Ensemble of 2D Residual Neural Networks Integrated with Atrous Spatial Pyramid Pooling Module for Myocardium Segmentation of Left Ventricle Cardiac MRI
Abstract
:1. Introduction
- The research work proposes integration of the 2-D residual neural network with the ASPP module. The designed network has different numbers of layers at the encoder and the decoder components. In addition, the ASPP module is added at the lowermost of the encoder and the decoder. The integrated framework captures multiscale information, and the ASPP module can be detect small objects with different shapes, sizes and orientations from MR heart images.
- The proposed model is designed to be robust because its training involves various hyperparameters. In addition, different models are brought together using a majority voting scheme to further enhance segmentation accuracy.
- A contrast enhancement method is proposed for preprocessing the input cardiac MRI dataset. Various performance metrics are used to relate the performance of recent models and the proposed model in deep learning contexts.
2. Related Work
3. Material and Methods
3.1. Dataset
3.2. Preprocessing of the Dataset
3.3. Proposed Model
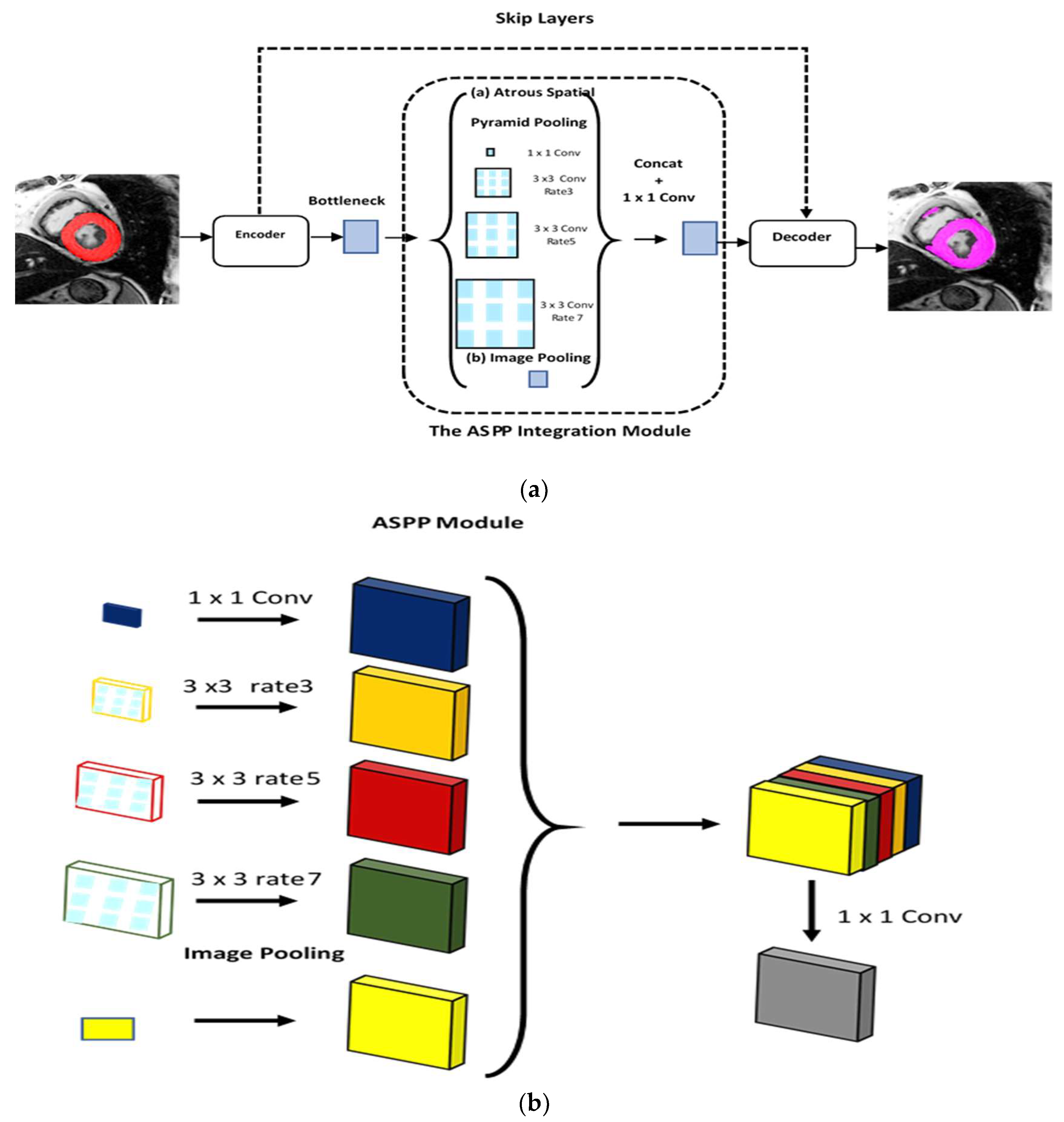
3.3.1. ASPP Module
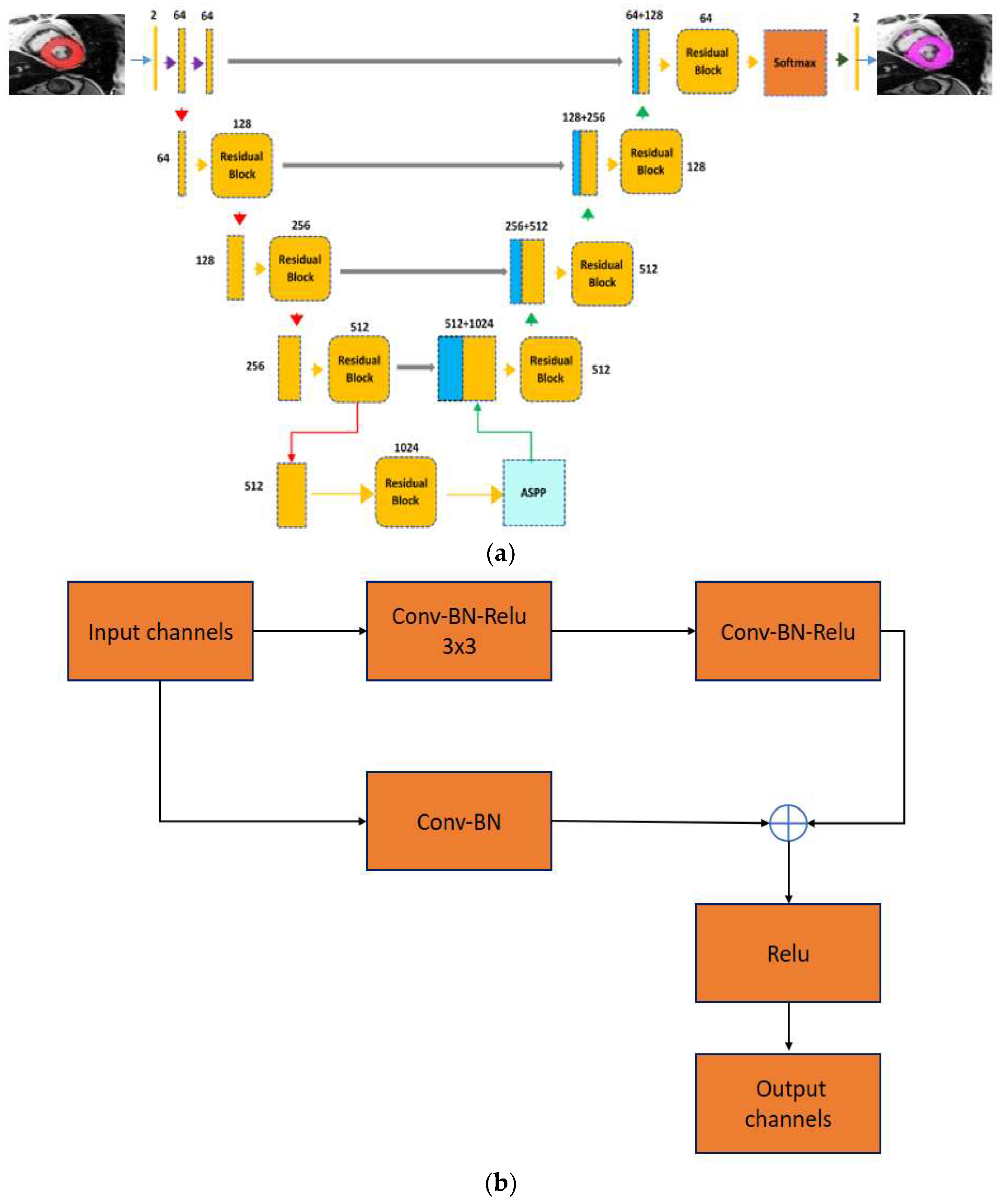
3.3.2. Proposed Hybrid Encoder-Decoder ASPP-RN Model
3.3.3. Ensemble of Proposed Models
3.3.4. Network Parameters or Configuration
3.4. Evaluation Criteria
4. Simulation
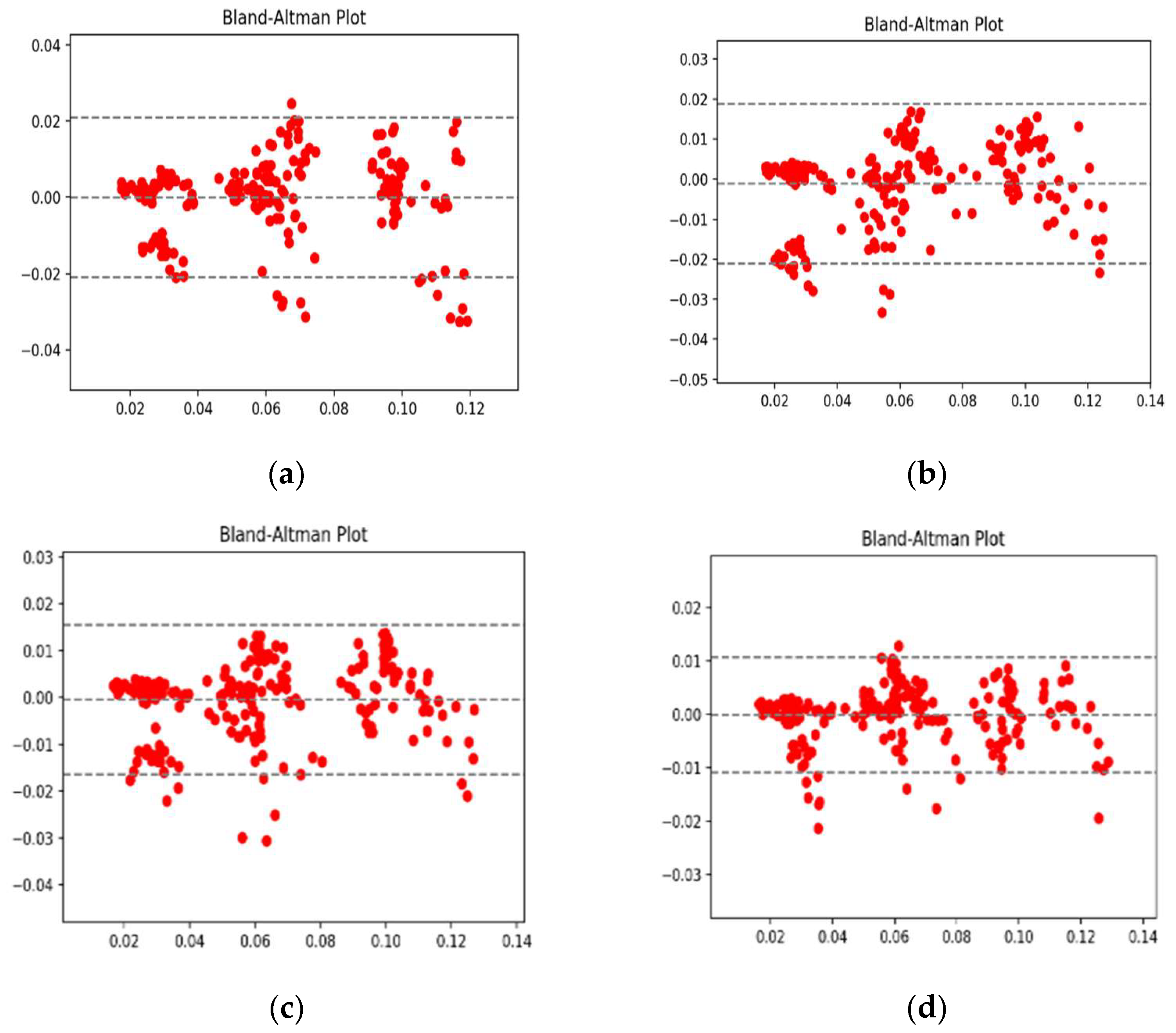
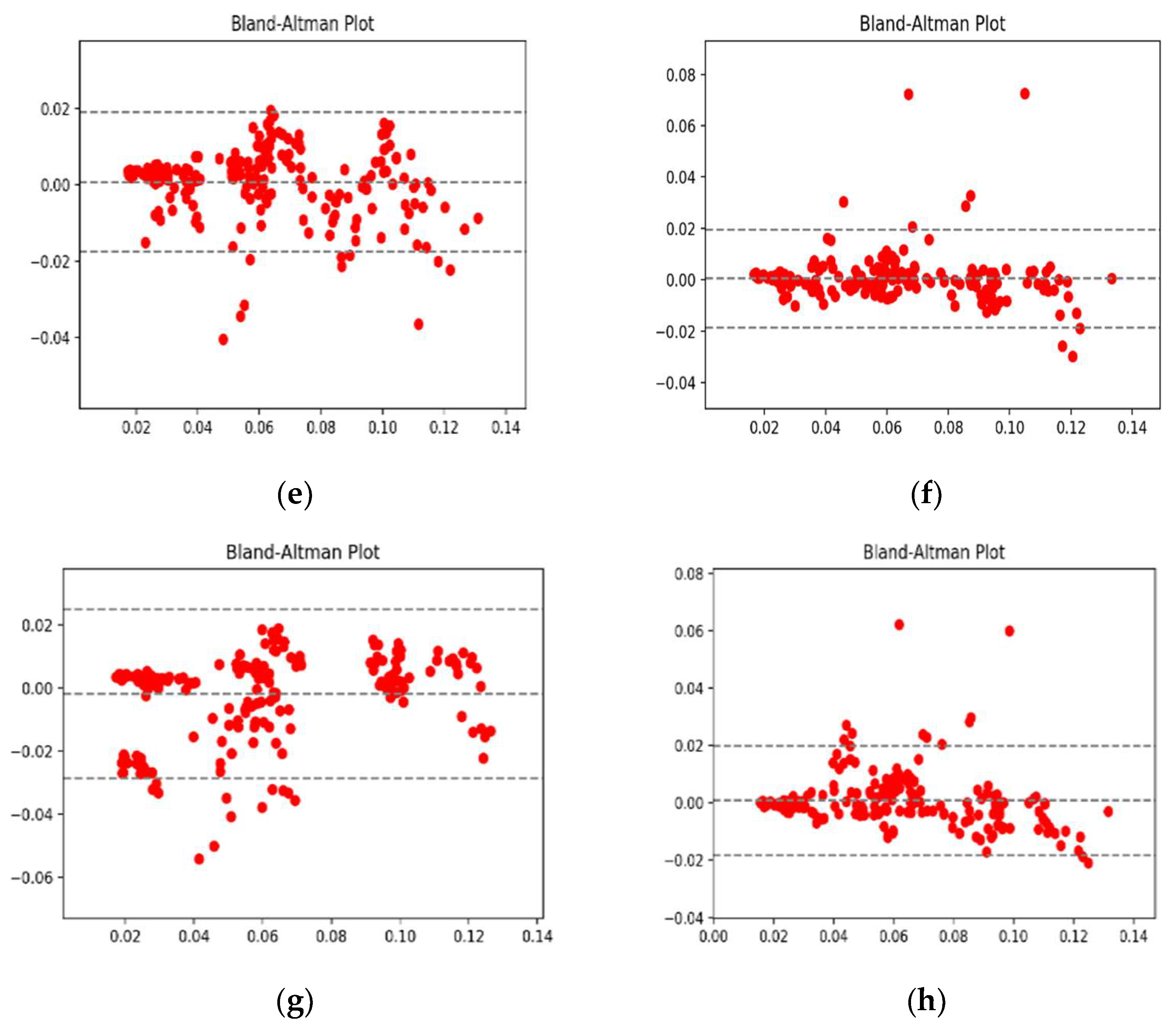
4.1. Performance Analysis Based on Performance Metrics
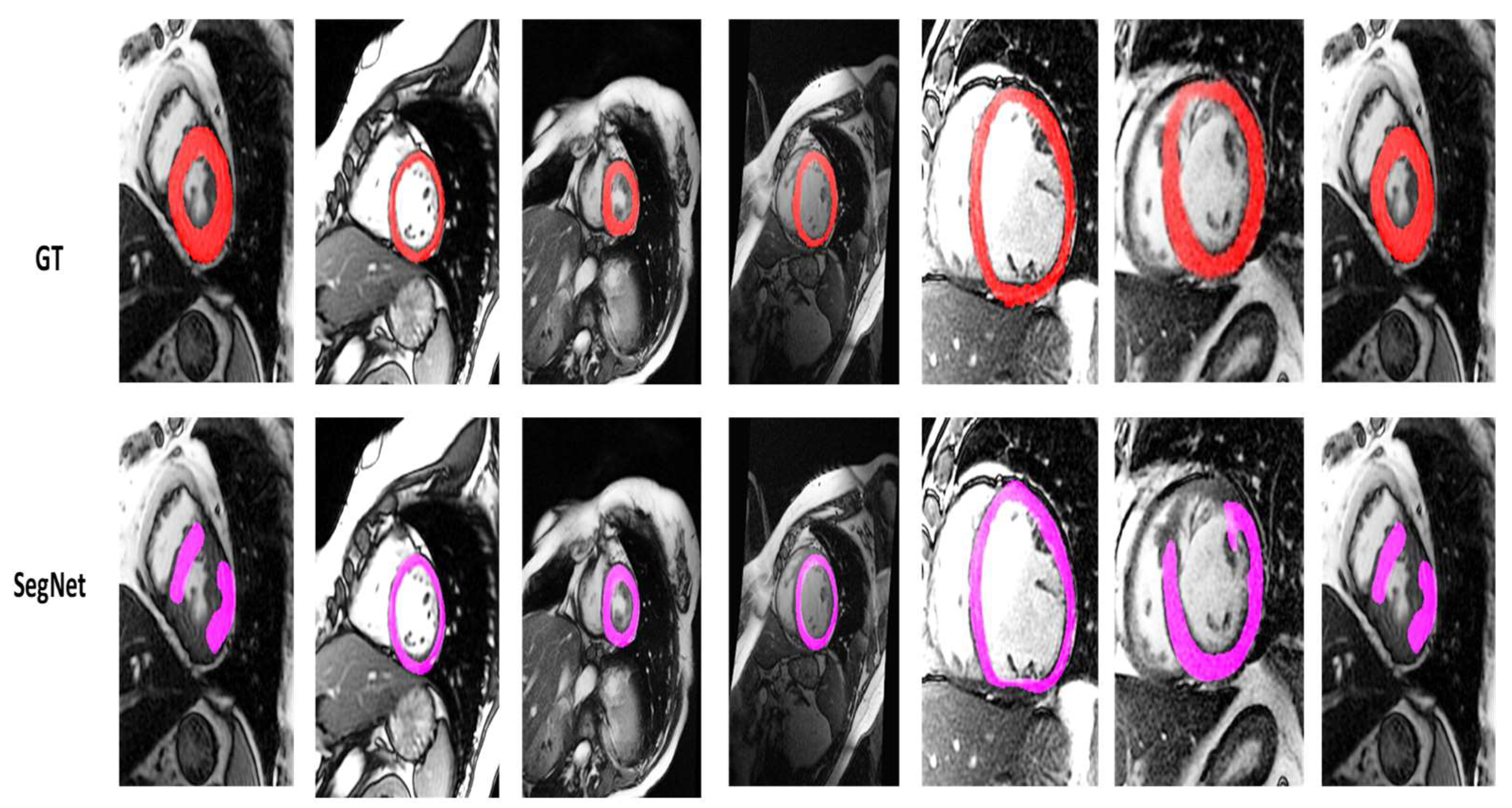
4.2. Visualization Segmentation Results
4.3. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karamitsos, T.D.; Francis, J.M.; Myerson, S.; Selvanayagam, J.B.; Neubauer, S. The role of cardiovascular magnetic resonance imaging in heart failure. J. Am. Coll. Cardiol. 