Abstract
Network representation learning is a machine learning method that maps network topology and node information into low-dimensional vector space. Network representation learning enables the reduction of temporal and spatial complexity in the downstream data mining of networks, such as node classification and graph clustering. Existing algorithms commonly ignore the global topological information of the network in network representation learning, leading to information loss. The complete subgraph in the network commonly has a community structure, or it is the component module of the community structure. We believe that the structure of the community serves as the revealed structure in the topology of the network and preserves global information. In this paper, we propose SF-NRL, a network representation learning algorithm based on complete subgraph folding. The algorithm preserves the global topological information of the original network completely, by finding complete subgraphs in the original network and folding them into the super nodes. We employ the network representation learning algorithm to study the node embeddings on the folded network, and then merge the embeddings of the folded network with those of the original network to obtain the final node embeddings. Experiments performed on four real-world networks prove the effectiveness of the SF-NRL algorithm. The proposed algorithm outperforms the baselines in evaluation metrics on community detection and multi-label classification tasks. The proposed algorithm can effectively generalize the global information of the network and provides excellent classification performance.
1. Introduction
With the development of deep learning techniques and increasing requirements for graph data mining, the study of Network Representation Learning (NRL) has attracted greater attention from scholars. There are limitations to the existing random walk-based methods in the research of NRL [1]. For instance, the DeepWalk algorithm [2] and the Node2Vec algorithm [3] estimate target node embedding based on a walking sequence with a finite step length set. In addition, the LINE algorithm [4] obtains neighborhood information of nodes based on their similarity, but only considers the second-order neighborhood, so the algorithm only considers the local topology information of the network and ignores its global topology information. A complete graph is a simple undirected graph in which there is one edge between each distinct pair of nodes. In numerous real-world networks, we find that these networks commonly contain many complete subgraphs consisting of several nodes [5], and some are even connected by complete subgraphs and some common connection nodes. In real-world networks, the nodes in the communities are closely connected, and the structure of complete subgraphs often appears in the communities. Therefore, it can be inferred that most of the nodes in the same complete subgraph are in the same community [6]. There is a greater first-order similarity between nodes in the complete subgraph. These nodes are closely connected and naturally form the structure of the community or become part of the community in the network. The American Football League Network [7] contains 115 nodes and 613 edges and there are a large number of complete subgraphs with k = 4. There are also 113 nodes in the complete subgraphs, with k = 4, and only two nodes; central Florida and Connecticut are isolated. These two nodes do not affect the acquisition of the global topology of the network. The complete subgraph in the network commonly has a community structure, or it is the component module of the community structure. We believe that the structure of the community serves as the revealed structure in the topology of the network and preserves its global information. The global structure of a network is the topology of the network that can provide valuable information for graph data mining tasks. Therefore, we assume that the community of the network is approximated by finding its complete subgraphs and we employ this approximate community information as the global topology information of the network to improve the performance of NRL.
Existing algorithms fail to completely cover the global topology information of the network, which leads to information loss. To solve this problem, we propose a Network Representation Learning Algorithm Based on Complete Subgraph Folding (SF-NRL). We first find the complete subgraphs in the network and fold the original network by treating the complete subgraphs as folding units and applying pre-defined folding rules. We employ this method to find global topological information on the network and merge the representation of the original network with the coarsened network representation. Finally, we obtain the node embeddings that consider both the global topology and the local topological information.
Overall, our paper makes the following contributions:
- (1)
- The proposed algorithm solves the problem that existing NRL methods fail to balance global and local network structures.
- (2)
- The algorithm enables the design of a graph-coarsening approach based on defined graph folding rules to obtain the global topological structure information of the network.
- (3)
- The experimental results on four network datasets show that the proposed algorithm can significantly improve the quality of node embeddings, and the effectiveness of the algorithm is demonstrated by community detection and multi-label classification tasks.
