Abstract
Transcription regulation plays an important role in bacterial activity. The operon concept coined by François Jacob and Jacques Monod has had a considerable effect on investigations into gene expression regulation, including modeling. However, most such studies have considered the regulation models devised manually for one or several operons. For that reason, the objective of the present study was automated genome model reconstruction for different bacteria. The suggested algorithm accounted for all possible interactions of transcription factors and their binding sites in an operon’s promoter region. Transcription factor enumeration was performed using the deep-first search technique. The obtained models are of interest for those involved in the research of transcription factor regulatory effects on bacterial gene expression in microbiology and biotechnology.
Keywords:
mathematical frame model; Hill’s generalized functions; bacterial transcription regulation MSC:
93A30; 62P10; 92B99
1. Introduction
The mathematical modeling of gene expression regulation (transcription and translation) has been greatly affected by the operon concept coined by François Jacob and Jacques Monod, who considered it to be a technique of organizing the transcribed genes united within one or several promoters (DNA region to launch RNA synthesis) [1]. Mathematical studies from researchers investigating the regulation effect on gene expression started to appear just a few years after the concept had been published [2,3,4].
Most such models describe the behavior of the most well-studied lactose and tryptophan operons of Escherichia coli [5,6]. Currently, models are built using standard differential [7,8,9] and delay differential equations [10,11,12,13], Hill’s equations [12,14], and their generalized version [15,16,17], stochastic equations [18,19], Boolean network models [20], etc. Such models can describe the interaction of a transcription factor (TF) and a promoter of a certain operon [21,22], TF binding/separation rates in relation to a number of them interacting [23], promoter activity [24,25], impact of transcriptional read-through on gene expression [26], etc.
However, most of the mentioned models have been built manually for one or several operons of certain model organisms. In this paper, we suggest an approach to the automated generation of genome-scale mathematical frame models of bacterial transcription regulation. Such models are not without interest, because they enable investigation of TF effects on bacterial gene expression.
For the sake of demonstration, we applied the suggested approach to the well-known lactose operon and observed how the biological information is converted into mathematical information (Figure 1). The lactose operon has been chosen for being the first discovered and the most-studied of their kind. The figure demonstrates how cistrons (coding proteins of DNA sequences) are transcribed by mRNA followed by proteins (different kinds of lactose).
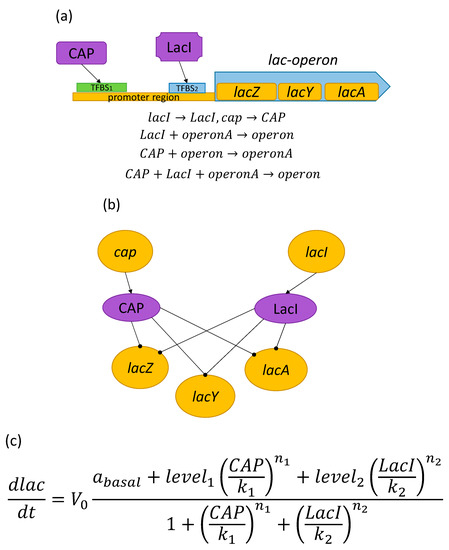
Figure 1.
Lac-operon: from the biochemical scheme to a mathematical model. (a) Depiction of the operon and promoter’s structure. Here, CAP and LacI are transcription factors (TFs); TFBS is a transcription factors biding site (DNA region in the promoter to which TF binds); lacZ, lacY, and lacA are the genes included in the operon; operon and operonA are the operon’s inactive and active forms, respectively. Repressor (LacI) binding converts the active into the inactive form, and the activator (CAP) acts in the opposite way, and simultaneous CAP and LacI binding leaves the operon inactive. (b) Graphical representation of a transcription regulation network (TRN) where the yellow and purple nodes are genes and TFs, respectively, and the arrowed and capped edges mark synthesis and regulation. (c) A mathematical TRN-based model describing lac-operon regulation.
