Proteome Mapping of South African Cassava Mosaic Virus-Infected Susceptible and Tolerant Landraces of Cassava
Abstract
1. Introduction
2. Materials and Methods
2.1. Cassava Infection with SACMV
2.2. Protein Extraction and Isolation by TCA/Acetone Precipitation
2.3. On-Bead Hydrophilic Interaction Liquid Chromatography (HILIC) Digest
2.4. Liquid Chromatography-Mass Spectrometry (LC-MS)
2.5. Viral Load
2.6. Changes in Gene Expression Levels
2.7. Bioinformatics Analyses
2.8. CMD Symptom Severity Scoring
3. Results
3.1. SACMV Infection Development and Viral Load
3.2. Protein Identification and Quantification
3.3. Gene Ontology Analysis
3.4. KEGG, Protein–Protein Interaction Network and Reactome Pathway Analysis
3.5. Differentially Expressed Protein Groups in Response to SACMV Infection at 67 dpi
3.5.1. Metabolic Pathways
3.5.2. Biosynthesis of Secondary Metabolites
3.5.3. Glycolysis/Gluconeogenesis
3.5.4. Ribosome
3.5.5. Proteasome
3.5.6. Transcription of Selected Differentially Expressed Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eze, A.V.; Nwibo, S.U. Economic and technical efficiency of cassava production in Ika North East Local Government Area of Delta State, Nigeria. J. Dev. Agric. Econ. 2014, 6, 429–436. [Google Scholar] [CrossRef]
- Berrie, L.C.; Palmer, K.; Rybicki, E.P.; Hiyadat, S.H.; Maxwell, D.P.; Rey, M.E.C. A new isolate of African cassava mosaic virus in South Africa. Afr. J. Root Tuber Crop. 1997, 2, 49–52. [Google Scholar]
- Rey, C.; Vanderschuren, H. Cassava Mosaic and Brown Streak Diseases: Current Perspectives and Beyond. Annu. Rev. Virol. 2017, 4, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.F.; Fiallo-Olivé, E.; Briddon, R.W.; Hernández-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, A.L.; Duffy, S.; Sseruwagi, P. Whitefly-transmitted viruses threatening cassava production in Africa. Curr. Opin. Virol. 2018, 33, 167–176. [Google Scholar] [CrossRef]
- Pradhan, B.; van Tien, V.; Dey, N.; Mukherjee, S.K. Molecular Biology of Geminivirus DNA Replication. Avid Sci. 2017, 2–35. [Google Scholar]
- Gutierrez, C. Geminivirus DNA replication. Cell. Mol. Life Sci. 1999, 56, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Alabi, O.; Kumar, P.; Naidu, R. Cassava mosaic disease: A curse to food security in sub-Saharan Africa. APSnet Features 2011, 1–16. [Google Scholar] [CrossRef]
- Kumar, R.V. Plant antiviral immunity against geminiviruses and viral counter-defense for survival. Front. Microbiol. 2019, 10, 1460. [Google Scholar] [CrossRef]
- Saeed, S.T.; Samad, A. Emerging threats of begomoviruses to the cultivation of medicinal and aromatic crops and their management strategies. Virus Dis. 2017, 28, 1–17. [Google Scholar] [CrossRef]
- Owor, B.; Legg, J.P.; Okao-Okuja, G.; Obonyo, R.; Ogenga-Latigo, M.W. The effect of cassava mosaic geminiviruses on symptom severity, growth and root yield of a cassava mosaic virus disease-susceptible cultivar in Uganda. Ann. Appl. Biol. 2004, 145, 331–337. [Google Scholar] [CrossRef]
- Raji, S.N.; Subhash, N.; Ravi, V.; Saravanan, R.; Mohanan, C.N.; MakeshKumar, T.; Nita, S. Detection and Classification of Mosaic Virus Disease in Cassava Plants by Proximal Sensing of Photochemical Reflectance Index. J. Indian Soc. Remote Sens. 2016, 44, 875–883. [Google Scholar] [CrossRef]
- Houngue, J.A.; Zandjanakou-Tachin, M.; Ngalle, H.B.; Pita, J.S.; Cacai, H.T.; Ngatat, S.E.; Bell, J.M.; Ahanhanzo, C. Evaluation of resistance to cassava mosaic disease in selected African cassava cultivars using combined molecular and greenhouse grafting tools. Physiol. Mol. Plant Pathol. 2019, 105, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Allie, F.; Pierce, E.J.; Okoniewski, M.J.; Rey, C. Transcriptional analysis of South African cassava mosaic virus-infected susceptible and tolerant landraces of cassava highlights differences in resistance, basal defense and cell wall associated genes during infection. BMC Genom. 2014, 15, 1006. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Sanchez, J.-C.; Gooley, A.A.; Appel, R.D.; Humphery-Smith, I.; Hochstrasser, D.F.; Williams, K.L. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 1996, 13, 19–50. [Google Scholar] [CrossRef]
- Eldakak, M.; Milad, S.I.M.; Nawar, A.I.; Rohila, J.S. Proteomics: A biotechnology tool for crop improvement. Front. Plant Sci. 2013, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Acero, F.J.F.; Carbú, M.; El-Akhal, M.R.; Garrido, C.; González-Rodríguez, V.E.; Cantoral, J.M. Development of proteomics-based fungicides:New strategies for environmentally friendly control of fungal plant diseases. Int. J. Mol. Sci. 2011, 12, 795–816. [Google Scholar] [CrossRef]