2. Related Work
NRL methods are widely employed in network data mining tasks, including community detection [8], node classification [9], and link prediction [10]. These methods enable capturing node relationships in complex networks and obtaining embedding of that nodes’ representations by mapping the topological structure information of the network and node attribute information into a low-dimensional vector space [11,12]. Random walk-based methods have received significant attention in recent years, and researchers have proposed algorithms such as DeepWalk and Node2Vec. Salamat et al. [13] defined a notion of a nodes network neighborhood and designed a balanced random walk procedure, which adapts to the graph topology. In addition, to balance the ability to obtain local and global information of the network topology, Chen et al. [14] proposed star-coarsening and edge-coarsening strategies for improving the quality of node embeddings. Liang et al. [15] designed novel edge-coarsening strategies considering the weighting problem of edge-coarsening. Bartal et al. [16] proposed that it is important to consider global exposure to information by non-neighbors when modeling information spread in online social networks. To balance the local and global network topology information, Liang et al. [15], inspired by the HARP algorithm [14], proposed the MILE algorithm, which merges nodes with similar local structures into a super node and considers the weights on the coarsened connected edges. Deng et al. [17] found that node embedding, community detection, and community embedding create a closed-loop by investigating the relationship between community embedding and node embedding. Node embedding contributes to the result of community detection, community detection helps to obtain community embedding, and community embedding contributes to the result of node embedding.
3. Methods
3.1. Folding Rules
In this paper, we define novel folding rules to find the global topology of the network. Folding is employed to preserve the original network structure by transforming each folding unit in the network into a sub node, while the edges between the nodes contained in the folding unit and those not in the folding unit are still retained.
The complete subgraph is a concept of graph theory [18]. If any two nodes in a subgraph have a connected edge, then the subgraph is called a complete subgraph and can also be called a clique. In general, if the number of nodes in a complete subgraph is k, it is called the K-complete subgraph. The K-complete subgraph with k nodes has k(k−1)/2 edges.
Each K-complete subgraph in the network is treated as a sub node, and these nodes preserve the K-complete subgraphs in the network. As shown in Figure 1, nodes 1, 2, 3, and 4 form a complete graph, which, as a whole, is connected with nodes 5 and 6. We fold the nodes (node 1, 2, 3, and 4) in the complete subgraph into a sub node to preserve the original topology of the network. Node 5 and node 6 are connected with the sub node.
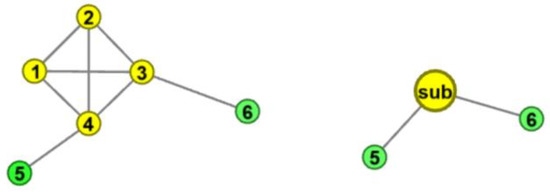
Figure 1.
Example of Complete subgraph folding.
The real-world network is often not as simple as in Figure 1; the relationships between the nodes are complex and there maybe a complete subgraph that is interconnected. When different complete subgraphs are closely connected, it is unreasonable to collapse all connected complete subgraphs into one node. Therefore, some rules should be designed for network folding to make the network structure more reasonable after folding.
There are two kinds of nodes in the original network: complete subgraph nodes and incomplete subgraph nodes. If we consider the complete subgraph as the folding unit, the connection between incomplete subgraph nodes and the connection between incomplete subgraphs and complete subgraphs is simple. However, there may be common nodes between complete subgraphs, and the number of common nodes is very important for the folding process. If there is a common node in two complete subgraphs and they are folded into one node, it is likely to make the folded network very small, leading to over-coarsening of the network [19]. In this way, the global information fails to be preserved completely. Therefore, for the different number of common nodes, the folding rules are designed.
The number of common nodes can be divided into two cases. When the number of common nodes between the complete subgraphs is 1, the complete subgraphs are folded into one node, respectively, and then the folded nodes are connected. When the number of common nodes between complete subgraphs is greater than 1, the complete subgraphs with common nodes are folded into one node. The following two cases are illustrated.