In our approach, transcription regulation was described by Hill’s generalized functions [27], whereas the general models were represented through standard differential equations. The basis of the model was the transcription regulation network (TRN) of a considered bacterium. The TRN is a combination of all the inter-related elements such as TFs, target genes, and the regulatory interactions between them [28,29]. In their graphical representation, the target genes were nodes and the regulatory interactions were non-weighted edges directed either from a TF to a gene (in case of regulation) or from a gene to a TF (in the case of synthesis). The networks were reconstructed based on available information about the structures of operons and their regulators.
This paper presents an algorithm for the reconstruction of mathematical frame models and its software implementation named Operon_Equation.
2. Materials and Methods
First, we considered a TF-assisted transcription regulation process. In such a case, one TF may correspond to several binding cites in several promoters, or one promoter may have several binding sites (TFBSs), i.e., the synthesis can be regulated by either one or several TFs. Commonly, promoters are relatively short; thus, the binding sites sometimes overlap, and sometimes they do not (Figure 2). In the first case, the TFs, which bind to their sites, will not be able to bind simultaneously, and will have to compete for the site.
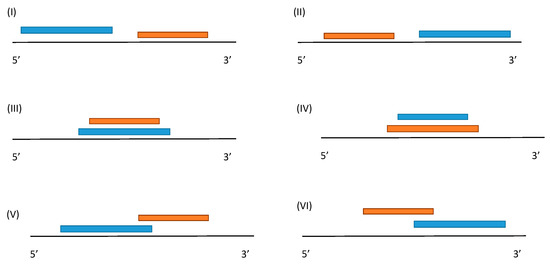
Figure 2.
Possible location of two neighboring TFBSs. The first TFBS is marked blue; the second is marked red. The black line is DNA, where the 5′ to 3′ direction of strand DNA is called the leading strand. (I,II) cases where two TFBS are located in the promoter region that do not overlap, (III,IV) cases are where one TFBS can be embedded in the other, and (V,VI) cases are where one TFBS partially overlaps the other.
For that reason, the modeling should account for all possible regulation scenarios and consider both the number of TFs and whether they bind free or competed binding sites.
Our solution utilized the deep-first search (DFS) [30] technique for the enumeration of TFs and their possible binding combinations. We organized the enumeration in such a way that the subtree can be traversed as far as possible. Figure 3 demonstrates trees for a case of three TFs for a single operon. First, the branch with the first TF is traversed, and then the same procedure is implemented for the second TF (except for the variants that were accounted for in the first branch involving the first TF), and so on. Figure 3a shows the complete tree for the three TFs, whereas in Figure 3b there are not two vertices, because the sites of the second and third TFs overlap. The number of enumeration variations for every operon depends on possible TFBS positions. During enumeration, the TFBS position relative to a transcription start site is neglected; in other words, it is not important from what TF enumeration begins: the results will be the same.
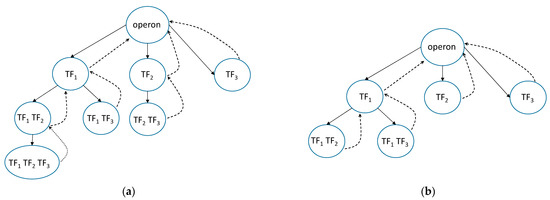
Figure 3.
DFS schemes where the solid arrows mark a direct traversal, and the dotted arrows mark a reverse traversal: (a) all kinds of bindings for 3 TFs, whose TBFSs do not overlap; (b) all kinds of bindings for 3 TFs with overlapping TF2 and TF3 binding sites.
For the mathematical models in question, Hill’s generalized functions were selected [27] because they enable minimization of the description complexity of modeled processes in an absence of detailed knowledge of their development. We used them to describe interactions between TFs, which combinatorially regulate synthesis from operons. This type of function enables us to take parameters into account, such as the effect of a particular TF, a group of TFs, or to consider the activity of a TF, without further complicating the construction of the mathematical formula.
Hill’s generalized functions are a kind of rational non-negative function which can generally be expressed as:
where is an efficiency coefficient that determines the generalized efficiency of the factor group on a given process; is Hill’s coefficient describing the nonlinearity degree of the effect of on a process; and is an activity coefficient that determines a type of the effect of on a process. All the parameters in the equation are non-negative. Parameter is dimensional, its dimensionality is consistent with , and parameters and are dimensionless values.
