021, 23, 108. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, X.; Gao, L.; Gupta, S.M. A genetic simulated annealing algorithm for parallel partial disassembly line balancing problem. Appl. Soft Comput. 2021, 107, 107404. [Google Scholar] [CrossRef]
- Li, G.; Li, J. An improved tabu search algorithm for the stochastic vehicle routing problem with soft time windows. IEEE Access 2020, 8, 158115–158124. [Google Scholar] [CrossRef]
- Vinod, C.; Anand, H. Nature inspired meta heuristic algorithms for optimization problems. Computing 2021, 104, 251–269. [Google Scholar]
- Tang, J.; Liu, G.; Pan, Q. A Review on representative swarm intelligence algorithms for solving optimization problems: Applications and trends. IEEE/CAA J. Autom. Sin. 2021, 8, 17. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, L. High-quality face image generation using particle swarm optimization-based generative adversarial networks. Future Gener. Comput. Syst. 2021, 122, 98–104. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, D.; Zhang, J.; Zhang, T.; Zhu, H. A path planning method based on the particle swarm optimization trained fuzzy neural network algorithm. Clust. Comput. 2021, 24, 1901–1915. [Google Scholar] [CrossRef]
- Mohan, S.; Bhattacharya, S.; Kaluri, R.; Guang, F.; Benny, L. Multi-modal prediction of breast cancer using particle swarm optimization with non-dominating sorting. Int. J. Distrib. Sens. Netw. 2020, 16, 1–12. [Google Scholar]
- Shen, Y.; Cai, W.; Kang, H.; Sun, X.; Chen, Q. A particle swarm algorithm based on a multi-stage search strategy. Entropy 2021, 23, 1200. [Google Scholar] [CrossRef]
- Xue, J.; Shen, B. A novel swarm intelligence optimization approach: Sparrow search algorithm. Syst. Sci. Control. Eng. 2020, 8, 22–34. [Google Scholar] [CrossRef]
- Dong, J.; Dou, Z.; Si, S.; Wang, Z.; Liu, L. Optimization of capacity configuration of wind–solar–diesel–storage using improved sparrow search algorithm. J. Electr. Eng. Technol. 2021, 17, 1–14. [Google Scholar] [CrossRef]
- Zhang, Z.; He, R.; Yang, K. A bioinspired path planning approach for mobile robots based on improved sparrow search algorithm. Adv. Manuf. 2021, 10, 114–130. [Google Scholar] [CrossRef]
- Li, G.; Hu, T.; Bai, D. BP neural network improved by sparrow search algorithm in predicting debonding strain of FRP-strengthened RC beams. Adv. Civ. Eng. 2021, 2021, 9979028. [Google Scholar] [CrossRef]
- Tang, Y.; Li, C.; Li, S.; Cao, B.; Chen, C. A fusion crossover mutation sparrow search algorithm. Math. Probl. Eng. 2021, 2021, 9952606. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, Y.; Yang, H. Research on economic optimization of microgrid cluster based on chaos sparrow search algorithm. Comput. Intell. Neurosci. 2021, 2021, 5556780. [Google Scholar] [CrossRef]
- Li, Y.; Han, M.; Guo, Q. Modified whale optimization algorithm based on tent chaotic mapping and its application in structural optimization. KSCE J. Civ. Eng. 2020, 24, 3703–3713. [Google Scholar] [CrossRef]
- Sun, J.; Yang, Y.; Wang, Y.; Wang, L.; Song, X.; Zhao, X. Survival risk prediction of esophageal cancer based on self-organizing maps clustering and support vector machine ensembles. IEEE Access 2020, 8, 131449–131460. [Google Scholar] [CrossRef]
