A Two-Step Data Normalization Approach for Improving Classification Accuracy in the Medical Diagnosis Domain
Abstract
1. Introduction
- The existence of historical data of different volumes intended for analysis;
- The need to analyze both enormous and tiny datasets that are difficult for humans to handle;
- A large number of features that may affect the patient’s diagnosis and are difficult or impossible for doctors to take into account during diagnosis;
- Complex, usually hidden, nonlinear interdependencies between the features of a particular dataset, which are very difficult to identify at first glance but are easily identified and taken into account by a specific machine learning model;
- The high classification or prediction accuracy of machine learning models, which exclude human factors and subjectivism and can serve as a source of additional information to the doctor.
- We develop a new two-step method for tabular data normalization that considers the interdependencies between the features of each observation and the absolute values of each of these features. The proposed method reduces the number of extrapolation problems for vectors at a distance from the training sample;
- We demonstrate the high efficiency of Decision Tree and Extra Trees classifiers based on the developed data normalization method for both binary and multiclass classification tasks using different medical datasets;
- We experimentally establish an increase in the classification accuracy based on several machine learning methods that use the developed two-step data normalization method compared with other existing methods.
2. The State-of-the-Art
3. Materials and Methods
The Proposed Two-Step Data Normalization Method
- Initial normalization for each -th column () of a given set of tabular data, according to the scheme of the maximal value of the absolute element in each column, according to the following formula:
- 2.
- Calculation of the norm of each vector using from (2) according to the following expression:
- 3.
- Normalization of each separate vector from the dataset, taking into account its norm according to the expression:
- Then, we expand each vector (3) of the dataset using each corresponding norm (2):
- We perform for each extended vector (5) transformations similar to procedure (3):
- A normalized dataset for each column and each row;
- A dataset that has been extended by one additional feature compared with the original, non-normalized dataset;
- A dataset that considers both the interdependencies between the features of each separate vector and their absolute values.
4. Modeling and Results
4.1. Datasets Used for the Modeling
4.2. Results
- Accuracy score;
- Precision score;
- Recall score; and
- F1-score
5. Comparison and Discussion
- Vector Scaler;
- Max Abs Scaler;
- Min Max Scaler;
- Standard Scaler;
- Robust Scaler.
- The Max Abs Scaler, Min-Max Scaler, Standard Scaler, and Robust Scaler do not provide a significant difference in the accuracy of the investigated classifiers;
- The proposed data normalization method provides an increase in the classification accuracy of 1 to 5% compared with the existing methods;
- The proposed data normalization method increases the classification accuracy from 1% to 3% compared with the most similar data normalization method (the Vector Scaler).
- The Max Abs Scaler, Min-Max Scaler, Standard Scaler, and Robust Scaler affect the accuracy of the investigated classifiers;
- The proposed data normalization method provides both a significant (1% to 6%) increase in the accuracy of the classifiers compared with the above-mentioned methods for normalization and the same level of accuracy as the Vector Scaler;
- The proposed data normalization method improves the accuracy of the classifiers compared with the most similar data normalization method (the Vector Scaler).
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| ML-Based Classifier | Parameters |
|---|---|
| AdaBoost Classifier | base_estimator = None, n_estimators = 100, learning_rate = 1.0, algorithm = ‘SAMME.R’, random_state = None |
| Bagging Classifier | base_estimator = None, n_estimators = 100, max_samples = 1.0, max_features = 1.0, bootstrap = True, bootstrap_features = False, oob_score = False, warm_start = False, n_jobs = None, random_state = None, verbose = 0 |
| Decision Tree Classifier | max_depth = None, min_samples_split = 2, random_state = 0 |
| Extra Trees Classifier | n_estimators = 100, max_depth = None, min_samples_split = 2, random_state = 0 |