In 2.2 of [31], a formal description of the fractional functions of various regulators is presented in more detail, including the TF.
3. Results
3.1. Algorithm Description
The idea behind the suggested algorithm is to account for all possible TF/binding site interactions in an operon’s promotor region and to automate this procedure for every operon in a genome In general, the algorithm is executed as follows such is presented on the Figure 4:
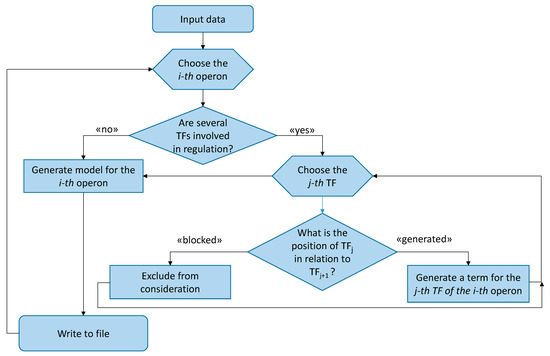
For the purposes of discussion, the algorithm can be subdivided into the following steps:
- Select an operon;
- Set the and parameters;
- Define the number of TFs. If it is one, generate a model; otherwise continue with step 4;
- At TF ≥ 2, check if the binding sites intercross. If they do, neglect this option; otherwise, a summand is generated;
- Repeat step 4 for every TF defined;
- Generate the general formula;
- Return to step 1.
The number of TFs is directly determined from the transcriptional regulatory network, which is fed into the input in text format. An example of such a file can be seen in Supplementary File S1. Depending on how much of the bacterium in question has been studied, different numbers may be known.
The general formula is generated as follows. First, for each TF, we generate a summand for the numerator and then for the denominator, i.e., we expand the fraction:
where and are summands of the numerator and denominator, respectively. These summands are consistently generated for the during one iteration of the algorithm. On the next iteration, the algorithm generates then for interactions and , and so on.
3.2. Approbation
To demonstrate how the algorithm is executed, consider the operon of fabA E. coli taken from RegulonDB (https://regulondb.ccg.unam.mx/index.jsp (accessed on 7 October 2022)). The operon’s description is given in Table 1. It has two TFs (FabR and FadR) whose sites do not intercross.

Table 1.
Transcription regulation of the fabA E. coli operon from RegulonDB where TFBSs mark the absolute positions of binding sites in a genome.
Theoretically, there are three binding options for this operon: FabR binds with a site (Figure 5a); FadR binds with a site (Figure 5b); or they bind simultaneously (Figure 5c).
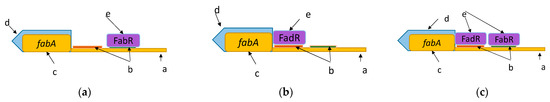
Figure 5.
Binding options for a TF where a—operon’s promotor region; b—TFBSs; c—gene; d—operon; e—TF: (a) FabR binds to a site; (b) FadR binds to a site; (c) FabR and FadR bind simultaneously.
At the first step, the parameter determining the level of the operon’s constitutive expression is set together with , the operon’s initial synthesis rate. Then, the TFBSs are checked for overlapping. In our case, the binding sites did not intercross; thus, we assumed that FabR and FadR could bind simultaneously. Then, we considered the first case (Figure 5a), when just FabR affects the synthesis. In this case, FabR is attributed parameters and ; then, the summand is generated first for the numerator:
where is a parameter to control a transcription activation level via FabR; is the efficiency constant of FabR’s effect on the transcription activation level; and is Hill’s coefficient to characterize the nonlinearity degree of FabR’s effect on the transcription activation level.
Then, for the denominator, the corresponding summand is generated in the following form:
Thus, the summands (3a) and (3b) as the parts of the Equation (3c) nominator and denominator, respectively, form the result of the first iteration of the algorithm (2), describing the regulation of FabR only.
