A Novel Stacking-Based Deterministic Ensemble Model for Infectious Disease Prediction
Abstract
:1. Introduction
- Developing a weighted-stacked ensemble model using linear and nonlinear statistical models.
- Enhancing the prediction accuracy of the proposed model by optimally training each base model.
- Predicting the future occurrences of infectious diseases viewed at some point as epidemics, namely, dengue, influenza, and tuberculosis.
2. Related Work
3. Materials and Methods
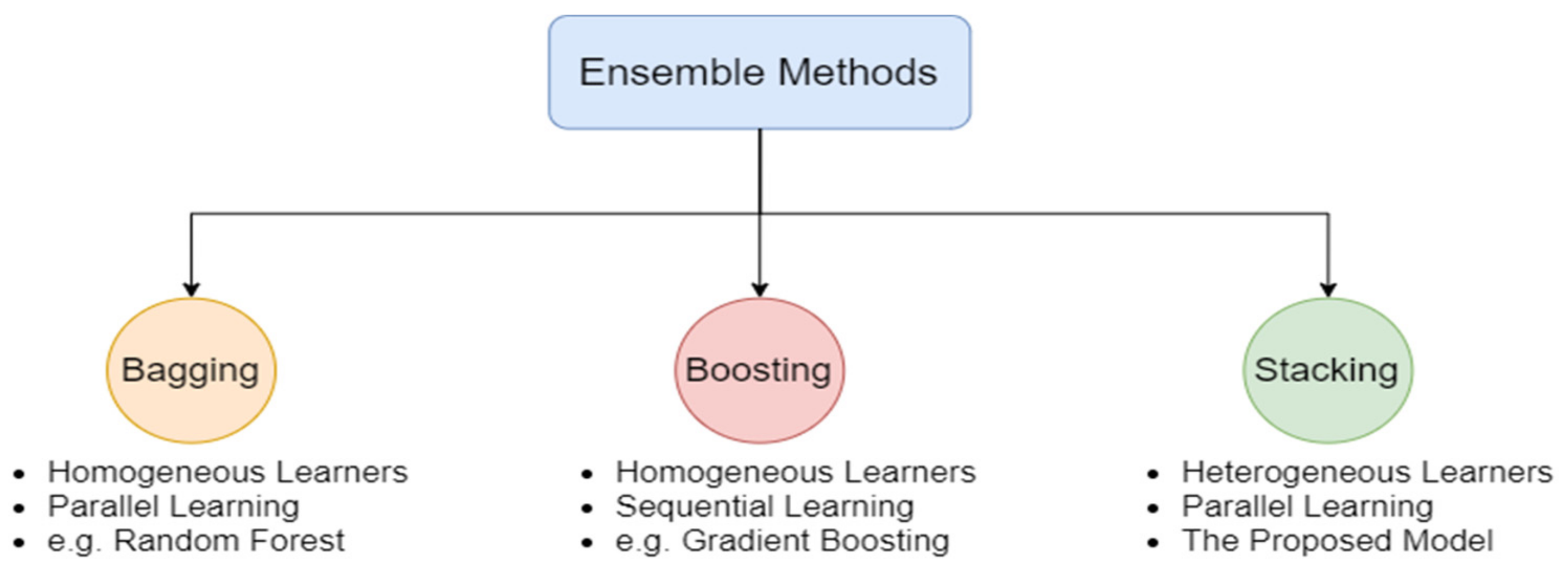
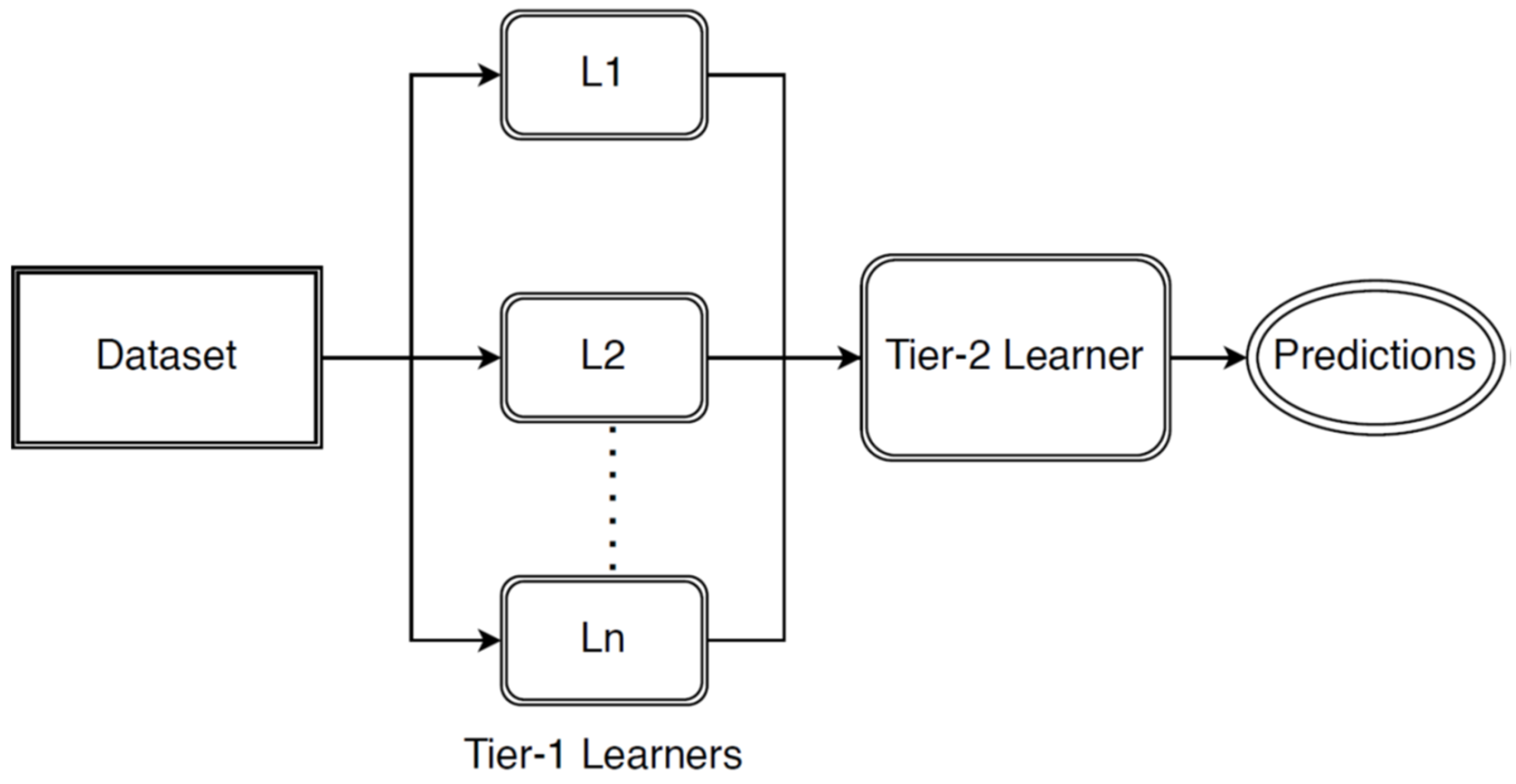
3.1. Development of Stacked Ensemble Model
- Collect the monthly datasets for each dengue, influenza, and tuberculosis disease.
- Divide each dataset into a training set and a testing set. Each dataset comprises ten years of monthly reported cases, of which 80% of the data (from the year 2010 to 2017) are taken as the training set and 20% (the years 2018 and 2019) are taken as the testing set.
- The datasets are not skewed much and are ordinarily distributed; hence, no data transformation steps are required.
- Each training set is then passed as input to the ARIMA, ETS, and NNAR models in parallel, and the models are trained until they generate minimum training errors. As the datasets have seasonal dependencies, these are removed by differencing the datasets according to their seasonality, after which they are fed to the base models.
- The fitted values from each model are then combined using the weighted average technique. The weights are assigned manually based on the training accuracy of each model. The model with higher training accuracy is given a higher weight. This step is performed so that the model whose fitted and actual values do not differ much is given more weightage than others to improve the accuracy of the stacked model.
- The fitted values resulting from the above step are then fed to the gradient boosting algorithm. The algorithm’s parameters, the number of times the algorithm is executed (nround), and the learning rate of the model (eta) are manually tuned. Tuning of the algorithm increases the overall performance and hence generates fewer errors.