| Gradient Boosting Classifier | loss = ‘deviance’, learning_rate = 0.1, n_estimators = 100, subsample = 1.0, criterion = ‘friedman_mse’, min_samples_split = 2, min_samples_leaf = 1, min_weight_fraction_leaf = 0.0, max_depth = 3, min_impurity_decrease = 0.0, init = None, random_state = None, max_features = None, verbose = 0, max_leaf_nodes = None, warm_start = False, validation_fraction = 0.1, n_iter_no_change = None, tol = 0.0001, ccp_alpha = 0.0 |
| Random Forest Classifier | n_estimators = 100, max_depth = None, min_samples_split = 2, random_state = 0 |
| Dataset Title | Classifier | Proposed Scaler | Vector Scaler | Max Abs Scaler | Min Max Scaler | Standard Scaler | Robust Scaler |
|---|---|---|---|---|---|---|---|
| Heart Attack Analysis & Prediction Dataset | AdaBoost | 0.770 | 0.770 | 0.787 | 0.787 | 0.770 | 0.787 |
| Bagging | 0.787 | 0.787 | 0.852 | 0.836 | 0.836 | 0.852 | |
| Decision Tree | 0.754 | 0.705 | 0.738 | 0.738 | 0.738 | 0.738 | |
| Extra Trees | 0.803 | 0.787 | 0.770 | 0.770 | 0.770 | 0.770 | |
| Gradient Boosting | 0.770 | 0.770 | 0.787 | 0.787 | 0.787 | 0.787 | |
| Random Forest | 0.787 | 0.803 | 0.820 | 0.820 | 0.820 | 0.836 | |
| Blood Transfusion Service Center Dataset | AdaBoost | 0.707 | 0.707 | 0.733 | 0.733 | 0.727 | 0.733 |
| Bagging | 0.713 | 0.700 | 0.700 | 0.713 | 0.713 | 0.720 | |
| Decision Tree | 0.680 | 0.680 | 0.667 | 0.667 | 0.667 | 0.673 | |
| Extra Trees | 0.740 | 0.740 | 0.687 | 0.687 | 0.687 | 0.687 | |
| Gradient Boosting | 0.707 | 0.707 | 0.753 | 0.753 | 0.753 | 0.753 | |
| Random Forest | 0.720 | 0.720 | 0.713 | 0.707 | 0.720 | 0.720 | |
| Heart Failure Prediction Dataset | AdaBoost | 0.800 | 0.800 | 0.750 | 0.750 | 0.750 | 0.750 |
| Bagging | 0.767 | 0.833 | 0.767 | 0.767 | 0.800 | 0.783 | |
| Decision Tree | 0.750 | 0.733 | 0.700 | 0.700 | 0.700 | 0.700 | |
| Extra Trees | 0.733 | 0.767 | 0.767 | 0.767 | 0.767 | 0.767 | |
| Gradient Boosting | 0.800 | 0.800 | 0.800 | 0.800 | 0.800 | 0.800 | |
| Random Forest | 0.817 | 0.833 | 0.800 | 0.800 | 0.817 | 0.817 | |
| Maternal Health Risk Dataset | AdaBoost | 0.562 | 0.562 | 0.690 | 0.690 | 0.690 | 0.690 |
| Bagging | 0.852 | 0.837 | 0.833 | 0.837 | 0.833 | 0.833 | |
| Decision Tree | 0.857 | 0.842 | 0.818 | 0.818 | 0.818 | 0.818 | |
| Extra Trees | 0.857 | 0.842 | 0.852 | 0.852 | 0.852 | 0.852 | |
| Gradient Boosting | 0.783 | 0.788 | 0.783 | 0.783 | 0.783 | 0.783 | |
| Random Forest | 0.837 | 0.828 | 0.833 | 0.837 | 0.842 | 0.833 | |
| Breast Tissue Dataset | AdaBoost | 0.429 | 0.429 | 0.524 | 0.524 | 0.524 | 0.524 |
| Bagging | 0.667 | 0.619 | 0.524 | 0.571 | 0.619 | 0.571 | |
| Decision Tree | 0.571 | 0.476 | 0.619 | 0.619 | 0.619 | 0.619 | |
| Extra Trees | 0.667 | 0.619 | 0.476 | 0.476 | 0.476 | 0.476 | |
| Gradient Boosting | 0.571 | 0.571 | 0.619 | 0.619 | 0.619 | 0.619 | |
| Random Forest | 0.571 | 0.667 | 0.571 | 0.571 | 0.571 | 0.571 | |
| Contraceptive Method Choice Dataset | AdaBoost | 0.475 | 0.475 | 0.512 | 0.512 | 0.512 | 0.512 |
| Bagging | 0.508 | 0.502 | 0.495 | 0.485 | 0.498 | 0.492 | |
| Decision Tree | 0.451 | 0.447 | 0.475 | 0.475 | 0.475 | 0.471 | |
| Extra Trees | 0.485 | 0.478 | 0.475 | 0.475 | 0.475 | 0.475 | |
| Gradient Boosting | 0.539 | 0.539 | 0.532 | 0.532 | 0.532 | 0.532 | |
| Random Forest | 0.502 | 0.515 | 0.502 | 0.495 | 0.498 | 0.508 |
| Dataset Title | Classifier | Proposed Scaler | Vector Scaler | Max Abs Scaler | Min Max Scaler | Standard Scaler | Robust Scaler |
|---|---|---|---|---|---|---|---|
| Heart Attack Analysis & Prediction Dataset | AdaBoost | 0.794 | 0.794 | 0.817 | 0.817 | 0.806 | 0.817 |
| Bagging | 0.827 | 0.827 | 0.883 | 0.868 | 0.865 | 0.880 | |
| Decision Tree | 0.795 | 0.757 | 0.778 | 0.778 | 0.778 | 0.778 | |
| Extra Trees | 0.842 | 0.827 | 0.800 | 0.800 | 0.800 | 0.800 | |
| Gradient Boosting | 0.806 | 0.806 | 0.827 | 0.827 | 0.827 | 0.827 | |
| Random Forest | 0.822 | 0.838 | 0.849 | 0.849 | 0.849 | 0.865 | |
| Blood Transfusion Service Center Dataset | AdaBoost | 0.817 | 0.817 | 0.831 | 0.831 | 0.826 | 0.831 |
| Bagging | 0.812 | 0.800 | 0.805 | 0.812 | 0.809 | 0.817 | |
| Decision Tree | 0.788 | 0.784 | 0.779 | 0.779 | 0.779 | 0.784 | |
| Extra Trees | 0.831 | 0.831 | 0.797 | 0.797 | 0.797 | 0.797 | |
| Gradient Boosting | 0.810 | 0.810 | 0.843 | 0.843 | 0.843 | 0.843 | |
| Random Forest | 0.814 | 0.814 | 0.811 | 0.805 | 0.816 | 0.816 | |
| Heart Failure Prediction Dataset | AdaBoost | 0.625 | 0.625 | 0.595 | 0.595 | 0.595 | 0.595 |
| Bagging | 0.533 | 0.667 | 0.533 | 0.533 | 0.625 | 0.581 | |
| Decision Tree | 0.571 | 0.556 | 0.400 | 0.400 | 0.400 | 0.400 | |