2009, 54, 1407–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, P.; Lekadir, K.; Gooya, A.; Shao, L.; Petersen, S.E.; Frangi, A.F. A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging. Magn. Reson. Mater. Phys. Biol. Med. 2016, 29, 155–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayed, I.B.; Chen, H.M.; Punithakumar, K.; Ross, I.; Li, S. Max-flow segmentation of the left ventricle by recovering subject-specific distributions via a bound of the Bhattacharyya measure. Med. Image Anal. 2012, 16, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Hayit, G.; Van Ginneken, B.; Summers, R.M. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 2016, 35, 1153–1159. [Google Scholar]
- Avendi, M.R.; Kheradvar, A.; Jafarkhani, H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med. Image Anal. 2016, 30, 108–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, T.A.; Lu, Z.; Carneiro, G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med. Image Anal. 2017, 35, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.V. A fully convolutional neural network for cardiac segmentation in short-axis MRI. arXiv 2016, arXiv:1604.00494. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Išgum, I. Automatic segmentation and disease classification using cardiac cine MR images. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 101–110. [Google Scholar]
- Mortazi, A.; Burt, J.; Bagci, U. Multi-planar deep segmentation networks for cardiac substructures from MRI and CT. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2017; pp. 199–206. [Google Scholar]
- Zotti, C.; Luo, Z.; Lalande, A.; Humbert, O.; Jodoin, P.M. Novel deep convolution neural network applied to MRI cardiac segmentation. arXiv 2017, arXiv:1705.08943. [Google Scholar]
- Tan, L.K.; Liew, Y.M.; Lim, E.; McLaughlin, R.A. Convolutional neural network regression for short-axis left ventricle segmentation in cardiac cine MR sequences. Med. Image Anal. 2017, 39, 78–86. [Google Scholar] [CrossRef]
- Brahim, K.; Qayyum, A.; Lalande, A.; Boucher, A.; Sakly, A.; Meriaudeau, F. A 3D deep learning approach based on Shape Prior for automatic segmentation of myocardial diseases. In Proceedings of the 2020 Tenth International Conference on Image Processing Theory, Tools and Applications (IPTA), Paris, France, 9–12 November 2020; IEEE: Piscataway, NJ, USA; pp. 1–6. [Google Scholar]