- The accuracy of the proposed model is then estimated by evaluating its performance metrics in terms of errors. After the model is trained, the proposed model is used to predict 2018 and 2019. The predicted and the test set values are then compared to calculate the errors.
| Algorithm 1 Generating Stacked Ensemble Model |
| Input: Disease Time Series Data D = {d1, d2, ……, dn} Total number of observations n = 120 Sampling Frequency f = 12 Base Learners Predictions B = {B1, B2, ……, Br} where B = avg(B1(d), B2(d), ……, Br(d)) Meta Learner Predictions M(B) Output: Ḿ (Prediction for unknown/test data) 1. Disease dataset is collected and sampled based on the frequency f. 2. Sampled dataset is divided into train and test sets: Train = n * 0.8 Test = n * 0.2 3. STL decomposition is done for training dataset: For i = 1 to Train do Td ← decompose(di) //Decompose the data into trend, seasonal and random components //Stacked Ensemble Learning 4. Decomposed data is fed to Base Learners: For i = 1 to r do For j = 1 to Train do Bi = B(dj) 5. Integrating the predictions from base learners: For i = 1 to r do WP ← Σ wi * Bi //Integrating predictions by weighted average technique // wi is the weight assigned to each base learner 6. Training of Meta-Learner: M ← M(WP) 7. Making predictions or forecasting for test data: For i = 1 to Test do Ḿ ← M(di) |
3.1.1. Training of Auto-Regressive Integrated Moving Average Model
3.1.2. Training of Exponential Smoothing Model
3.1.3. Training of Neural Network AutoRegression Model
3.1.4. Construction of Tier-2 Learner Algorithm, GBRM
3.2. Performance Analysis
4. Result Analysis
4.1. Data Collection and Preprocessing
4.2. Results
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Report. Available online: https://www.who.int/whr/1996/media_centre/press_release/en/ (accessed on 26 January 2022).
- Infectious Diseases. Available online: https://www.who.int/topics/infectious_diseases/en/ (accessed on 27 January 2022).
- Chowell, G.; Luo, R.; Sunb, K.; Roosa, K.; Tariq, A.; Viboud, C. Real-time forecasting of epidemic trajectories using computational dynamic ensembles. Epidemics 2020, 30, 100379. [Google Scholar] [CrossRef] [PubMed]
- Shashvat, K.; Basu, R.; Bhondekar, P.A.; Kaur, A. An ensemble model for forecasting infectious diseases in India. Trop. Biomed. 2019, 36, 822–832. [Google Scholar] [PubMed]
- Ravì, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.Z. Deep Learning for Health Informatics. IEEE J. Biomed. Health Inform. 2017, 21, 4–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dengue Fever. Available online: https://www.chp.gov.hk/en/healthtopics/content/24/19.html (accessed on 22 August 2018).
- Statistics on Communicable Diseases. Available online: https://www.chp.gov.hk/en/statistics/submenu/26/index.html (accessed on 1 March 2022).
- Mahajan, A.; Rastogi, A.; Sharma, N. Annual Rainfall Prediction Using Time Series Forecasting. In Soft Computing: Theories and Applications; Pant, M., Kumar Sharma, T., Arya, R., Sahana, B., Zolfagharinia, H., Eds.; Advances in Intelligent Systems and Computing; Springer: Singapore, 2020; Volume 1154, pp. 69–79. [Google Scholar]
- Seasonal Influenza. Available online: https://www.chp.gov.hk/en/healthtopics/content/24/29.html (accessed on 24 April 2020).
- Influenza Virus Infections in Humans. Available online: https://www.who.int/influenza/human_animal_interface/virology_laboratoriesandvaccines/influenzavirusinfectionshumansOct18.pdf (accessed on 1 March 2022).
- Tuberculosis. Available online: https://www.chp.gov.hk/en/healthtopics/content/24/44.html (accessed on 10 April 2019).