| Extra Trees | 0.4 | 0.462 | 0.462 | 0.462 | 0.462 | 0.462 | |
| Gradient Boosting | 0.600 | 0.600 | 0.625 | 0.625 | 0.625 | 0.625 | |
| Random Forest | 0.621 | 0.667 | 0.625 | 0.625 | 0.645 | 0.645 | |
| Maternal Health Risk Dataset | AdaBoost | 0.563 | 0.563 | 0.692 | 0.692 | 0.692 | 0.692 |
| Bagging | 0.852 | 0.837 | 0.833 | 0.838 | 0.833 | 0.832 | |
| Decision Tree | 0.857 | 0.843 | 0.819 | 0.819 | 0.819 | 0.819 | |
| Extra Trees | 0.857 | 0.842 | 0.853 | 0.853 | 0.853 | 0.853 | |
| Gradient Boosting | 0.783 | 0.788 | 0.783 | 0.783 | 0.783 | 0.783 | |
| Random Forest | 0.837 | 0.828 | 0.833 | 0.838 | 0.843 | 0.833 | |
| Breast Tissue Dataset | AdaBoost | 0.324 | 0.376 | 0.475 | 0.475 | 0.475 | 0.475 |
| Bagging | 0.683 | 0.627 | 0.511 | 0.568 | 0.615 | 0.568 | |
| Decision Tree | 0.638 | 0.534 | 0.628 | 0.628 | 0.628 | 0.628 | |
| Extra Trees | 0.695 | 0.659 | 0.511 | 0.511 | 0.511 | 0.511 | |
| Gradient Boosting | 0.598 | 0.607 | 0.618 | 0.618 | 0.618 | 0.635 | |
| Random Forest | 0.591 | 0.678 | 0.568 | 0.568 | 0.568 | 0.568 | |
| Contraceptive Method Choice Dataset | AdaBoost | 0.476 | 0.476 | 0.510 | 0.510 | 0.510 | 0.510 |
| Bagging | 0.508 | 0.502 | 0.496 | 0.484 | 0.498 | 0.492 | |
| Decision Tree | 0.452 | 0.447 | 0.475 | 0.475 | 0.475 | 0.471 | |
| Extra Trees | 0.482 | 0.476 | 0.474 | 0.474 | 0.474 | 0.474 | |
| Gradient Boosting | 0.537 | 0.537 | 0.534 | 0.534 | 0.534 | 0.534 | |
| Random Forest | 0.501 | 0.514 | 0.502 | 0.494 | 0.499 | 0.509 |
References
- Kumar, P.; Kumar, Y.; Tawhid, M.A. (Eds.) Machine Learning, Big Data, and IoT for Medical Informatics; Intelligent Data Centric Systems; Academic Press: Cambridge, MA, USA, 2021; ISBN 978-0-12-821777-1. [Google Scholar]
- Hu, Z.; Tereykovski, I.A.; Tereykovska, L.O.; Pogorelov, V.V. Determination of Structural Parameters of Multilayer Perceptron Designed to Estimate Parameters of Technical Systems. IJISA 2017, 9, 57–62. [Google Scholar] [CrossRef]
- Shakhovska, N.; Yakovyna, V.; Kryvinska, N. An Improved Software Defect Prediction Algorithm Using Self-Organizing Maps Combined with Hierarchical Clustering and Data Preprocessing. In International Conference on Database and Expert Systems Applications; Hartmann, S., Küng, J., Kotsis, G., Tjoa, A.M., Khalil, I., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 414–424. [Google Scholar]
- Hu, Z.; Ivashchenko, M.; Lyushenko, L.; Klyushnyk, D. Artificial Neural Network Training Criterion Formulation Using Error Continuous Domain. IJMECS 2021, 13, 13–22. [Google Scholar] [CrossRef]
- Tlebaldinova, A.; Denissova, N.; Baklanova, O.; Krak, I.; Györök, G. Normalization of Vehicle License Plate Images Based on Analyzing of Its Specific Features for Improving the Quality Recognition. Acta Polytech. Hung. 2020, 17, 193–206. [Google Scholar] [CrossRef]
- Hu, Z.; Bodyanskiy, Y.V.; Kulishova, N.Y.; Tyshchenko, O.K. A Multidimensional Extended Neo-Fuzzy Neuron for Facial Expression Recognition. IJISA 2017, 9, 29–36. [Google Scholar] [CrossRef]
- Izonin, I.; Tkachenko, R. Universal Intraensemble Method Using Nonlinear AI Techniques for Regression Modeling of Small Medical Data Sets. In Cognitive and Soft Computing Techniques for the Analysis of Healthcare Data; Elsevier: Amsterdam, The Netherlands, 2022; pp. 123–150. ISBN 978-0-323-85751-2. [Google Scholar]
- Krak, I.; Barmak, O.; Manziuk, E. Using Visual Analytics to Develop Human and Machine-centric Models: A Review of Approaches and Proposed Information Technology. Comput. Intell. 2020, 1–26. [Google Scholar] [CrossRef]
- Krak, Y.V. Dynamics of Manipulation Robots: Numerical-Analytical Method of Formation and Investigation of Computational Complexity. J. Automat. Inf. Sci. 1999, 31, 121–128. [Google Scholar] [CrossRef]
- Babichev, S.; Lytvynenko, V.; Škvor, J.; Korobchynskyi, M.; Voronenko, M. Information Technology of Gene Expression Profiles Processing for Purpose of Gene Regulatory Networks Reconstruction. In Proceedings of the 2018 IEEE Second International Conference on Data Stream Mining Processing (DSMP), Lviv, Ukraine, 21–25 August 2018; pp. 336–341. [Google Scholar]
- Lytvynenko, V.; Wojcik, W.; Fefelov, A.; Lurie, I.; Savina, N.; Voronenko, M.; Boskin, O.; Smailova, S. Hybrid Methods of GMDH-Neural Networks Synthesis and Training for Solving Problems of Time Series Forecasting. In Lecture Notes in Computational Intelligence and Decision Making; Lytvynenko, V., Babichev, S., Wójcik, W., Vynokurova, O., Vyshemyrskaya, S., Radetskaya, S., Eds.; Advances in Intelligent Systems and Computing; Springer International Publishing: Cham, Switzerland, 2020; Volume 1020, pp. 513–531. ISBN 978-3-030-26473-4. [Google Scholar]