- Suinesiaputra, A.; Bluemke, D.A.; Cowan, B.R.; Friedrich, M.G.; Kramer, C.M.; Kwong, R.; Plein, S.; Schulz-Menger, J.; Westenberg, J.J.; Young, A.A.; et al. Quantification of LV function and mass by cardiovascular magnetic resonance: Multi-center variability and consensus contours. J. Cardiovasc. Magn. Reson. 2015, 17, 63. [Google Scholar] [CrossRef] [Green Version]
- Petitjean, C.; Dacher, J.N. A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 2011, 15, 169–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Wang, Y.; Jia, Y. Segmentation of the left ventricle in cardiac cine MRI using a shape-constrained snake model. Comput. Vis. Image Underst. 2013, 117, 990–1003. [Google Scholar] [CrossRef]
- Pednekar, A.; Kurkure, U.; Muthupillai, R.; Flamm, S.; Kakadiaris, I.A. Automated left ventricular segmentation in cardiac MRI. IEEE Trans. Biomed. Eng. 2006, 53, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Lötjönen, J.; Kivistö, S.; Koikkalainen, J.; Smutek, D.; Lauerma, K. Statistical shape model of atria, ventricles and epicardium from short-and long-axis MR images. Med. Image Anal. 2004, 8, 371–386. [Google Scholar] [CrossRef]
- Nachtomy, E.; Cooperstein, R.; Vaturi, M.; Bosak, E.; Vered, Z.; Akselrod, S. Automatic assessment of cardiac function from short-axis MRI: Procedure and clinical evaluation. Magn. Reson. Imaging 1998, 16, 365–376. [Google Scholar] [CrossRef]
- Qayyum, A.; Razzak, I.; Tanveer, M.; Kumar, A. Depth-wise dense neural network for automatic COVID19 infection detection and diagnosis. Ann. Oper. Res. 2021, 1–21, published online ahead of print. [Google Scholar] [CrossRef]
- Zhen, X.; Islam, A.; Bhaduri, M.; Chan, I.; Li, S. Direct and simultaneous four-chamber volume estimation by multi-output regression. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 669–676. [Google Scholar]
- Zhen, X.; Wang, Z.; Islam, A.; Bhaduri, M.; Chan, I.; Li, S. Direct estimation of cardiac bi-ventricular volumes with regression forests. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 586–593. [Google Scholar]