Before considering the second case (Figure 5c), when FabR and FadR jointly affect the operon, parameter , where index 1 describes the effect of FabR, index 2, describing that of FadR, is assigned. The summand is also generated first for the numerator like Equation (4a), then for the denominator like Equation (4b).
where is the parameter determining the level of joint FabR/FadR effect on the transcription; are the constants of FabR/FadR effect efficiency in relation to transcription level; are Hill’s coefficients characterizing the nonlinearity degree of the FabR/FadR effect; and is a parameter responsible for the joint FabR/FadR effect.
As a result, after the second iteration, the following summand is generated:
The summands (4a) and (4b), as the parts of the Equation (4c) nominator and denominator, respectively, describe the co-regulation of the operon of the two TFs: FabR and FadR.
At the following iteration, parameters and are assigned to FadR (Figure 5b) and the generated summands as (3a–3b). Thus, the general formula of operon fabA is written as:
where is used to reduce the formula’s arrangements, and the other parameters remain the same.
Hence, the general formula to describe transcription regulation in operon fabA is expressed as:
where is the mRNA concentration synthesized from operon fabA and is derived from Equation (5).
In order to demonstrate how the method works, we took the regulatory data of the bacterium Pseudomonas aeruginosa PAO1 (GenBank: AE004091.2) from the Prodoric database (https://www.prodoric.de/ (accessed on 7 October 2022)) [32]. For P. aeruginosa PAO1, information is stored on 45 confirmed TRN (including sigma factor 54), which regulates 220 operons. In total, 220 frame models were generated using our algorithm, which can be viewed in the Supplementary Materials (File S2 and Archive S1).
The considered algorithm was implemented as Operon_equations software written in Java, which is available at github.com/tlakhova/Operon_equations. As input data, these require a table-oriented file. Examples of the input and output files can be seen in the Supplementary Materials (File S1, Archive S2 and Supplementary Note).
We also tested the program with different input data. For this, we used a personal computer with the following characteristics: AMD FX-6330 Six-Core Processor, 16 GB RAM.
We considered how the computation time depends on the number of TFs that can bind to their binding sites in a single operon. We also considered when TFs can overlap and when they cannot. Detailed information is presented in Supplementary Table S1. Therefore, we set the upper limit in the program for the number of TFs per operon to 15.
The software enables one to build the models based on the information about biologically significant bacterial strains obtained from the Kurchatov Genomic Centre of the Institute of Cytology Genetics, Novosibirsk, Russian Federation.
4. Discussion
This paper presents an algorithm for the generation of frame models describing genetic regulation in bacteria. This solution is of high potential in terms of building a mathematical frame model of transcription regulation at genomic scale for different kinds of bacteria, including non-model ones. In the algorithm, one model describes transcription regulation in a single operon, which provides a close view on its behavioral patterns and changes in the metabolic pathways. The obtained model can be specified through the experimental data found in dedicated databases and publications.
Understanding the contribution of the transcription factors in the regulation will open an additional field of research for genetic engineers and biotechnologists when modifying bacterial strains.
5. Patents
The program for automatic reconstruction of mathematical models of microbial gene transcription regulation (MicroTranscriptMod). No 2022660245, 01.06.2022.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/math10234480/s1, The Supplementary Materials contain input and output text files both test data and for P. aeruginosa PAO1. We explain what is in these files in Note S1.