- Figure 2 shows what happens when the number of common nodes is 1. In the graph, nodes 1, 2, 3, and 4 form a complete subgraph, nodes 4, 5, 6, and 7 form other complete subgraphs, and node 4 is a common node. The two complete subgraphs are not considered to be closely connected here, and the two complete subgraphs are, respectively, folded into two sub nodes.
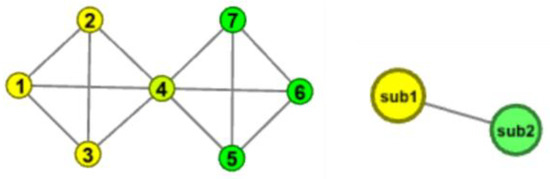 Figure 2. Example of Complete subgraph folding.
Figure 2. Example of Complete subgraph folding. - Figure 3 shows the situation when the number of common nodes is greater than 1. In the figure, nodes 1, 2, 3, 4 and 3, 4, 5, 6, respectively, form two complete subgraphs, and the number of common nodes is 2. Obviously, due to the existence of public nodes 3 and 4, nodes 2, 3, 4, 5 and nodes 1, 3, 4, 6, respectively, form an approximate complete subgraph, which makes the original two complete subgraphs connect very closely, so the nodes 1, 2, 3, 4, 5, 6 tend to be in the same community with a high probability, so it is reasonable that the two complete subgraphs are folded as a node.
 Figure 3. Example of complete subgraph folding.
Figure 3. Example of complete subgraph folding.
In the following, we define the selection strategy for k. Since complete subgraphs with k = 3 are frequent in the network and there are many edges between these complete subgraphs, adopting the complete subgraph with k = 3 as the folding unit can easily fold the network into an over-coarsening network, leading to over-coarsening of local and global information, which is not beneficial for information retention. In addition, complete subgraphs with k = 5 and k = 6 are not frequent in small-scale datasets, and both complete subgraphs contain complete subgraphs with k = 4. Therefore, we set the complete subgraphs with k = 4 as folded units. We first defined a set sub-set to preserve all folded sub nodes and we defined a set un-sub-set for preserving the nodes which are not in the complete subgraph. The steps of the folding process are as follows:
Step 1: From the original network G, we extracted all the complete subgraphs of k = 4 in the original network, and each complete subgraph was added to the sub-set as a sub node.
Step 2: The sub nodes were traversed in the sub-set. If the number of common nodes in subi and subj was greater than 1, the two nodes were merged into one node.
Step 3: The node set in the original network G was divided into set sub-set = {sub1, ..., subn} and set unsub-set = {v1, ..., vn}, where each element in the sub-set represents a complete subgraph, and each element in the unsub-set represents a non-complete subgraph node.
Step 4: We then rebuilt an empty graph M to represent the folding network, and every element in the sub-set was called a sub node. Each sub node in the folded sub-set and the node in unsub-set was added to the empty graph M.
Step 5: All the edges were traversed in the original network G. If there were edges between nodes in the unsub-set in the original network G, then an edge between nodes in unsub-set was added in the empty graph M. If there is an edge between the node in the sub and the node in unsub-set in the original network G, then an edge is added between the node in sub and the node in unsub-set in the empty graph M. If there is an edge between nodes subi and subj in the original network G, then edge between nodes subi and subj was added to the empty graph M until all edges were traversed.