- Zhen, X.; Wang, Z.; Islam, A.; Bhaduri, M.; Chan, I.; Li, S. Multi-scale deep networks and regression forests for direct bi-ventricular volume estimation. Med. Image Anal. 2016, 30, 120–129. [Google Scholar] [CrossRef]
- Kabani, A.; El-Sakka, M.R. Estimating ejection fraction and left ventricle volume using deep convolutional networks. In International Conference on Image Analysis and Recognition; Springer: Cham, Switzerland, 2016; pp. 678–686. [Google Scholar]
- Xue, W.; Islam, A.; Bhaduri, M.; Li, S. Direct multitype cardiac indices estimation via joint representation and regression learning. IEEE Trans. Med. Imaging 2017, 36, 2057–2067. [Google Scholar] [CrossRef] [Green Version]
- Xue, W.; Nachum, I.B.; Pandey, S.; Warrington, J.; Leung, S.; Li, S. Direct estimation of regional wall thicknesses via residual recurrent neural network. In International Conference on Information Processing in Medical Imaging; Springer: Cham, Switzerland, 2017; pp. 505–516. [Google Scholar]
- Afshin, M.; Ayed, I.B.; Punithakumar, K.; Law, M.; Islam, A.; Goela, A.; Peters, T.; Li, S. Regional assessment of cardiac left ventricular myocardial function via MRI statistical features. IEEE Trans. Med. Imaging 2013, 33, 481–494. [Google Scholar] [CrossRef]
- Wang, Z.; Salah, M.B.; Gu, B.; Islam, A.; Goela, A.; Li, S. Direct estimation of cardiac biventricular volumes with an adapted bayesian formulation. IEEE Trans. Biomed. Eng. 2014, 61, 1251–1260. [Google Scholar] [CrossRef]
- Qayyum, A.; Lalande, A.; Meriaudeau, F. Automatic segmentation of tumors and affected organs in the abdomen using a 3D hybrid model for computed tomography imaging. Comput. Biol. Med. 2020, 127, 104097. [Google Scholar] [CrossRef]
- Fritscher, K.; Raudaschl, P.; Zaffino, P.; Spadea, M.F.; Sharp, G.C.; Schubert, R. Deep neural networks for fast segmentation of 3D medical images. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2016; pp. 158–165. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2014; pp. 818–833. [Google Scholar]