- Oh, G.; Song, J.; Park, H.; Na, C. Evaluation of random forest in crime prediction: Comparing three-layered random forest and logistic regression. Deviant Behav. 2021, 1–14. [Google Scholar] [CrossRef]

| Type | Expressions | Parameters |
|---|---|---|
| Radial Basis Kernel | : Free parameters. | |
| Linear Kernel | : Constant | |
| Polynomial Kernel | : Slope : Constant : Number of polynomials | |
| Wavelet Kernel | : Wavelet expansion factor : Translation factor |
| Classification category | 1 | 2 | 3 | 4 | 5 |
| Number of samples | 82 | 76 | 53 | 50 | 79 |
| Kernel Function Type | Risk Level | (%) | (%) | ||||
|---|---|---|---|---|---|---|---|
| Radial basis function | Level 1 | 70.0 | 68.5 | 70.0 | 66.7 | 33.3 | 30.0 |
| Level 2 | 66.7 | ||||||
| Linear | level 1 | 60.0 | 53.4 | 60.0 | 45.5 | 54.5 | 40.0 |
| level 2 | 45.5 | ||||||
| Polynomial | level 1 | 65.0 | 54.8 | 65.0 | 42.4 | 57.6 | 35.0 |
| level 2 | 42.4 | ||||||
| Wavelet | level 1 | 37.5 | 43.8 | 37.5 | 51.5 | 48.5 | 62.5 |
| level 2 | 51.5 |
| Predictive Model | Risk Level | (%) | (%) | Running Time(s) | ||||
|---|---|---|---|---|---|---|---|---|
| KELM | level 1 | 70.0 | 68.5 | 70 | 66.7 | 33.3 | 30.0 | 3.20 |
| level 2 | 66.7 | |||||||
| SSA-KELM | level 1 | 90.0 | 89.0 | 90 | 87.8 | 12.1 | 10.0 | 15.38 |
| level 2 | 87.8 | |||||||
| PSO-KELM | level 1 | 92.5 | 84.9 | 93 | 75.6 | 24.2 | 7.5 | 17.56 |
| level 2 | 75.6 | |||||||
| CLPSO-KELM | level 1 | 95.0 | 89.0 | 95 | 81.8 | 18.2 | 5 | 14.12 |
| level 2 | 81.8 | |||||||
| AMSSA-KELM | level 1 | 95.0 | 91.8 | 95 | 87.8 | 12.1 | 5 | 10.26 |
| level 2 | 87.8 |
| Predictive Model | Risk Level | (%) | (%) | Running Time(s) | ||||
|---|---|---|---|---|---|---|---|---|
| AMSSA-KELM | level 1 | 95.0 | 91.8 | 95.0 | 87.8 | 12.1 | 5.0 | 10.26 |
| level 2 | 87.8 | |||||||
| ABC-SVM | level 1 | 87.5 | 81.8 | 87.5 | 72.7 | 27.3 | 12.5 | 10.38 |
| level 2 | 72.7 | |||||||
| TLRF | level 1 | 57.5 | 61.6 | 57.5 | 66.7 | 33.3 | 42.5 | 20.15 |
| level 2 | 66.7 | |||||||
| GP-SVM | level 1 | 90.0 | 83.6 | 90.0 | 75.6 | 24.2 | 10.0 | 30.50 |
| level 2 | 75.6 | |||||||
| Cox-LMM | level 1 | 62.5 | 61.6 | 62.5 | 60.6 | 39.4 | 37.5 | 15.41 |
| level 2 | 60.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, H.; Li, S.; Wang, L. Survival Risk Prediction of Esophageal Cancer Based on the Kohonen Network Clustering Algorithm and Kernel Extreme Learning Machine. Mathematics 2022, 10, 1367. https://doi.org/10.3390/math10091367
Wang Y, Wang H, Li S, Wang L. Survival Risk Prediction of Esophageal Cancer Based on the Kohonen Network Clustering Algorithm and Kernel Extreme Learning Machine. Mathematics. 2022; 10(9):1367. https://doi.org/10.3390/math10091367
Chicago/Turabian StyleWang, Yanfeng, Haohao Wang, Sanyi Li, and Lidong Wang. 2022. "Survival Risk Prediction of Esophageal Cancer Based on the Kohonen Network Clustering Algorithm and Kernel Extreme Learning Machine" Mathematics 10, no. 9: 1367. https://doi.org/10.3390/math10091367
APA StyleWang, Y., Wang, H., Li, S., & Wang, L. (2022). Survival Risk Prediction of Esophageal Cancer Based on the Kohonen Network Clustering Algorithm and Kernel Extreme Learning Machine. Mathematics, 10(9), 1367. https://doi.org/10.3390/math10091367