| Algorithm 1 FLOD-N | |
| Input: Network G Output: Folded network M | |
| 1 | Sub-set = k_clique_communities (G, k)// Obtain K-complete subgraphs. |
| 2 | forsubiinSub-setdo // Merges complete subgraphs with public nodes greater than 1. |
| 3 | for subj in Sub-set do |
| 4 | if len (subi. intersection(subj))>1 then |
| 5 | Sub-set.remove(subi, subj); |
| 6 | Sub-set.append(subi.union(subj)); |
| 7 | for subinSub-setdo |
| 8 | for node in sub do |
| 9 | if node not in UnSub-set then |
| 10 | tmp.append(node); |
| 11 | UnSub-set =G.nodes − tmp; |
| 12 | M = new Graph; //create a new network |
| 13 | fornodeinUnsub-set, Sub-setdo |
| 14 | M.add_nodes(node); |
| 15 | fornode1inUnSub-setdo |
| 16 | for sub in Sub-set do |
| 17 | for node2 in sub do |
| 18 | if (node1, node2) in G.edges then |
| 19 | M.add_edges (node1, sub): |
| 20 | fornode1inUnSub-setdo |
| 21 | for node2 in UnSub-set do |
| 22 | if (node1, node2) in G.edges then |
| 23 | M.add_edges(node1, sub); |
| 24 | forsubiinSub-setdo |
| 25 | for subj in Sub-set do |
| 26 | for node1 in subi do |
| 27 | for node2 in subj do |
| 28 | if (sub1, sub2) in G.edges then |
| 29 | M.add_edges(subi, subj); |
| 30 | return M |
As shown in Figure 4, the original network is shown in the example on the left of the figure, in which red nodes 1–6 are connected to gray nodes 14 and 17, yellow nodes 7–10 are connected to gray nodes 18, and have a common node with green nodes; green nodes 10–13 are connected to yellow nodes and gray nodes 16. Through the above folding process, the red node, the yellow node, and the green node, respectively, form three sub nodes: sub0, sub1, and sub2; the nodes 14–18 remain as the original topological structure. The folded network is shown in the example on the right of the figure. The sub node is separated from the node in the unsub-set and sub0 is connected to the original nodes 14 and 17 of the network. Since sub1 and sub2 have a common node, they are connected. Similarly, nodes 16 and 18 are also connected to sub1. The original network structure is not destroyed by folding, and the global topology information of the network is well preserved. The pseudo-code of this algorithm to obtain the folded network is described in Algorithm 1.
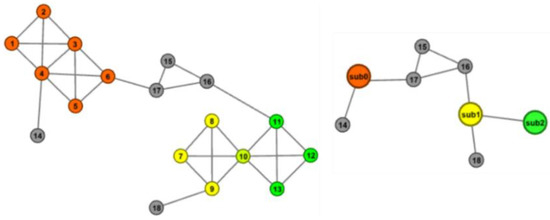
Figure 4.
Example of folding a network.
3.2. Algorithm Process
We fold the original network into a coarsened network M based on the defined graph folding rules, employing the complete subgraph as the folding unit, and find embeddings on the original network G and the small coarsened network M, respectively, with the NRL algorithms, and then the embeddings of the coarsened network M are fused with the embeddings of the original network G to learn the embeddings of the target network for downstream network data mining tasks. The whole procedure of the algorithm is shown in Figure 5.
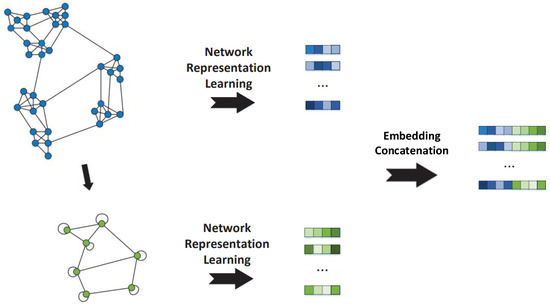
Figure 5.
The process of the SF-NRL algorithm.
4. Experiments
We employ the popular NRL algorithms DeepWalk, Node2Vec, and LINE as baselines to prove the effectiveness of our proposed algorithm and perform extensive experiments on community detection and multi-label classification tasks.
4.1. Datasets
In the community detection, we employed books about US Politics [20], American College Football [7], a jazz musicians network [21], and CORA citation network [22]. In the multi-label classification task, we employed books about US Politics, American College Football, and the CORA citation network. The statistics of the four datasets are shown in Table 1. The following is an introduction about the datasets:

Table 1.