- Ngo, T.A.; Carneiro, G. Left ventricle segmentation from cardiac MRI combining level set methods with deep belief networks. In Proceedings of the 2013 IEEE International Conference on Image Processing, Melbourne, Australia, 15–18 September 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 695–699. [Google Scholar]
- Poudel, R.P.; Lamata, P.; Montana, G. Recurrent fully convolutional neural networks for multi-slice MRI cardiac segmentation. In Reconstruction, Segmentation, and Analysis of Medical Images; Springer: Cham, Switzerland, 2017; pp. 83–94. [Google Scholar]
- Zheng, Q.; Yang, M.; Yang, J.; Zhang, Q.; Zhang, X. Improvement of Generalization Ability of Deep CNN via Implicit Regularization in Two-Stage Training Process. IEEE Access 2018, 6, 15844–15869. [Google Scholar] [CrossRef]
- Zheng, Q.; Yang, M.; Tian, X.; Jiang, N.; Wang, D. A Full Stage Data Augmentation Method in Deep Convolutional Neural Network for Natural Image Classification. Discret. Dyn. Nat. Soc. 2020, 2020, 4706576. [Google Scholar] [CrossRef]
- Liu, C.; Wang, K.; Ye, L.; Wang, Y.; Yuan, X. Deep learning with neighborhood preserving embedding regularization and its application for soft sensor in an industrial hydrocracking process. Inf. Sci. 2021, 567, 42–57. [Google Scholar] [CrossRef]
- Rucco, M.; Viticchi, G.; Falsetti, L. Towards Personalized Diagnosis of Glioblastoma in Fluid-Attenuated Inversion Recovery (FLAIR) by Topological Interpretable Machine Learning. Mathematics 2020, 8, 770. [Google Scholar] [CrossRef]
- Teso-Fz-Betoño, D.; Zulueta, E.; Sánchez-Chica, A.; Fernandez-Gamiz, U.; Saenz-Aguirre, A. Semantic Segmentation to Develop an Indoor Navigation System for an Autonomous Mobile Robot. Mathematics 2020, 8, 855. [Google Scholar] [CrossRef]
- Perez, H.; Tah, J.H.M. Improving the Accuracy of Convolutional Neural Networks by Identifying and Removing Outlier Images in Datasets Using t-SNE. Mathematics 2020, 8, 662. [Google Scholar] [CrossRef]