- Hassler, A.P.; Menasalvas, E.; García-García, F.J.; Rodríguez-Mañas, L.; Holzinger, A. Importance of Medical Data Preprocessing in Predictive Modeling and Risk Factor Discovery for the Frailty Syndrome. BMC Med. Inform. Decis. Mak. 2019, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Singh, B. Investigating the Impact of Data Normalization on Classification Performance. Appl. Soft Comput. 2020, 97, 105524. [Google Scholar] [CrossRef]
- Pandey, A.; Jain, A. Comparative Analysis of KNN Algorithm Using Various Normalization Techniques. IJCNIS 2017, 9, 36–42. [Google Scholar] [CrossRef]
- Alshdaifat, E.; Alshdaifat, D.; Alsarhan, A.; Hussein, F.; El-Salhi, S.M.F.S. The Effect of Preprocessing Techniques, Applied to Numeric Features, on Classification Algorithms’ Performance. Data 2021, 6, 11. [Google Scholar] [CrossRef]
- Polatgil, Mesut. Investigation of the Effect of Normalization Methods on ANFIS Success: Forestfire and Diabets Datasets. IJITCS 2022, 14, 1–8. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Mahmud, M.A.P.; Saha, P.K.; Gupta, K.D.; Siddique, Z. Effect of Data Scaling Methods on Machine Learning Algorithms and Model Performance. Technologies 2021, 9, 52. [Google Scholar] [CrossRef]
- Vafaei, N.; Ribeiro, R.A.; Camarinha-Matos, L.M. Normalization Techniques for Multi-Criteria Decision Making: Analytical Hierarchy Process Case Study. In Technological Innovation for Cyber-Physical Systems; Camarinha-Matos, L.M., Falcão, A.J., Vafaei, N., Najdi, S., Eds.; IFIP Advances in Information and Communication Technology; Springer International Publishing: Cham, Switzerland, 2016; Volume 470, pp. 261–269. ISBN 978-3-319-31164-7. [Google Scholar]
- Izonin, I.; Tkachenko, R.; Shakhovska, N.; Ilchyshyn, B.; Gregus, M.; Strauss, C. Towards Data Normalization Task for the Efficient Mining of Medical Data. In Proceedings of the 2022 12th International Conference on Advanced Computer Information Technologies, Spišská Kapitula, Slovakia, 26–28 September 2022; pp. 1–5. [Google Scholar]
- Nam, S.L.; de la Mata, A.P.; Dias, R.P.; Harynuk, J.J. Towards Standardization of Data Normalization Strategies to Improve Urinary Metabolomics Studies by GC×GC-TOFMS. Metabolites 2020, 10, 376. [Google Scholar] [CrossRef]
- Viallon, V.; His, M.; Rinaldi, S.; Breeur, M.; Gicquiau, A.; Hemon, B.; Overvad, K.; Tjønneland, A.; Rostgaard-Hansen, A.L.; Rothwell, J.A.; et al. A New Pipeline for the Normalization and Pooling of Metabolomics Data. Metabolites 2021, 11, 631. [Google Scholar] [CrossRef]
- Isaksson, F.; Lundy, L.; Hedström, A.; Székely, A.J.; Mohamed, N. Evaluating the Use of Alternative Normalization Approaches on SARS-CoV-2 Concentrations in Wastewater: Experiences from Two Catchments in Northern Sweden. Environments 2022, 9, 39. [Google Scholar] [CrossRef]
- Chumachenko, D.; Sokolov, O.; Yakovlev, S. Fuzzy Recurrent Mappings in Multiagent Simulation of Population Dynamics Systems. IJC 2020, 19, 290–297. [Google Scholar] [CrossRef]
- Strontsitska, A.-O.; Pavliuk, O.; Dunaev, R.; Derkachuk, R. Forecast of the Number of New Patients and Those Who Died from COVID-19 in Bahrain. In Proceedings of the 2020 International Conference on Decision Aid Sciences and Application (DASA), Sakheer, Bahrain, 8 November 2020; pp. 422–426. [Google Scholar]
- Mochurad, L.; Hladun, Y. Modeling of Psychomotor Reactions of a Person Based on Modification of the Tapping Test. Int. J. Comput. 2021, 20, 1–10, in press. [Google Scholar] [CrossRef]
- Pavliuk, O.; Strontsitska, A.-O. Combined Machine Learning Model for Covid-19 Analysis and Forecasting in Ukraine. In The International Conference on Artificial Intelligence and Logistics Engineering; Hu, Z., Zhang, Q., Petoukhov, S., He, M., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 16–26. [Google Scholar]
- Hovorushchenko, T.; Pavlova, O. Method of Activity of Ontology-Based Intelligent Agent for Evaluating Initial Stages of the Software Lifecycle. In Recent Developments in Data Science and Intelligent Analysis of Information; Chertov, O., Mylovanov, T., Kondratenko, Y., Kacprzyk, J., Kreinovich, V., Stefanuk, V., Eds.; Advances in Intelligent Systems and Computing; Springer International Publishing: Cham, Switzerland, 2019; Volume 836, pp. 169–178. ISBN 978-3-319-97884-0. [Google Scholar]
- API Reference. Available online: https://scikit-learn/stable/modules/classes.html (accessed on 8 May 2022).