Author Contributions
Conceptualization, S.A.L., N.A.K. and Y.G.M.; methodology, T.N.L. and F.V.K.; software, T.N.L., A.M.M., and F.V.K.; validation, S.A.L. and F.V.K.; formal analysis, S.A.L., Y.G.M. and F.V.K.; investigation, T.N.L.; resources, S.A.L. and N.A.K.; data curation, T.N.L., A.M.M. and F.V.K.; writing—original draft preparation, T.N.L.; writing—review and editing, S.A.L. and T.N.L.; visualization, T.N.L.; supervision, S.A.L.; project administration, S.A.L.; funding acquisition, S.A.L. and N.A.K. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Kurchatov Genomic Centre of the Institute of Cytology and Genetics SB RAS No. 075-15-2019-1662 and the Ministry of Science and Higher Education budget project No. FWNR-2022-0006.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Program and test data are available at github.com/tlakhova/Operon_equations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jacob, F.; Monod, J. Genetic Regulatory Mechanisms in the Synthesis of Proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, B.C. Oscillatory Behavior in Enzymatic Control Processes. Adv. Enzyme Regul. 1965, 3, 425–437. [Google Scholar] [CrossRef]
- Griffith, J.S. Mathematics of Cellular Control Processes II. Positive Feedback to One Gene. J. Theor. Biol. 1968, 20, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.S. Mathematics of Cellular Control Processes I. Negative Feedback to One Gene. J. Theor. Biol. 1968, 20, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Mackey, M.C.; Santillán, M.; Yildirim, N. Modeling Operon Dynamics: The Tryptophan and Lactose Operons as Paradigms. C. R. Biol. 2004, 327, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Mackey, M.C.; Santilĺan, M.; Tyran-Kamínska, M.; Zeron, E.S. The Utility of Simple Mathematical Models in Understanding Gene Regulatory Dynamics. In Silico Biol. 2015, 12, 23–53. [Google Scholar] [CrossRef] [PubMed]
- Zorzan, I.; Del Favero, S.; Giaretta, A.; Manganelli, R.; Di Camillo, B.; Schenato, L. Mathematical Modelling of SigE Regulatory Network Reveals New Insights into Bistability of Mycobacterial Stress Response. BMC Bioinform. 2021, 22, 558. [Google Scholar] [CrossRef] [PubMed]
- Gerosa, L.; Kochanowski, K.; Heinemann, M.; Sauer, U. Dissecting Specific and Global Transcriptional Regulation of Bacterial Gene Expression. Mol. Syst. Biol. 2013, 9, 658. [Google Scholar]
- Youlden, G.H.; Ricci, V.; Wang-Kan, X.; Piddock, L.J.V.; Jabbari, S.; King, J.R. Time Dependent Asymptotic Analysis of the Gene Regulatory Network of the AcrAB-TolC Efflux Pump System in Gram-Negative Bacteria. J. Math. Biol. 2021, 82, 31. [Google Scholar] [CrossRef] [PubMed]
- Chávez, J.; Barberena-Jonas, C.; Sotelo-Fonseca, J.E.; Alquicira-Hernández, J.; Salgado, H.; Collado-Torres, L.; Reyes, A. Programmatic Access to Bacterial Regulatory Networks with Regutools. Bioinformatics 2020, 36, 4532–4534. [Google Scholar] [CrossRef]
- Zamora-Chimal, C.; Santillán, M.; Rodríguez-González, J. Influence of the Feedback Loops in the Trp Operon of B. Subtilis on the System Dynamic Response and Noise Amplitude. J. Theor. Biol. 2012, 310, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Liu, H.; Yan, F. Oscillation Dynamic Mechanism Driven by Time Delays in the Competent Gene Regulatory Circuit of B. Subtilis. Int. J. Biomath. 2022, 15, 2250017. [Google Scholar] [CrossRef]
- Zhao, N.; Liu, H.; Yan, F. Oscillation Dynamics of MeKS Core Module Containing Positive and Negative Feedback Loops with Time Delay. Phys. A Stat. Mech. Its Appl. 2020, 538, 122729. [Google Scholar] [CrossRef]
- Bhartiya, S.; Rawool, S.; Venkatesh, K.V. Dynamic Model of Escherichia Coli Tryptophan Operon Shows an Optimal Structural Design. Eur. J. Biochem. 2003, 270, 2644–2651. [Google Scholar] [CrossRef] [PubMed]