- Hu, C.-S.; Lawson, A.; Chen, J.-S.; Chung, Y.-M.; Smyth, C.; Yang, S.-M. TopoResNet: A Hybrid Deep Learning Architecture and Its Application to Skin Lesion Classification. Mathematics 2021, 9, 2924. [Google Scholar] [CrossRef]
- Joshi, G.P.; Alenezi, F.; Thirumoorthy, G.; Dutta, A.K.; You, J. Ensemble of Deep Learning-Based Multimodal Remote Sensing Image Classification Model on Unmanned Aerial Vehicle Networks. Mathematics 2021, 9, 2984. [Google Scholar] [CrossRef]
- Kang, J.; Gwak, J. Ensemble Learning of Lightweight Deep Learning Models Using Knowledge Distillation for Image Classification. Mathematics 2020, 8, 1652. [Google Scholar] [CrossRef]
- Ganaie, M.A.; Hu, M.; Tanveer, M.; Suganthan, P.N. Ensemble deep learning: A review. arXiv 2021, arXiv:2104.02395. [Google Scholar]
- Neshir, G.; Rauber, A.; Atnafu, S. Meta-Learner for Amharic Sentiment Classification. Appl. Sci. 2021, 11, 8489. [Google Scholar] [CrossRef]
- Xue, W.; Brahm, G.; Pandey, S.; Leung, S.; Li, S. Full left ventricle quantification via deep multitask relationships learning. Med. Image Anal. 2018, 43, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Lum, A.; Mercado, A.; Landis, M.; Warrington, J.; Li, S. Full quantification of left ventricle via deep multitask learning network respecting intra-and inter-task relatedness. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017; pp. 276–284. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Ni, Q.; Zhang, L.; Li, L. A Heterogeneous Ensemble Approach for Activity Recognition with Integration of Change Point-Based Data Segmentation. Appl. Sci. 2018, 8, 1695. [Google Scholar] [CrossRef] [Green Version]
- Ju, C.; Bibaut, A.; van der Laan, M. The relative performance of ensemble methods with deep convolutional neural networks for image classification. J. Appl. Stat. 2018, 45, 2800–2818. [Google Scholar] [CrossRef]
- Christ, P.F.; Ettlinger, F.; Grün, F.; Elshaera, M.E.A.; Lipkova, J.; Schlecht, S.; Ahmaddy, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; et al. Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv 2017, arXiv:1702.05970. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 770–778. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Larsson, G.; Maire, M.; Shakhnarovich, G. Fractalnet: Ultra-deep neural networks without residuals. arXiv 2016, arXiv:1605.07648. [Google Scholar]