- Babenko, V.; Panchyshyn, A.; Zomchak, L.; Nehrey, M.; Artym-Drohomyretska, Z.; Lahotskyi, T. Classical Machine Learning Methods in Economics Research: Macro and Micro Level Examples. Wseas Trans. Bus. Econ. 2021, 18, 209–217. [Google Scholar] [CrossRef]
- Rabcan, J.; Levashenko, V.; Zaitseva, E.; Kvassay, M.; Subbotin, S. Application of Fuzzy Decision Tree for Signal Classification. IEEE Trans. Ind. Inf. 2019, 15, 5425–5434. [Google Scholar] [CrossRef]
- Rawat, B.; Dwivedi, S.K. Selecting Appropriate Metrics for Evaluation of Recommender Systems. IJITCS 2019, 11, 14–23. [Google Scholar] [CrossRef]
- Aamir, M.; Rahman, Z.; Ahmed Abro, W.; Tahir, M.; Mustajar Ahmed, S. An Optimized Architecture of Image Classification Using Convolutional Neural Network. IJIGSP 2019, 11, 30–39. [Google Scholar] [CrossRef]
- Khavalko, V.; Tsmots, I.; Kostyniuk, A.; Strauss, C. Classification and Recognition of Medical Images Based on the SGTM Neuroparadigm. In Proceedings of the 2nd International Workshop on Informatics & Data-Driven Medicine (IDDM 2019), Lviv, Ukraine, 11–13 November 2019; Volume 2488, pp. 234–245. [Google Scholar]
- Bodyanskiy, Y.; Vynokurova, O.; Savvo, V.; Tverdokhlib, T.; Mulesa, P. Hybrid Clustering-Classification Neural Network in the Medical Diagnostics of the Reactive Arthritis. IJISA 2016, 8, 1–9. [Google Scholar] [CrossRef]
- Perova, I.; Pliss, I. Deep Hybrid System of Computational Intelligence with Architecture Adaptation for Medical Fuzzy Diagnostics. IJISA 2017, 9, 12–21. [Google Scholar] [CrossRef][Green Version]
- Dhar, P.; Rahman, M.S.; Abedin, Z. Classification of Leaf Disease Using Global and Local Features. IJITCS 2022, 14, 43–57. [Google Scholar] [CrossRef]
- Singh, A.K.; Shukla, V.P.; Biradar, S.R.; Tiwari, S. Enhanced Performance of Multi Class Classification of Anonymous Noisy Images. IJIGSP 2014, 6, 27–34. [Google Scholar] [CrossRef][Green Version]
- Heart Attack Analysis & Prediction Dataset. Available online: https://www.kaggle.com/rashikrahmanpritom/heart-attack-analysis-prediction-dataset (accessed on 8 May 2022).
- Datopian Blood Transfusion Service Center. Available online: https://datahub.io/machine-learning/blood-transfusion-service-center#data (accessed on 6 April 2022).