- Likhoshvai, V.A.; Stepanova, T.Y.; Zadorozhnyi, A.V.; Tikunova, N.V.; Khlebodarova, T.M. Escherichia Coli Dps Gene Expression in Response to Toxic Agents: Analysis and Mathematical Modeling. VOGiS Her. 2009, 13, 731–740. (In Russian) [Google Scholar]
- Khlebodarova, T.M.; Oshchepkov, D.Y.; Tikunova, N.V.; Babkin, I.V.; Gruzdev, A.D.; Likhoshvai, V.A. Reconstruction of the Mechanisms That Regulate the Expression of the Escherihia Coli YfiA Gene under Stress Conditions. Russ. J. Genet. Appl. Res. 2013, 3, 271–278. [Google Scholar] [CrossRef]
- Khlebodarova, T.M.; Stepanova, T.Y.; Oshchepkov, D.Y.; Babkin, I.V.; Tikunova, N.V.; Likhoshvai, V.A. Mechanisms Regulating Escherichia coli Dps Gene Expression under Stress: Reconstruction on Kinetic Data. Math. Biol. Bioinforma. 2015, 10, 1–14. [Google Scholar] [CrossRef]
- Zeron, E.S.; Santillán, M. Distributions for Negative-Feedback-Regulated Stochastic Gene Expression: Dimension Reduction and Numerical Solution of the Chemical Master Equation. J. Theor. Biol. 2010, 264, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, A.; Rybakova, K.N.; Bruggeman, F.J. Transcription Stochasticity of Complex Gene Regulation Models. Biophys. J. 2012, 103, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Kerr, R.; Jabbari, S.; Blair, J.M.A.; Johnston, I.G. Dynamic Boolean Modelling Reveals the Influence of Energy Supply on Bacterial Efflux Pump Expression. J. R. Soc. Interface 2022, 19, 20210771. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Zabet, N.R.; Mitavskiy, B. Models of Transcription Factor Binding: Sensitivity of Activation Functions to Model Assumptions. J. Theor. Biol. 2009, 257, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.; Razo-Mejia, M.; Phillips, R. Reconciling Kinetic and Thermodynamic Models of Bacterial Transcription. PLoS Comput. Biol. 2021, 17, e1008572. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, V.P. Mathematical Aspects of the Regulation of Gene Transcription by Promoters. Math. Biosci. 2017, 283, 84–90. [Google Scholar] [CrossRef]
- Lee, S.B.; Bailey, J.E. Genetically Structured Models for Lac Promoter–Operator Function in the Chromosome and in Multicopy Plasmids: Lac Promoter Function. Biotechnol. Bioeng. 1984, 26, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Berthoumieux, S.; De Jong, H.; Baptist, G.; Pinel, C.; Ranquet, C.; Ropers, D.; Geiselmann, J. Shared Control of Gene Expression in Bacteria by Transcription Factors and Global Physiology of the Cell. Mol. Syst. Biol. 2013, 9, 634. [Google Scholar] [CrossRef] [PubMed]
- Nagy-Staron, A.; Tomasek, K.; Caruso Carter, C.; Sonnleitner, E.; Kavčič, B.; Paixão, T.; Guet, C.C. Local Genetic Context Shapes the Function of a Gene Regulatory Network. eLife 2021, 10, e65993. [Google Scholar] [CrossRef] [PubMed]
- Likhoshvai, V.; Ratushny, A. Generalized Hill Function Method for Modeling Molecular Processes. J. Bioinform. Comput. Biol. 2007, 5, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M.; Luscombe, N.M.; Aravind, L.; Gerstein, M.; Teichmann, S.A. Structure and Evolution of Transcriptional Regulatory Networks. Curr. Opin. Struct. Biol. 2004, 14, 283–291. [Google Scholar] [CrossRef]
- Saint-André, V. Computational Biology Approaches for Mapping Transcriptional Regulatory Networks. Comput. Struct. Biotechnol. J. 2021, 19, 4884–4895. [Google Scholar] [CrossRef] [PubMed]
- Skiena, S.S. Graph Traversal. In The Algorithm Design Manual; Springer: London, UK, 2012; pp. 145–190. ISBN 978-1-84800-070-4. [Google Scholar]
- Ratushny, A.V.; Ramsey, S.A.; Aitchison, J.D. Mathematical Modeling of Biomolecular Network Dynamics. In Network Biology: Methods and Applications; Cagney, G., Emili, A., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; Volume 781, pp. 415–433. ISBN 978-1-61779-275-5. [Google Scholar]
- Dudek, C.-A.; Jahn, D. PRODORIC: State-of-the-Art Database of Prokaryotic Gene Regulation. Nucleic Acids Res. 2022, 50, D295–D302. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).