- Lalande, A.; Chen, Z.; Decourselle, T.; Qayyum, A.; Pommier, T.; Lorgis, L.; de la Rosa, E.; Cochet, A.; Cottin, Y.; Ginhac, D.; et al. Emidec: A database usable for the automatic evaluation of myocardial infarction from delayed-enhancement cardiac MRI. Data 2020, 5, 89. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W.; Wang, K.; Ye, C.; Luo, G. An automatic cardiac segmentation framework based on multi-sequence MR image. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2019; pp. 220–227. [Google Scholar]
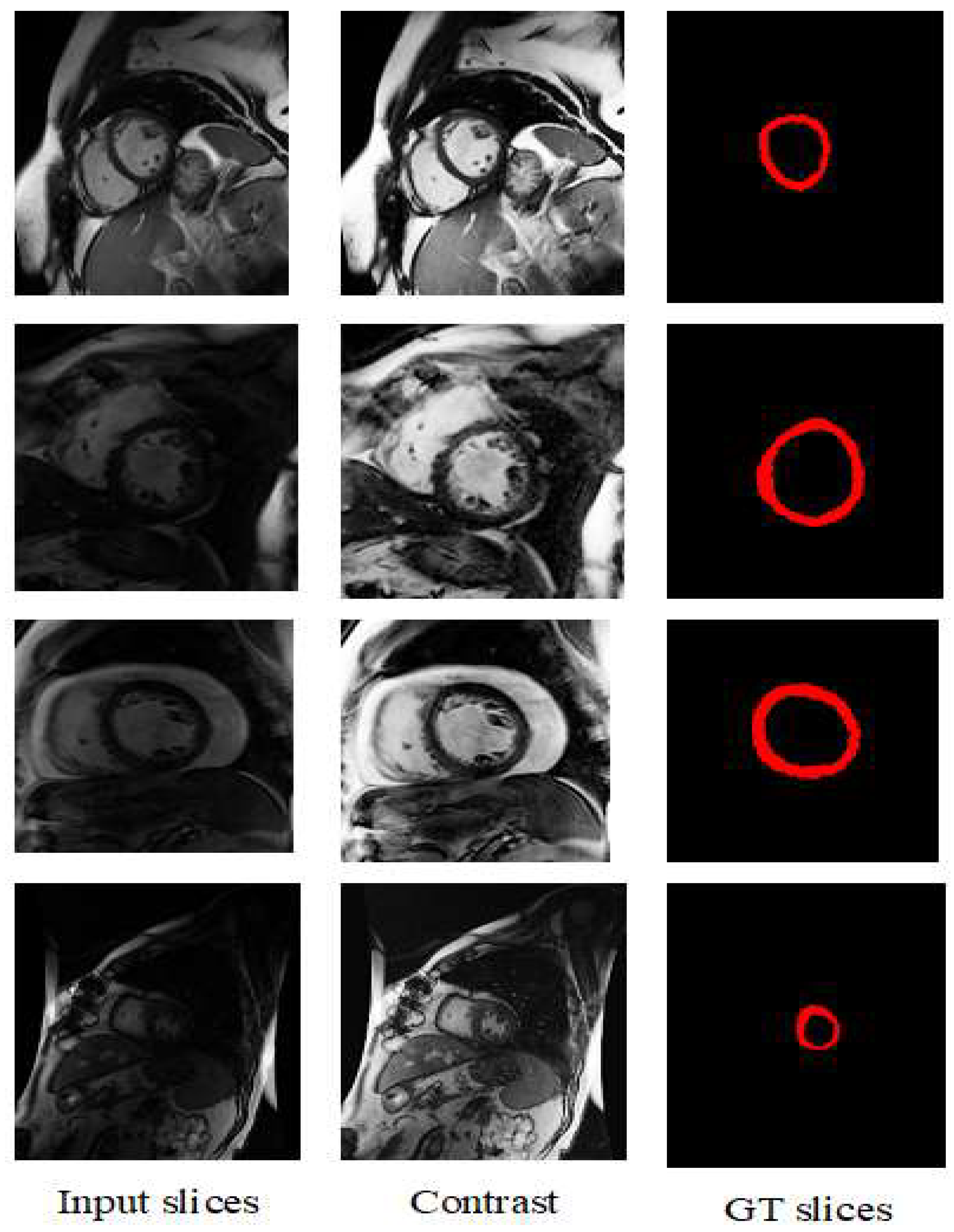

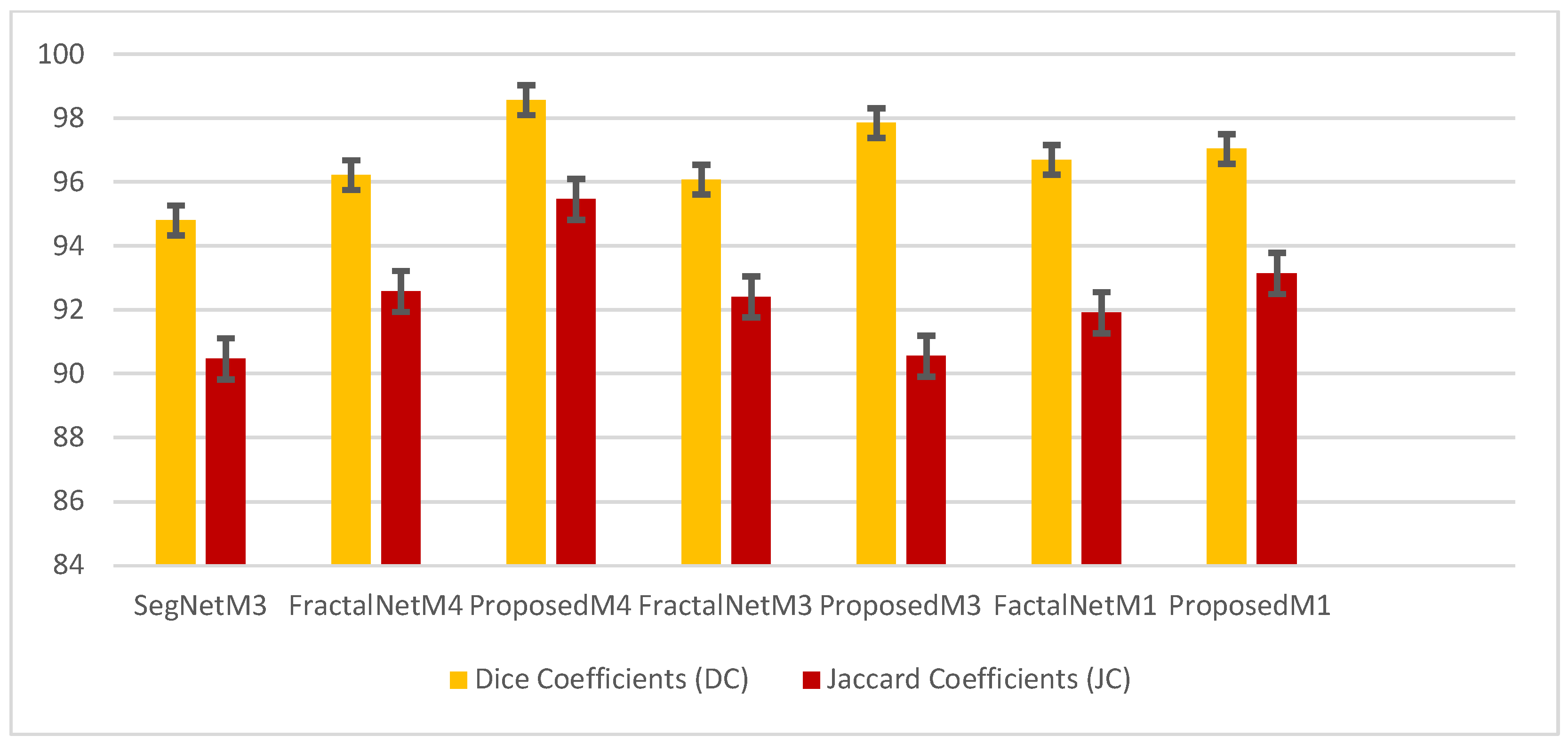
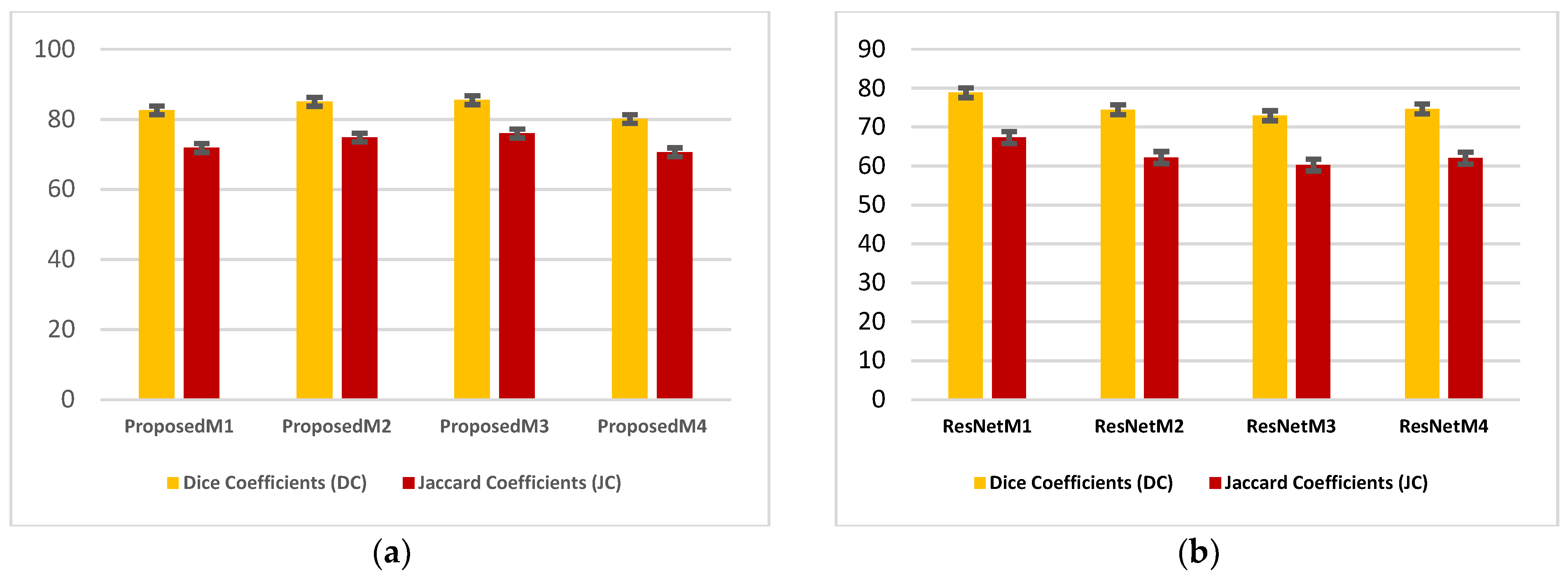
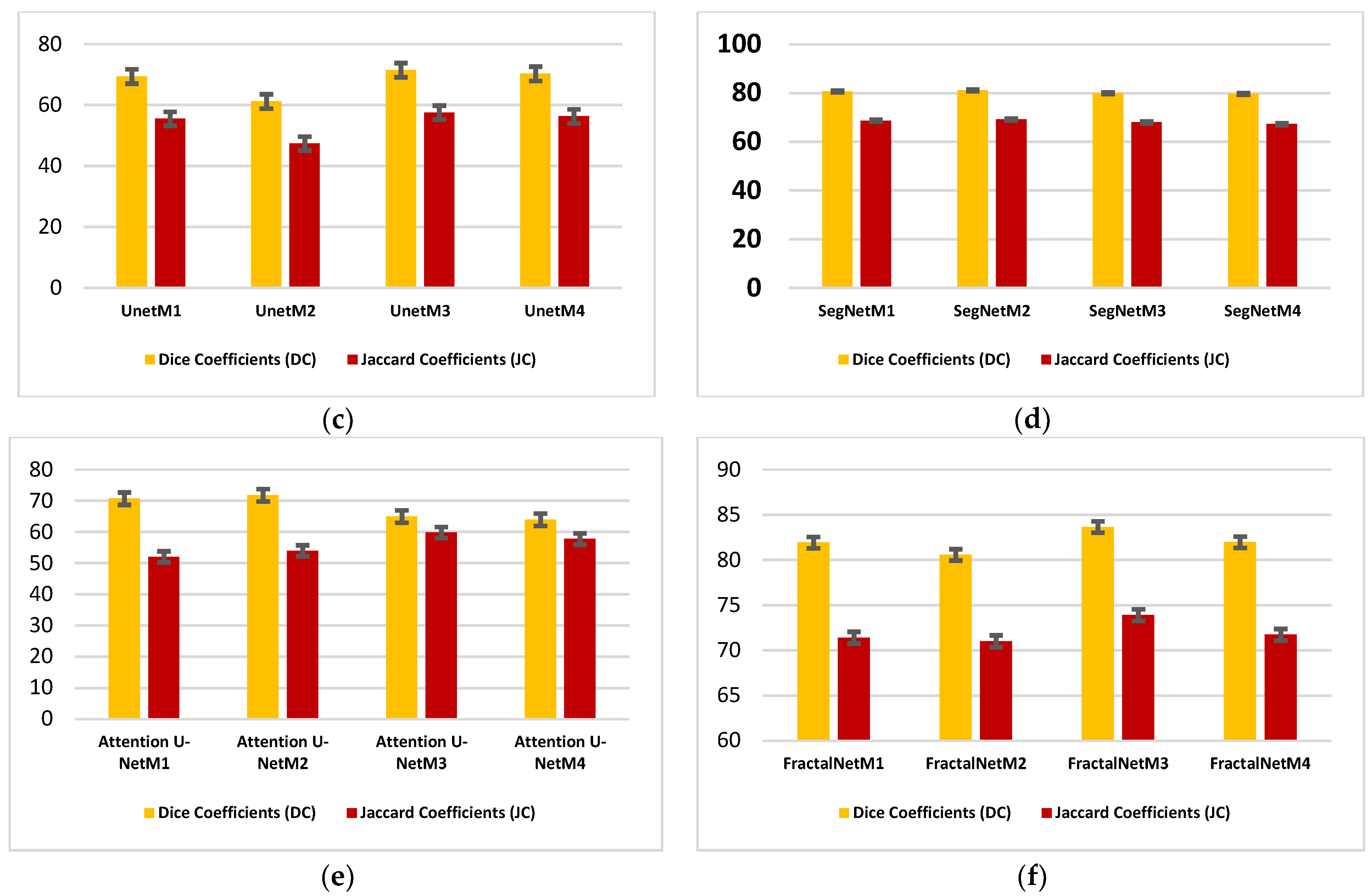


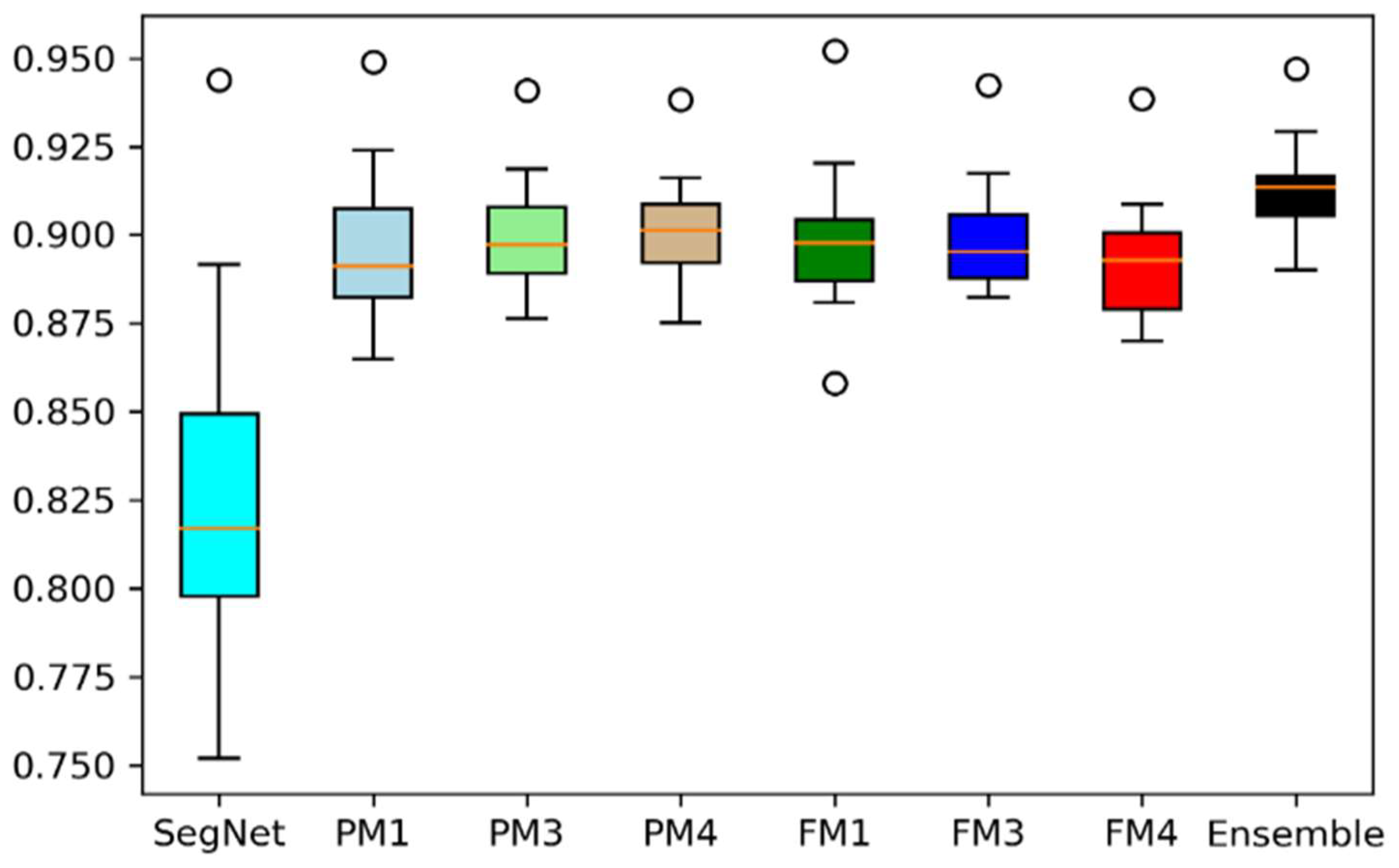
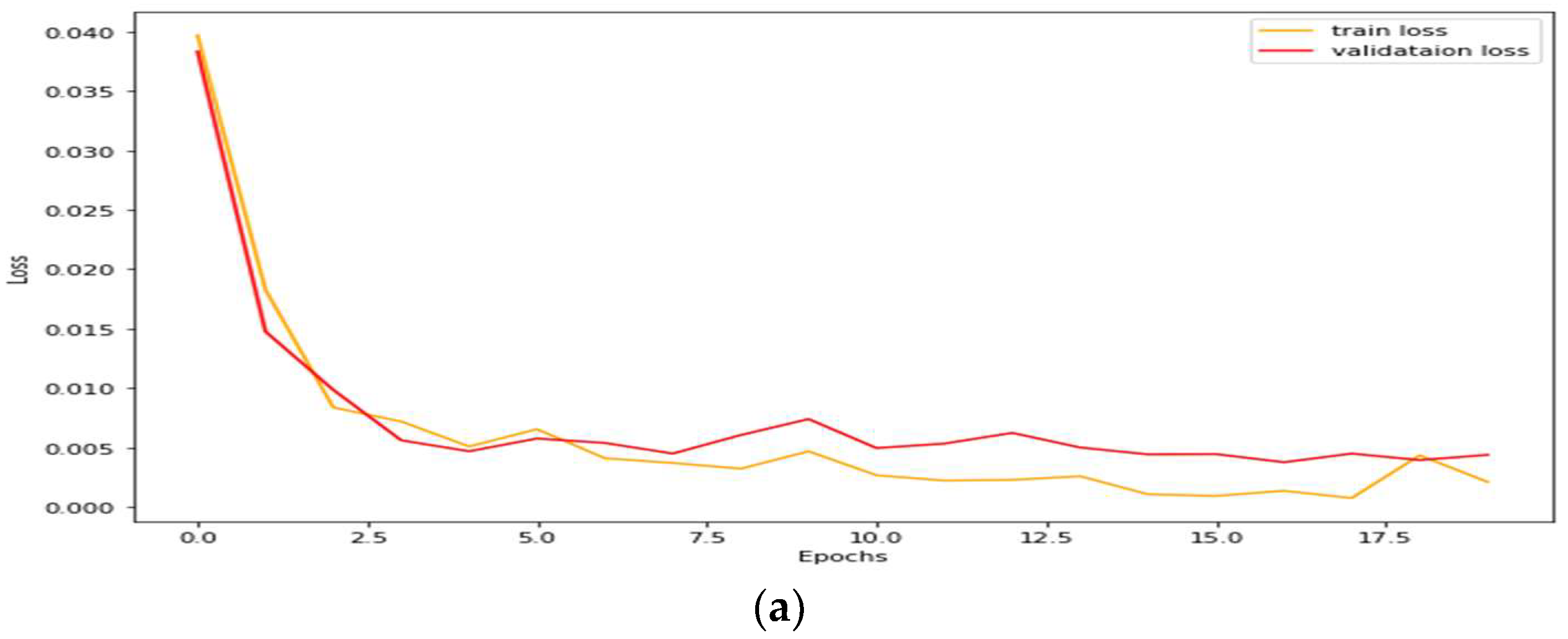
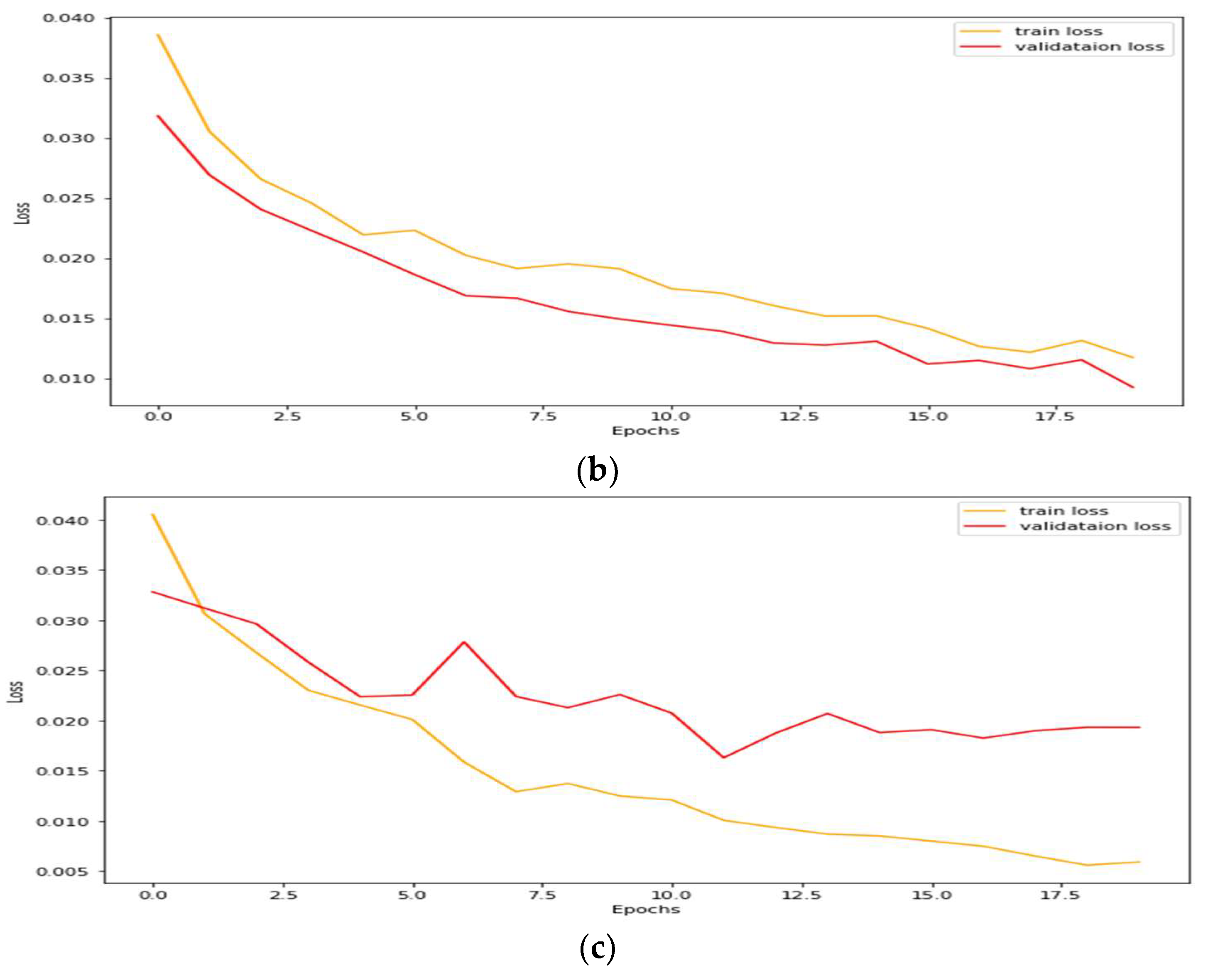
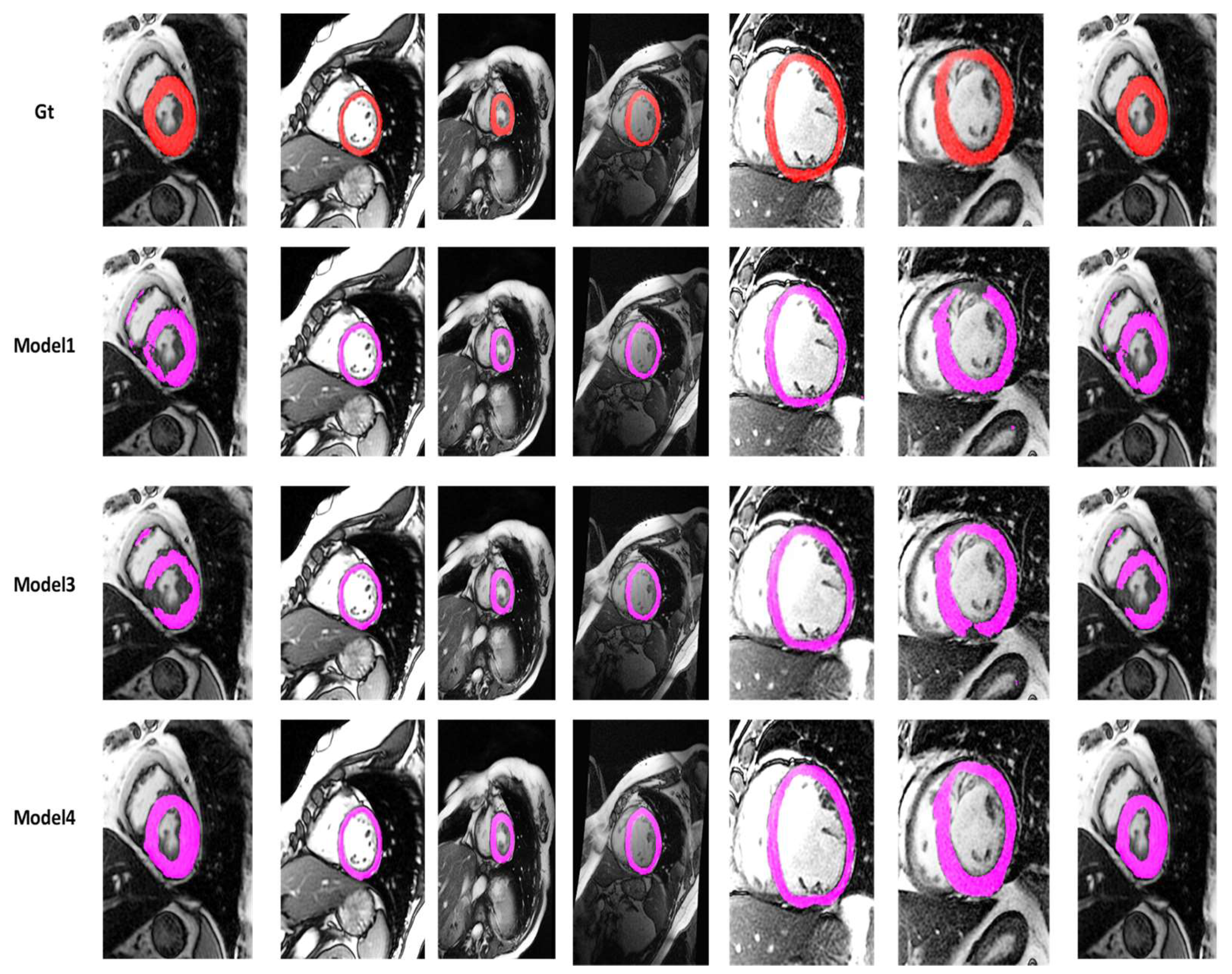
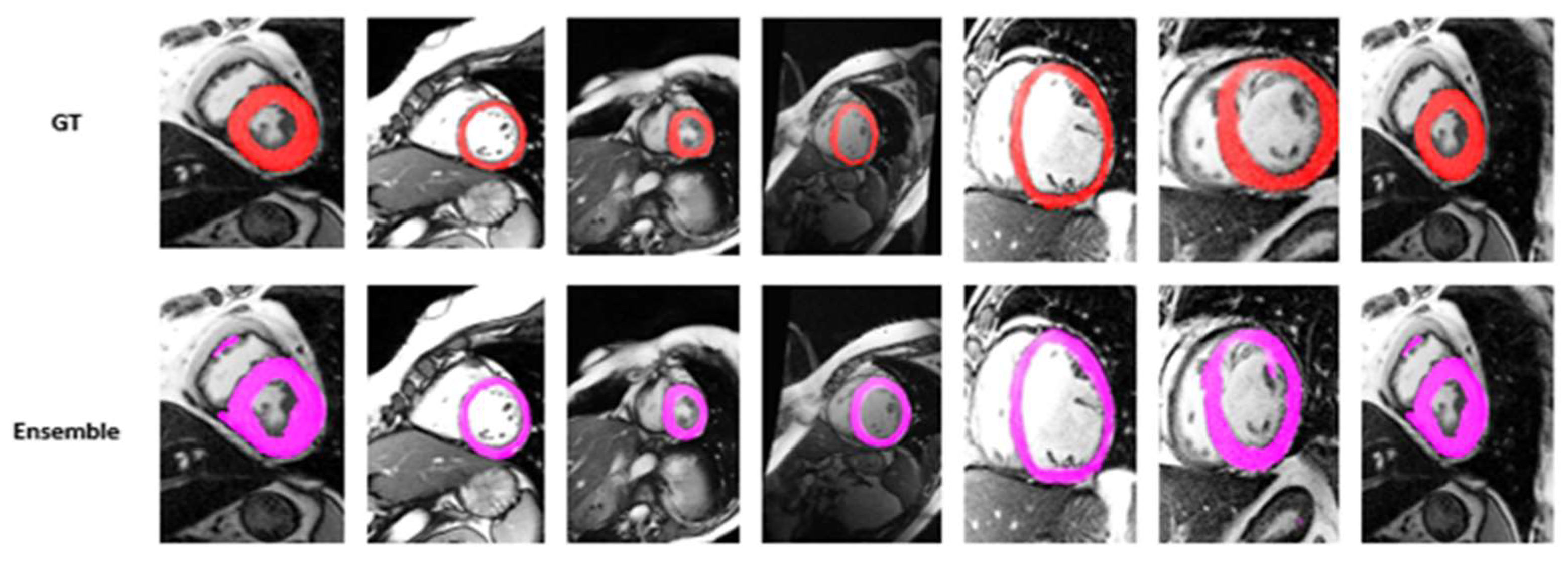
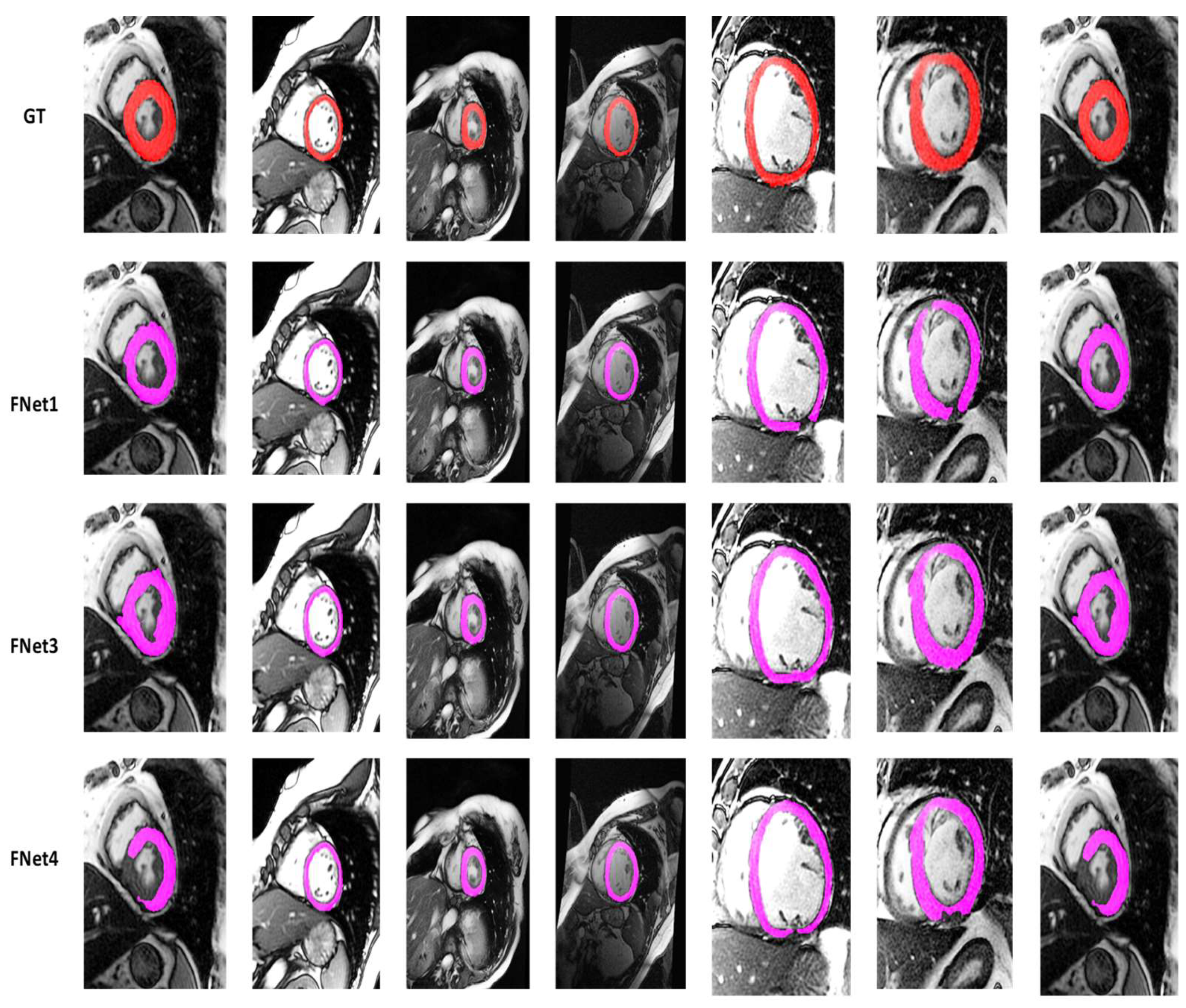
| Models | DC | JC | Hd95 | HD | Specificity | Sensitivity | VOE | ASSD | RVD |
|---|---|---|---|---|---|---|---|---|---|
| SegNetM3 | 81.00 | 69.10 | 8.5227 | 13.6867 | 98.63 | 84.87 | 0.3089 | 2.5484 | 0.1201 |
| FractalNetM4 | 81.95 | 71.71 | 9.0721 | 16.3544 | 98.72 | 89.72 | 0.2828 | 2.4974 | 0.3443 |
| ProposedM4 | 85.43 | 75.93 | 6.1702 | 10.0142 | 99.14 | 85.51 | 0.2406 | 1.7823 | 0.0141 |
| FractalNetM3 | 83.63 | 73.88 | 13.1938 | 20.2024 | 99.04 | 83.18 | 0.2611 | 2.9740 | 0.0169 |
| ProposedM3 | 84.96 | 74.78 | 11.3915 | 24.4692 | 98.97 | 87.48 | 0.2521 | 2.3937 | 0.0795 |
| FactalNetM1 | 81.91 | 71.38 | 9.2453 | 30.4901 | 98.60 | 85.92 | 0.2861 | 2.80443 | 0.1231 |
| ProposedM1 | 82.51 | 71.80 | 11.4752 | 26.905 | 98.76 | 87.22 | 0.2819 | 2.7359 | 0.1746 |
| Ensemble Model | 84.99 | 75.17 | 5.6782 | 12.6781 | 97.99 | 90.78 | 0.2790 | 1.8932 | 0.0674 |
| Hyperparameters | Proposed Model | SegNet [52] | ResNet [53] | Unet-Base [54] | Attention with Unet [55] | FractalNet [56] |