- Heart Failure Prediction. Available online: https://www.kaggle.com/andrewmvd/heart-failure-clinical-data (accessed on 8 May 2022).
- UCI Machine Learning Repository: Maternal Health Risk Data Set Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Maternal+Health+Risk+Data+Set (accessed on 8 May 2022).
- UCI Machine Learning Repository: Breast Tissue Data Set. Available online: http://archive.ics.uci.edu/ml/datasets/breast+tissue (accessed on 8 May 2022).
- UCI Machine Learning Repository: Contraceptive Method Choice Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Contraceptive+Method+Choice (accessed on 8 May 2022).
- Oliinyk, A.; Fedorchenko, I.; Stepanenko, A.; Rud, M.; Goncharenko, D. Implementation of Evolutionary Methods of Solving the Travelling Salesman Problem in a Robotic Warehouse. In Data-Centric Business and Applications; Radivilova, T., Ageyev, D., Kryvinska, N., Eds.; Lecture Notes on Data Engineering and Communications Technologies; Springer International Publishing: Cham, Switzerland, 2021; Volume 48, pp. 263–292. ISBN 978-3-030-43069-6. [Google Scholar]
- Kumar, M.B.P.; Amaresh Savadatti, D.M. Virobot the Artificial Assistant Nurse for Health Monitoring, Telemedicine and Sterilization through the Internet. IJWMT 2020, 10, 16–26. [Google Scholar] [CrossRef]
- Hu, Z.; Khokhlachova, Y.; Sydorenk, V.; Opirskyy, I. Method for Optimization of Information Security Systems Behavior under Conditions of Influences. IJISA 2017, 9, 46–58. [Google Scholar] [CrossRef]
- Bykov, M.M.; Kovtun, V.V.; Smolarz, A.; Junisbekov, M.; Targeusizova, A.; Satymbekov, M. Research of Neural Network Classifier in Speaker Recognition Module for Automated System of Critical Use. In Photonics Applications in Astronomy, Communications, Industry, and High Energy Physics Experiments; Romaniuk, R.S., Linczuk, M., Eds.; International Society for Optics and Photonics: Wilga, Poland, 2017; p. 1044521. [Google Scholar]
- Teslyuk, V.; Kazarian, A.; Kryvinska, N.; Tsmots, I. Optimal Artificial Neural Network Type Selection Method for Usage in Smart House Systems. Sensors 2021, 21, 47. [Google Scholar] [CrossRef]