Statistics of network datasets. Nodes and Edges represent the number of nodes and edges of the network, respectively, <k> indicates the average degree, and <d> denotes the average shortest path length of the network.
PolBooks: Based on the network of Books about US Politics on Amazon, the entire web is divided into three categories according to political affiliation. The nodes in the network represent political books, and the edges between nodes represent the same buyers of both books.
Football: Statistics for games between school teams during the 2000 American football season included a total of 115 teams in 12 divisions. The nodes represent teams from different schools, and the edges represent two schools that have played a match.
Jazz: A network created by Arenas and other 197 collaborators. The edge of the data set represents the cooperative relationship between the two musicians. There is a total of 198 nodes and 2742 edges in the network.
CORA: This data set is a citation network composed of 2708 papers. Each node in the network represents a paper, and the edges represent the citation relationship between papers, which can be divided into seven categories.
4.2. Community Detection
The above four datasets with real labels, PolBooks, Football, Jazz, and CORA, are used in the community detection task. In the experiment, the SF-NRL algorithm and the baseline were used to obtain the network representation vectors, respectively, and the k-means clustering algorithm [23] was used to cluster the obtained vectors.
The Normalized Mutual Information (NMI) value was calculated to compare the clustering result with the real community situation. The higher the NMI value, the more similar the clustering result was to the real community situation, which can prove that the clustering effect is better. Furthermore, it can be shown that the higher quality of the network representation obtained by the algorithm can indicate the effectiveness of the algorithm.
where A denotes the truth community labels and B denotes the community detection results obtained using the algorithm, respectively, CA and CB denote the number of communities in A and B, respectively, Nij denotes the elements in the confusion matrix, Ni and Nj denote the sum of the elements in row i and column j of the confusion matrix, respectively. n denotes the number of nodes in the network.
The experiment compares three classic NRL algorithms, DeepWalk, Node2Vec, and LINE with the proposed algorithm, respectively, that is, SF-NRL(DW) with DeepWalk, SF-NRL(N2V) with Node2Vec, and SF-NRL(Line) with LINE. Simultaneously, the experimental environment and algorithm parameters of each group of comparative experiments are kept consistent. The experimental results are shown in Table 2. We bold the best results.

Table 2.
NMI value between SF-NRL algorithm and the comparing algorithms.
It can be seen from Table 2 that the SF-NRL algorithm has certain advantages in the comparative experiment in terms of NMI value, among which the DeepWalk algorithm and Node2Vec algorithm based on Word2Vec have small improvement, while for the LINE algorithm the result is significantly improved.
Among them, the jazz dataset has the least improvement under the DeepWalk algorithm and Node2Vec algorithm and is at a disadvantage in comparison with LINE. The result is due to the tight structure of the jazz dataset, which is very dense with 198 nodes and 2742 edges, and so holds a high average degree, resulting in dramatic data folding. Although global information can be captured, the folding granularity is large, and the over-coarsening phenomenon occurs, which contributes to insignificant improvement.
4.3. Multi-Label Classification
In multi-label classification, PolBooks, Football, and CORA were employed for verification. A total of 10–90% of the original data were selected as training data, and the rest were used as verification data. The classification results were determined by two indexes, Macro-F1 value, and Micro-F1 value. The higher the Macro-F1 value and Micro-F1 value, the better the classification effect. We provide the definitions of Macro-F1 and Micro-F1 in Equations (2)–(5).
where n denotes the number of classes, TP denotes true positive, FP denotes false positive, and FN denotes false negative.
Corresponding to NRL algorithms DeepWalk, Node2Vec, and LINE, this paper proposed SF-NRL(DW), SF-NRL(N2V), SF-NRL(LINE). We compared SF-NRL(DW) with DeepWalk, SF-NRL(N2V) with Node2Vec, and SF-NRL(LINE) with LINE, respectively.