- Zhang, X.; Zhang, T.; Young, A.A.; Li, X. Applications and Comparisons of Four Time Series Models in Epidemiological Surveillance Data. PLoS ONE 2014, 9, e88075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, K.; Chen, Y.; Wu, C.H.J.; Ben-Arieh, D. A Memetic Algorithm for Solving Optimal Control Problems of Zika Virus Epidemic with Equilibriums and Backward Bifurcation Analysis. Commun. Nonlinear Sci. Numer. Simul. 2020, 84, 105176. [Google Scholar] [CrossRef]
- Bi, K.; Chen, Y.; Wu, C.H.J.; Ben-Arieh, D. Learning-based impulse control with event-triggered conditions for an epidemic dynamic system. Commun. Nonlinear Sci. Numer. Simul. 2021, 108, 106204. [Google Scholar] [CrossRef]
- Mahalle, P.N.; Sable, N.P.; Mahalle, N.P.; Shinde, G.R. Data Analytics: COVID-19 Prediction Using Multimodal Data. In Intelligent Systems and Methods to Combat Covid-19; Springer: Singapore, 2020; pp. 1–10. [Google Scholar]
- Xi, G.; Yin, L.; Li, Y.; Mei, S. A Deep Residual Network Integrating Spatial-temporal Properties to Predict Influenza Trends at an Intra-urban Scale. In Proceedings of the 2nd ACM SIGSPATIAL International Workshop on AI for Geographic Knowledge Discovery (GeoAI′18), Seattle, WA, USA, 6 November 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 19–28. [Google Scholar]
- Zhang, T.; Ma, Y.; Xiao, X.; Lin, Y.; Zhang, X.; Yin, F.; Li, X. Dynamic Bayesian network in infectious diseases surveillance: A simulation study. Sci. Rep. 2019, 9, 10376. [Google Scholar] [CrossRef] [PubMed]
- Siriyasatien, P.; Chadsuthi, S.; Jampachaisri, K.; Kesorn, K. Dengue Epidemics Prediction: A Survey of the State-of-the-Art Based on Data Science Processes. IEEE Access 2018, 6, 53757–53795. [Google Scholar] [CrossRef]
- Wang, M.; Wang, H.; Wang, J.; Liu, H.; Lu, R.; Duan, T.; Gong, X.; Feng, S.; Liu, Y.; Cui, Z.; et al. A novel model for malaria prediction based on ensemble algorithms. PLoS ONE 2019, 14, e0226910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Tian, Y.; Qiu, R.G. Long Short-Term Memory Recurrent Neural Networks for Multiple Diseases Risk Prediction by Leveraging Longitudinal Medical Records. IEEE J. Biomed. Health Inform. 2020, 24, 2337–2346. [Google Scholar] [CrossRef] [PubMed]
- Mehrmolaei, S.; Keyvanpour, M.R. Time series forecasting using improved ARIMA. In Proceedings of the Artificial Intelligence and Robotics (IRAN OPEN), Qazvin, Iran, 9 April 2016; pp. 92–97. [Google Scholar]
- Song, X.; Xiao, J.; Deng, J.; Kang, Q.; Zhang, Y.; Xu, J. Time series analysis of influenza incidence in Chinese provinces from 2004 to 2011. Medicine 2016, 95, e3929. [Google Scholar] [CrossRef] [PubMed]
- Hyndman, R.J.; Koehler, A.B.; Snyder, R.D.; Grose, S. A state space framework for automatic forecasting using exponential smoothing methods. Int. J. Forecast. 2002, 18, 439–454. [Google Scholar] [CrossRef] [Green Version]
- Xuan, P.; Sun, C.; Zhang, T.; Ye, Y.; Shen, T.; Dong, Y. Gradient Boosting Decision Tree-Based Method for Predicting Interactions Between Target Genes and Drugs. Front. Genet. 2019, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Bath, P.A. The Use of Data Mining Methods for the Prediction of Dementia: Evidence from the English Longitudinal Study of Aging. IEEE J. Biomed. Health Inform. 2020, 24, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Ray, E.L.; Reich, N.G. Prediction of infectious disease epidemics via weighted density ensembles. PLoS Comput. Biol. 2018, 14, e1005910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamana, T.K.; Kandula, S.; Shaman, J. Superensemble forecasts of dengue outbreaks. J. R. Soc. Interface 2016, 13, 20160410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centre for Health Protection (CHP) of the Department of Health. The Government of the Hong Kong Special Administrative Region. Available online: https://www.chp.gov.hk/en/healthtopics/24/index.html (accessed on 1 March 2022).
- Seasonal ARIMA Models. Available online: https://otexts.com/fpp2/seasonal-arima.html (accessed on 1 March 2022).