- Tkachenko, R. An Integral Software Solution of the SGTM Neural-Like Structures Implementation for Solving Different Data Mining Tasks. In International Scientific Conference “Intellectual Systems of Decision Making and Problem of Computational Intelligence; Babichev, S., Lytvynenko, V., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 696–713. [Google Scholar]
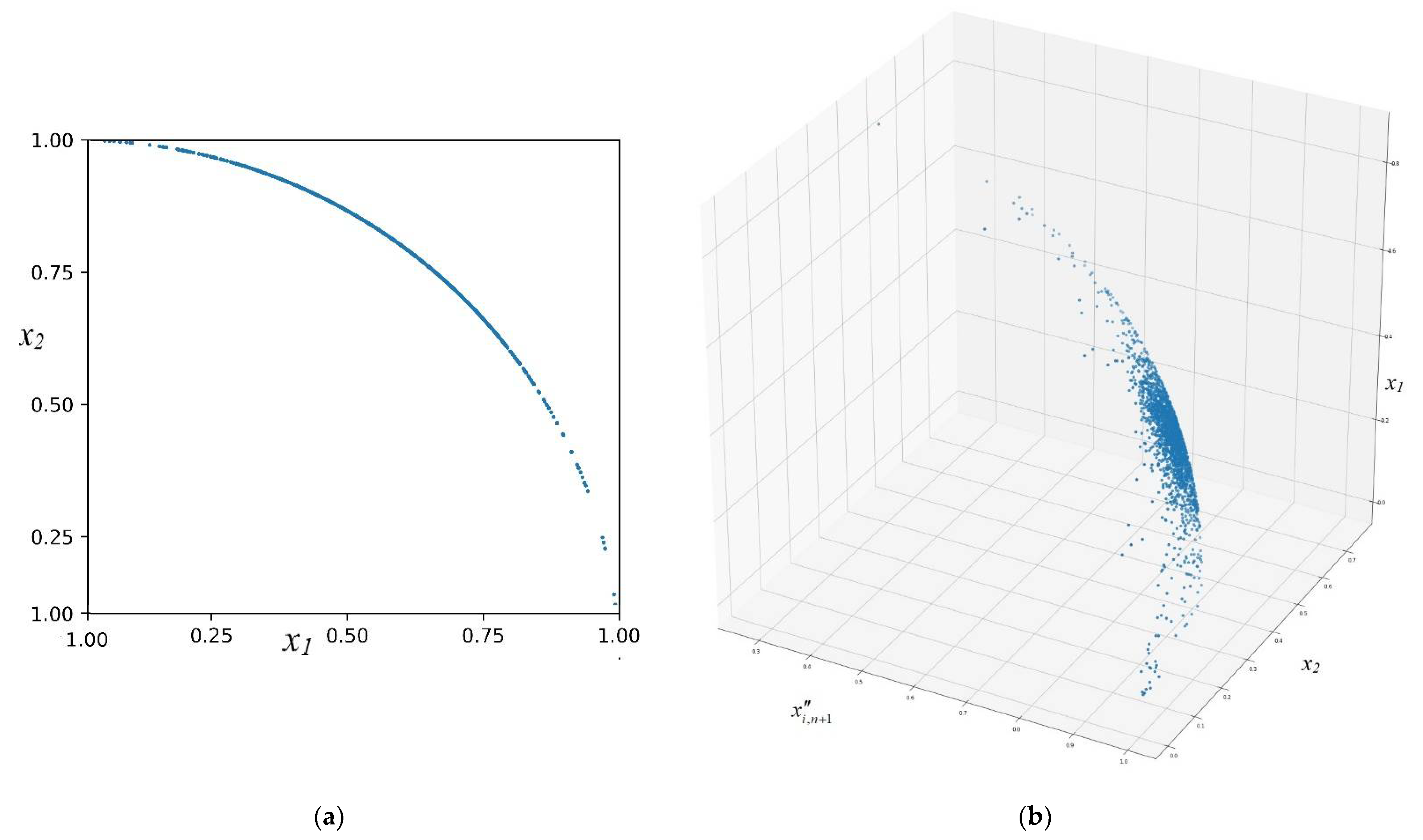
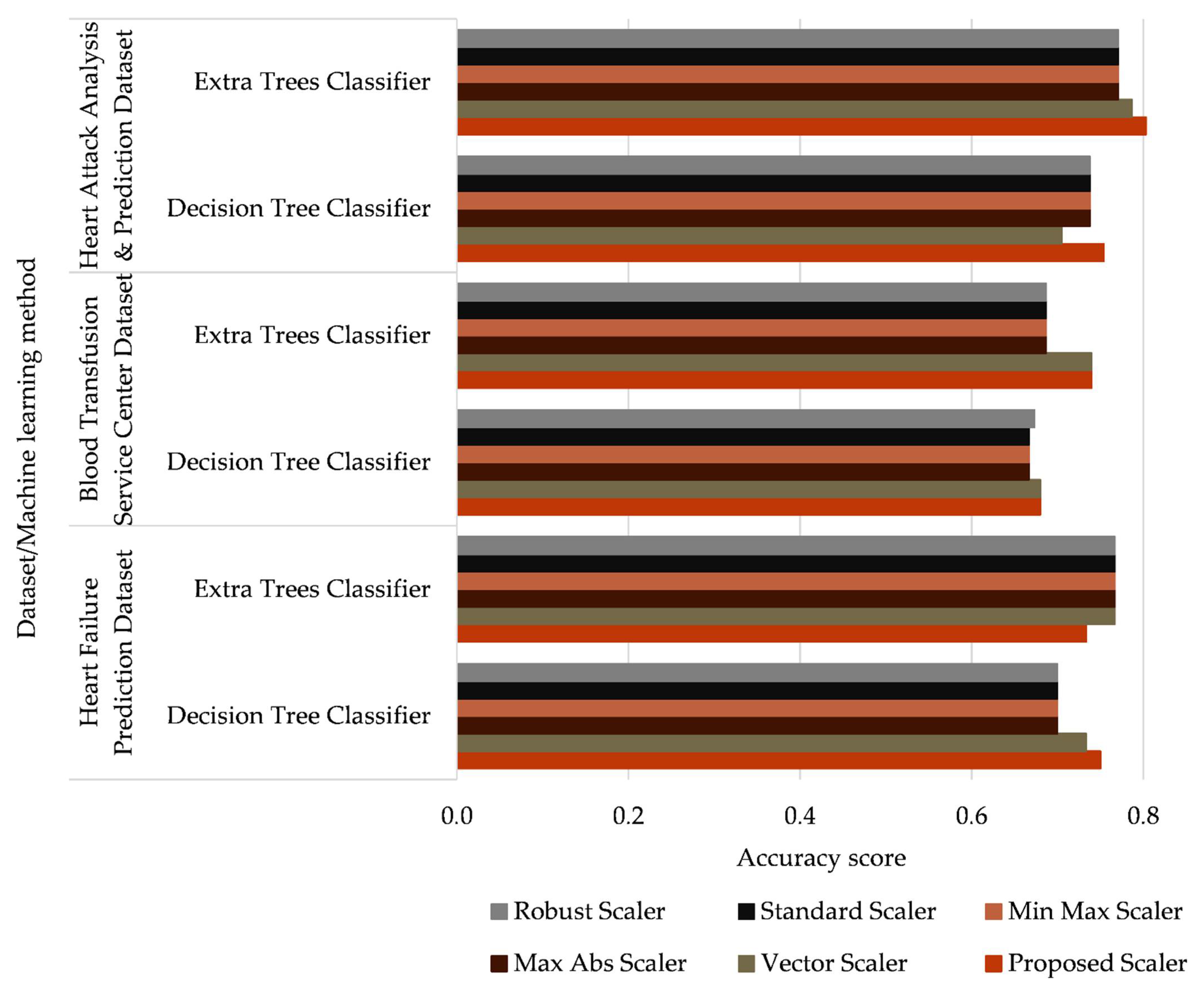
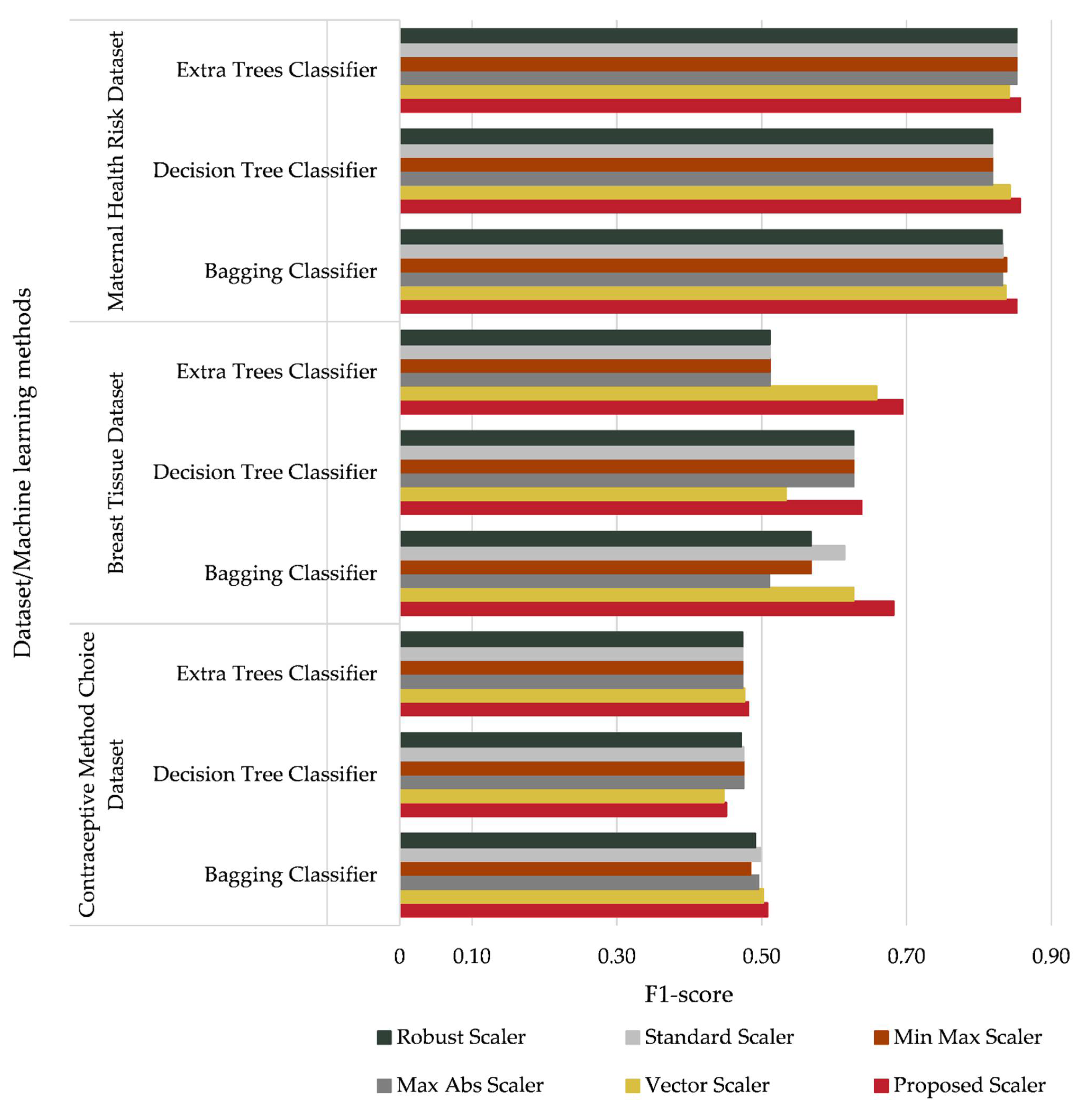
| # | Data Normalization Method | Mathematical Expression |
|---|---|---|
| 1 | Min Max Scaler | |
| 2 | Max Abs Scaler | |
| 3 | Robust Scaler | |
| 4 | Standard Scaler | |
| 5 | Vector Scaler |
| Dataset Title | Problem | Attributes | Vectors | Classes | Reference |
|---|---|---|---|---|---|
| Heart Attack Analysis & Prediction Dataset | Binary classification | 13 | 303 | 2 | [38] |
| Blood Transfusion Service Center Dataset | Binary classification | 4 | 748 | 2 | [39] |
| Heart Failure Prediction Dataset | Binary classification | 12 | 299 | 2 | [40] |
| Maternal Health Risk Dataset | Multiclass classification | 6 | 1014 | 3 | [41] |
| Breast Tissue Dataset | Multiclass classification | 9 | 212 | 6 | [42] |
| Contraceptive Method Choice Dataset | Multiclass classification | 9 | 1473 | 3 | [43] |
| Dataset Title | Classifier | Accuracy Score | Precision Score * | Recall Score * | F1-Score * |
|---|---|---|---|---|---|
| Heart Attack Analysis & Prediction Dataset | AdaBoost | 0.770 | 0.844 | 0.750 | 0.794 |
| Bagging | 0.787 | 0.795 | 0.861 | 0.827 | |
| Decision Tree | 0.754 | 0.784 | 0.806 | 0.795 | |
| Extra Trees | 0.803 | 0.800 | 0.889 | 0.842 | |
| Gradient Boosting | 0.770 | 0.806 | 0.806 | 0.806 | |
| Random Forest | 0.787 | 0.811 | 0.833 | 0.822 | |
| Blood Transfusion Service Center Dataset | AdaBoost | 0.707 | 0.731 | 0.925 | 0.817 |
| Bagging | 0.713 | 0.756 | 0.877 | 0.812 | |
| Decision Tree | 0.680 | 0.742 | 0.840 | 0.788 | |
| Extra Trees | 0.740 | 0.768 | 0.906 | 0.831 | |
| Gradient Boosting | 0.707 | 0.746 | 0.887 | 0.810 | |
| Random Forest | 0.720 | 0.767 | 0.868 | 0.814 | |
| Heart Failure Prediction Dataset | AdaBoost | 0.800 | 0.667 | 0.588 | 0.625 |
| Bagging | 0.767 | 0.615 | 0.471 | 0.533 | |
| Decision Tree | 0.750 | 0.556 | 0.588 | 0.571 | |
| Extra Trees | 0.733 | 0.625 | 0.294 | 0.4 | |