As shown in Figure 6, the F1 values of the comparison experiment between SF-NRL(DW) and DeepWalk are presented in the form of a polyline graph, with Micro-F1 values on the left and Macro-F1 values on the right. From Figure 6, the Micro-F1 of SF-NRL(DW) on the Football dataset is lower than that of DeepWalk when the training set is 10%, and for Macro-F1, the training set is 60%. In addition, the Micro-F1 and Macro-F1 of the SF-NRL(DW) are superior to DeepWalk in all other cases.
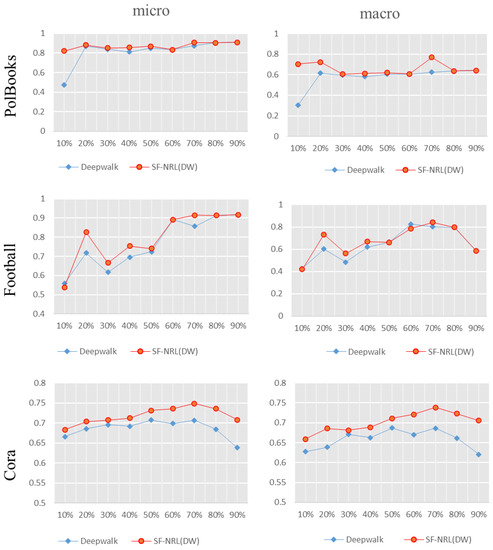
Figure 6.
Comparison of F1 value of the multi-label classification experiment results. The red polyline is the F1 value of SF-NRL(DW) and the blue one is that of DeepWalk.
Figure 7 exhibits the results between SF-NRL(N2V) and Node2Vec. As can be seen from Figure 7, the Macro-F1 value SF-NRL(N2V) on the PolBooks dataset is significantly higher than that of Node2Vec when the training data are 10% and 70%, while the difference is trivial for the remaining proportion of training data. In other cases, the algorithm proposed in this paper is superior to the comparative algorithm Node2Vec.
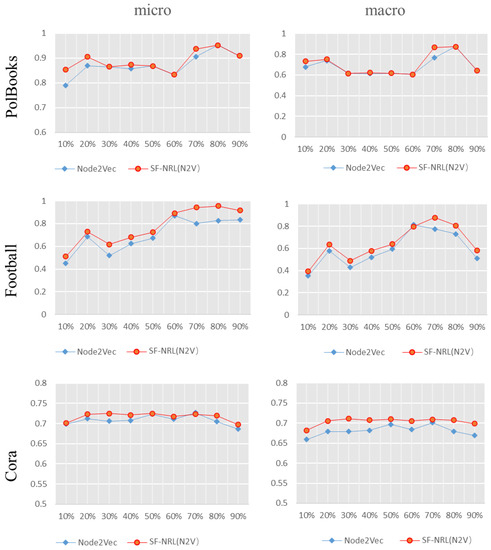
Figure 7.
Comparison of F1 value of the multi-label classification experiment results. The red polyline is the F1 value of SF-NRL(N2V) and the blue one is that of Node2Vec.
Similarly, Figure 8 depicts the comparison between SF-NRL(LINE) and LINE; the left side is Micro-F1 value. From top to bottom, the results on datasets of PolBooks, Football, and CORA are, respectively, shown where the red polyline represents SF-NRL(LINE) and the blue one indicates LINE. In this group of experiments, compared with the NMI value in the community discovery experiment, SF-NRL(LINE) has been greatly improved, and it still performs well in the multi-label classification task. Compared with the comparative algorithm LINE, SF-NRL(LINE) obtains greater improvement in both Micro-F1 and Macro-F1 values.