- Exponential Smoothing Models. Available online: https://robjhyndman.com/talks/ABS1.pdf (accessed on 1 March 2022).
- Neural Network Models. Available online: https://otexts.com/fpp2/nnetar.html (accessed on 1 March 2022).
- Azeez, A.; Obaromi, D.; Odeyemi, A.; Ndege, J.; Muntabayi, R. Seasonality and Trend Forecasting of Tuberculosis Prevalence Data in Eastern Cape, South Africa, Using a Hybrid Model. Int. J. Environ. Res. Public Health 2016, 13, 757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.; Xie, X. Predicting Hospital Readmission: A Joint Ensemble-Learning Model. IEEE J. Biomed. Health Inform. 2020, 24, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Regression Error Metrics. Available online: https://towardsdatascience.com/regression-an-explanation-of-regression-metrics-and-what-can-go-wrong-a39a9793d914 (accessed on 1 March 2022).
- Withanage, G.P.; Viswakula, S.D.; Nilmini Silva Gunawardena, Y.I.; Hapugoda, M.D. A forecasting model for dengue incidence in the District of Gampaha, Sri Lanka. Parasites Vectors 2018, 11, 262. [Google Scholar] [CrossRef] [PubMed]





| Dengue | Influenza | Tuberculosis | |
|---|---|---|---|
| ARIMA | 11.42 | 13.55 | 36.23 |
| ETS | 9.75 | 12.23 | 31.23 |
| NNAR | 9.99 | 17.60 | 36.43 |
| Datasets | Naïve | SNaive | SES | Holt’s Winter | ETS | ARIMA | NNAR |
|---|---|---|---|---|---|---|---|
| Dengue | 7.39 | 6.62 | 6.38 | 5.59 | 4.60 | 5.56 | 5.11 |
| Influenza | 8.06 | 10.81 | 7.89 | 8.20 | 6.76 | 7.06 | 6.21 |
| Tuberculosis | 49.48 | 40.10 | 40.31 | 33.86 | 26.77 | 33.57 | 0.99 |
| Models | Dengue | Influenza | Tuberculosis |
|---|---|---|---|
| Random Forest | 14.71 | 13.41 | 30.94 |
| XG Boost | 13.75 | 8.82 | 28.64 |
| Dengue | Influenza | Tuberculosis | ||||
|---|---|---|---|---|---|---|
| MAE | RMSE | MAE | RMSE | MAE | RMSE | |
| SVM | 10.98 | 14.80 | 12.19 | 13.11 | 32.60 | 40.29 |
| RF | 16.5 | 18.94 | 12.19 | 13.41 | 25.30 | 30.94 |
| XGB | 11.75 | 14.90 | 6.30 | 8.83 | 22.18 | 28.64 |
| ENSEMBLE | 6.99 | 10.33 | 5.21 | 6.71 | 17.82 | 21.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahajan, A.; Sharma, N.; Aparicio-Obregon, S.; Alyami, H.; Alharbi, A.; Anand, D.; Sharma, M.; Goyal, N. A Novel Stacking-Based Deterministic Ensemble Model for Infectious Disease Prediction. Mathematics 2022, 10, 1714. https://doi.org/10.3390/math10101714
Mahajan A, Sharma N, Aparicio-Obregon S, Alyami H, Alharbi A, Anand D, Sharma M, Goyal N. A Novel Stacking-Based Deterministic Ensemble Model for Infectious Disease Prediction. Mathematics. 2022; 10(10):1714. https://doi.org/10.3390/math10101714
Chicago/Turabian StyleMahajan, Asmita, Nonita Sharma, Silvia Aparicio-Obregon, Hashem Alyami, Abdullah Alharbi, Divya Anand, Manish Sharma, and Nitin Goyal. 2022. "A Novel Stacking-Based Deterministic Ensemble Model for Infectious Disease Prediction" Mathematics 10, no. 10: 1714. https://doi.org/10.3390/math10101714
APA StyleMahajan, A., Sharma, N., Aparicio-Obregon, S., Alyami, H., Alharbi, A., Anand, D., Sharma, M., & Goyal, N. (2022). A Novel Stacking-Based Deterministic Ensemble Model for Infectious Disease Prediction. Mathematics, 10(10), 1714. https://doi.org/10.3390/math10101714