| Gradient Boosting | 0.800 | 0.692 | 0.529 | 0.600 | |
| Random Forest | 0.817 | 0.750 | 0.529 | 0.621 | |
| Maternal Health Risk Dataset | AdaBoost | 0.562 | 0.589 | 0.562 | 0.563 |
| Bagging | 0.852 | 0.854 | 0.852 | 0.852 | |
| Decision Tree | 0.857 | 0.859 | 0.857 | 0.857 | |
| Extra Trees | 0.857 | 0.858 | 0.857 | 0.857 | |
| Gradient Boosting | 0.783 | 0.785 | 0.783 | 0.783 | |
| Random Forest | 0.837 | 0.840 | 0.837 | 0.837 | |
| Breast Tissue Dataset | AdaBoost | 0.429 | 0.313 | 0.429 | 0.324 |
| Bagging | 0.667 | 0.829 | 0.667 | 0.683 | |
| Decision Tree | 0.571 | 0.786 | 0.571 | 0.638 | |
| Extra Trees | 0.667 | 0.749 | 0.667 | 0.695 | |
| Gradient Boosting | 0.571 | 0.683 | 0.571 | 0.598 | |
| Random Forest | 0.571 | 0.698 | 0.571 | 0.591 | |
| Contraceptive Method Choice Dataset | AdaBoost | 0.475 | 0.482 | 0.475 | 0.476 |
| Bagging | 0.508 | 0.510 | 0.508 | 0.508 | |
| Decision Tree | 0.451 | 0.453 | 0.451 | 0.452 | |
| Extra Trees | 0.485 | 0.483 | 0.485 | 0.482 | |
| Gradient Boosting | 0.539 | 0.541 | 0.539 | 0.537 | |
| Random Forest | 0.502 | 0.503 | 0.502 | 0.501 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izonin, I.; Tkachenko, R.; Shakhovska, N.; Ilchyshyn, B.; Singh, K.K. A Two-Step Data Normalization Approach for Improving Classification Accuracy in the Medical Diagnosis Domain. Mathematics 2022, 10, 1942. https://doi.org/10.3390/math10111942
Izonin I, Tkachenko R, Shakhovska N, Ilchyshyn B, Singh KK. A Two-Step Data Normalization Approach for Improving Classification Accuracy in the Medical Diagnosis Domain. Mathematics. 2022; 10(11):1942. https://doi.org/10.3390/math10111942
Chicago/Turabian StyleIzonin, Ivan, Roman Tkachenko, Nataliya Shakhovska, Bohdan Ilchyshyn, and Krishna Kant Singh. 2022. "A Two-Step Data Normalization Approach for Improving Classification Accuracy in the Medical Diagnosis Domain" Mathematics 10, no. 11: 1942. https://doi.org/10.3390/math10111942
APA StyleIzonin, I., Tkachenko, R., Shakhovska, N., Ilchyshyn, B., & Singh, K. K. (2022). A Two-Step Data Normalization Approach for Improving Classification Accuracy in the Medical Diagnosis Domain. Mathematics, 10(11), 1942. https://doi.org/10.3390/math10111942