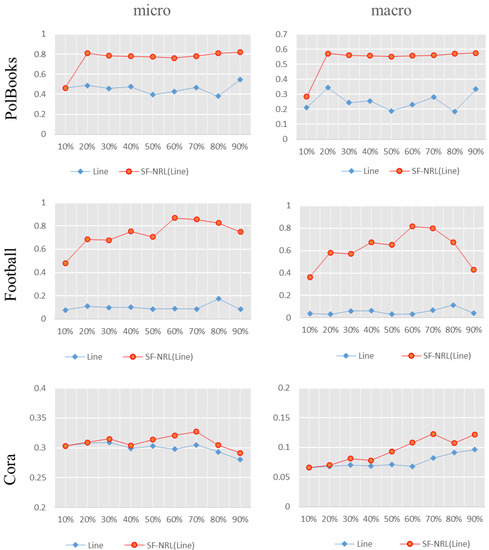
Figure 8.
Comparison of F1 value of the multi-label classification experiment results. The red polyline is the F1 value of SF-NRL(LINE) and the blue one is that of LINE.
According to the experimental results of multi-label classification, we found that the proposed method improves the classification performance of the existing NRL algorithm effectively, and this usage of global information can enable the NRL algorithm to learn more information about the network, rather than just focusing on local information. The proposed method can consider more valuable information based on the original algorithm, and this idea can help scholars to design more effective algorithms to implement graph data mining.
5. Conclusions
These NRL algorithms, which estimate the destination node embedding by obtaining neighborhood information, ignore the problem of global structural information loss. We propose the SF-NRL algorithm to fold the original network into a small-scale network by finding the complete subgraphs in the network, and we employ the complete subgraph as the folding unit to obtain the global topology information of the original network with the folding rules The original network and the folded network are, respectively, found for network representation, and the two kinds of representations are merged to obtain a network representation incorporating both global and local information.
The effectiveness of the SF-NRL algorithm is verified by the community discovery test and the multi-label classification test. Three groups of comparative experiments SF-NRL(DW) vs. DeepWalk, SF-NRL(N2V) vs. Node2Vec, and SF-NRL(LINE) vs. LINE are set up. The experimental results and analysis show the outperformance of the proposed algorithm in community discovery and multi-label classification. Distinguishingly, the comparison between SF-NRL(LINE) and LINE shows nontrivial advantages of the SF-NRL algorithm. Therefore, the effectiveness of the SF-NRL algorithm is verified. The proposed algorithm adequately employs the information that the network topology can provide to improve the performance of existing algorithms. The proposed idea can provide a novel method for researchers to further applications of network global information [24,25]. In future work, we will focus on designing more efficient graph folding strategies and designing more accurate NRL algorithms.
Author Contributions
Conceptualization, D.C., J.Y. and D.W.; Formal analysis, M.N. and Q.G.; Funding acquisition, D.C. and D.W.; Methodology, M.N. and J.Y.; Project administration, D.C., M.N. and D.W.; Resources, M.N.; Software, J.Y.; Supervision, D.C. and D.W.; Validation, D.C.; Visualization, M.N. and Q.G.; Writing—original draft, M.N., J.Y. and Q.G.; Writing—review and editing, D.C. and D.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Qi, J.S.; Liang, X.; Li, Z.Y.; Chen, Y.F.; Xu, Y. Representation Learning of Large-Scale Complex Information Network: Concepts, Methods and Challenges. Jisuanji Xuebao/Chin. J. Comput. 2018, 41, 2394–2420. [Google Scholar] [CrossRef]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. DeepWalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 701–710. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Tang, J.; Qu, M.; Wang, M.; Zhang, M.; Yan, J.; Mei, Q. Line: Large-scale information network embedding. In Proceedings of the 24th International Conference on World Wide Web, Florence, IT, USA, 18–22 May 2015; pp. 1067–1077. [Google Scholar]
- Gregori, E.; Lenzini, L.; Orsini, C. k-clique Communities in the Internet AS-level Topology Graph. In Proceedings of the 2011 31st International Conference on Distributed Computing Systems Workshops, Minneapolis, MN, USA, 20–24 June 2011; pp. 134–139. [Google Scholar]