|---|---|---|---|---|---|---|
| Learning rate | 3 × 10−4, 1 × 10−4, 2 × 10−4, 1 × 10−4 | - | - | - | - | - |
| Optimizers | Adam | - | - | - | - | - |
| Batch size | 8, 12, 16, 20 | - | - | - | - | - |
| Number of epochs | 100, 200, 300, 500 | - | - | - | - | - |
| Datasets | DC | JC | Hd95 | HD |
|---|---|---|---|---|
| Xue et al. [45] | 84.99 | 75.17 | 5.6782 | 12.6781 |
| Alain et al. [57] | 79.67 | 72.09 | 8.33 | 10.33 |
| Liu et al. [58] | 78.33 | 70.01 | 8.34 | 14.22 |
| Models | Trainable Parameters | Number of Flops |
|---|---|---|
| Model1 | 27,889,221 | 334,516,681 |
| Model2 | 27,414,339 | 333,277,610 |
| Model3 | 27,228,939 | 337,723,411 |
| Model4 | 27,907,333 | 337,612,235 |
| Model5 | 27,221,797 | 332,236,130 |
| Model6 | 27,698,773 | 332,112,244 |
| Model7 | 27,896,532 | 338,333,759 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, I.; Qayyum, A.; Gupta, B.B.; Alassafi, M.O.; AlGhamdi, R.A. Ensemble of 2D Residual Neural Networks Integrated with Atrous Spatial Pyramid Pooling Module for Myocardium Segmentation of Left Ventricle Cardiac MRI. Mathematics 2022, 10, 627. https://doi.org/10.3390/math10040627
Ahmad I, Qayyum A, Gupta BB, Alassafi MO, AlGhamdi RA. Ensemble of 2D Residual Neural Networks Integrated with Atrous Spatial Pyramid Pooling Module for Myocardium Segmentation of Left Ventricle Cardiac MRI. Mathematics. 2022; 10(4):627. https://doi.org/10.3390/math10040627
Chicago/Turabian StyleAhmad, Iftikhar, Abdul Qayyum, Brij B. Gupta, Madini O. Alassafi, and Rayed A. AlGhamdi. 2022. "Ensemble of 2D Residual Neural Networks Integrated with Atrous Spatial Pyramid Pooling Module for Myocardium Segmentation of Left Ventricle Cardiac MRI" Mathematics 10, no. 4: 627. https://doi.org/10.3390/math10040627
APA StyleAhmad, I., Qayyum, A., Gupta, B. B., Alassafi, M. O., & AlGhamdi, R. A. (2022). Ensemble of 2D Residual Neural Networks Integrated with Atrous Spatial Pyramid Pooling Module for Myocardium Segmentation of Left Ventricle Cardiac MRI. Mathematics, 10(4), 627. https://doi.org/10.3390/math10040627