- Chen, Q.; Wu, T.-T.; Fang, M. Detecting local community structures in complex networks based on local degree central nodes. Phys. A Stat. Mech. Its Appl. 2013, 392, 529–537. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wang, C.; Li, D. MINC-NRL: An Information-Based Approach for Community Detection. Algorithms 2022, 15, 20. [Google Scholar] [CrossRef]
- Lin, G.; Wang, J.; Liao, K.; Zhao, F.; Chen, W. Structure Fusion Based on Graph Convolutional Networks for Node Classification in Citation Networks. Electronics 2020, 9, 432. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Chen, Y. Link prediction based on graph neural networks. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montréal, QC, Canada, 3–8 December 2018; pp. 5171–5181. [Google Scholar]
- Chen, D.; Nie, M.; Wang, J.; Kong, Y.; Wang, D.; Huang, X. Community Detection Based on Graph Representation Learning in Evolutionary Networks. Appl. Sci. 2021, 11, 4497. [Google Scholar] [CrossRef]
- Huang, X.; Chen, D.; Ren, T.; Wang, D. A survey of community detection methods in multilayer networks. Data Min. Knowl. Discov. 2021, 35, 1–45. [Google Scholar] [CrossRef]
- Salamat, A.; Luo, X.; Jafari, A. BalNode2Vec: Balanced Random Walk based Versatile Feature Learning for Networks. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar]
- Chen, H.; Perozzi, B.; Hu, Y.; Skiena, S. Harp: Hierarchical representation learning for networks. arXiv 2017, arXiv:1706.07845. [Google Scholar]
- Liang, J.; Gurukar, S.; Parthasarathy, S. Mile: A multi-level framework for scalable graph embedding. arXiv 2018, arXiv:1802.09612. [Google Scholar]
- Bartal, A.; Jagodnik, K.M. Role-Aware Information Spread in Online Social Networks. Entropy 2021, 23, 1542. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Zhao, Z.; Wang, Y.; Zhang, Z.; Feng, Z. Graphzoom: A multi-level spectral approach for accurate and scalable graph embedding. arXiv 2019, arXiv:1910.02370. [Google Scholar]
- Fang, L.; Zhai, M.; Wang, B. Complete subgraphs in connected graphs and its application to spectral moment. Discret. Appl. Math. 2021, 291, 36–42. [Google Scholar] [CrossRef]
- Khomami, M.M.D.; Rezvanian, A.; Meybodi, M.R.; Bagheri, A. CFIN: A community-based algorithm for finding influential nodes in complex social networks. J. Supercomput. 2021, 77, 2207–2236. [Google Scholar] [CrossRef]
- Adamic, L.A.; Glance, N. The political blogosphere and the 2004 US election: Divided they blog. In Proceedings of the 3rd International Workshop on Link Discovery, Chicago, LI, USA, 21–24 August 2005; pp. 36–43. [Google Scholar]
- Gleiser, P.M.; Danon, L. Community structure in jazz. Adv. Complex Syst. 2003, 6, 565–573. [Google Scholar] [CrossRef] [Green Version]
- Sen, P.; Namata, G.; Bilgic, M.; Getoor, L.; Galligher, B.; Eliassi-Rad, T. Collective classification in network data. AI Mag. 2008, 29, 93. [Google Scholar] [CrossRef] [Green Version]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Society. Ser. C Appl. Stat. 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Cao, S.; Lu, W.; Xu, Q. Grarep: Learning graph representations with global structural information. In Proceedings of the 24th ACM International on Conference on Information and Knowledge Management, Melbourne, VIC, Australia, 19–23 October 2015; pp. 891–900. [Google Scholar]
- Mavromatis, C.; Karypis, G. Graph InfoClust: Leveraging cluster-level node information for unsupervised graph representation learning. arXiv 2020, arXiv:2009.06946. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).