- Vanderschuren, H.; Lentz, E.; Zainuddin, I.; Gruissem, W. Proteomics of model and crop plant species: Status, current limitations and strategic advances for crop improvement. J. Proteom. 2013, 93, 5–19. [Google Scholar] [CrossRef]
- Sant’Ana, D.V.P.; Lefsrud, M. Tomato proteomics: Tomato as a model for crop proteomics. Sci. Hortic. 2018, 239, 224–233. [Google Scholar] [CrossRef]
- Casado-Vela, J.; Sellés, S.; Martínez, R.B. Proteomic analysis of tobacco mosaic virus-infected tomato (Lycopersicon esculentum M.) fruits and detection of viral coat protein. Proteomics 2006, 6, S196–S206. [Google Scholar] [CrossRef]
- Saboki Ebrahim, K.U.; Singh, B. Pathogenesis related (PR) proteins in plant defense mechanism. Sci. Against Microb. Pathog. 2011, 2, 1043–1054. [Google Scholar]
- Alexander, M.M.; Cilia, M. A molecular tug-of-war: Global plant proteome changes during viral infection. Curr. Plant Biol. 2016, 5, 13–24. [Google Scholar] [CrossRef]
- Di Carli, M.; Villani, M.E.; Bianco, L.; Lombardi, R.; Perrotta, G.; Benvenuto, E.; Donini, M. Proteomic analysis of the plant-virus interaction in cucumber mosaic virus (CMV) resistant transgenic tomato. J. Proteome Res. 2010, 9, 5684–5697. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ma, H.Y.; Huang, W.; Wang, F.; Xu, Z.S.; Xiong, A.S. Comparative proteomic analysis provides novel insight into the interaction between resistant vs susceptible tomato cultivars and TYLCV infection. BMC Plant Biol. 2016, 16, 162. [Google Scholar] [CrossRef] [PubMed]
- Carmo, L.S.T.; Murad, A.M.; Resende, R.O.; Boiteux, L.S.; Ribeiro, S.G.; Jorrin-Novo, J.V.; Mehta, A. Plant responses to tomato chlorotic mottle virus: Proteomic view of the resistance mechanisms to a bipartite begomovirus in tomato. J. Proteom. 2017, 151, 284–292. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, L.; Tie, W.; Yan, Y.; Wu, C.; Hu, W.; Zhang, J. Extensive Post-Transcriptional Regulation Revealed by Transcriptomic and Proteomic Integrative Analysis in Cassava under Drought. J. Agric. Food Chem. 2019, 67, 3521–3534. [Google Scholar] [CrossRef]
- Chang, L.; Wang, L.; Peng, C.; Tong, Z.; Wang, D.; Ding, G.; Xiao, J.; Guo, A.; Wang, X. The chloroplast proteome response to drought stress in cassava leaves. Plant Physiol. Biochem. 2019, 142, 351–362. [Google Scholar] [CrossRef]
- Shan, Z.; Luo, X.; Wei, M.; Huang, T.; Khan, A.; Zhu, Y. Physiological and proteomic analysis on long-term drought resistance of cassava (Manihot esculenta Crantz). Sci. Rep. 2018, 8, 17982. [Google Scholar] [CrossRef]
- Vanderschuren, H.; Nyaboga, E.; Poon, J.S.; Baerenfaller, K.; Grossmann, J.; Hirsch-Hoffmann, M.; Kirchgessner, N.; Nanni, P.; Gruissen, W. Large-scale proteomics of the cassava storage root and identification of a target gene to reduce postharvest deterioration. Plant Cell 2014, 26, 1913–1924. [Google Scholar] [CrossRef]
- An, F.; Fan, J.; Li, J.; Li, Q.X.; Zhu, W.; Wen, F.; Carvalho, L.J.C.B.; Chen, S. Comparison of Leaf Proteomes of Cassava (Manihot esculenta Crantz) Cultivar NZ199 Diploid and Autotetraploid Genotypes. PLoS ONE 2014, 9, e85991. [Google Scholar]
- Mitprasat, M.; Roytrakul, S.; Jiemsup, S.; Boonseng, O.; Yokthongwattana, K. Leaf proteomic analysis in cassava (Manihot esculenta, Crantz) during plant development, from planting of stem cutting to storage root formation. Planta 2011, 233, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Duraisamy, R.; Natesan, S.; Muthrajan, R.; Gandhi, K.; Lakshmanan, P.; Janaky, J.G.; Karuppusamy, N.; Chokkappan, M. Proteomic Analysis of Cassava Mosaic Virus (CMV) Responsive Proteins in Cassava Leaf. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2988–3005. [Google Scholar] [CrossRef]
- Schulze, W.X.; Altenbuchinger, M.; He, M.; Kränzlein, M.; Zörb, C. Proteome profiling of repeated drought stress reveals genotype-specific responses and memory effects in maize. Plant Physiol. Biochem. 2021, 159, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Awasthi, S.; Indoliya, Y.; Chauhan, A.S.; Mishra, S.; Agrawal, L.; Srivastava, S.; Dwivedi, S.; Singh, P.C.; Mallick, S.; et al. Transcriptome and proteome analyses reveal selenium mediated amelioration of arsenic toxicity in rice (Oryza sativa L.). J. Hazard. Mater. 2020, 390, 122122. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Shi, C.; Su, C.; Liu, Y. Complementary analyses of the transcriptome and iTRAQ proteome revealed mechanism of ethylene dependent salt response in bread wheat (Triticum aestivum L.). Food Chem. 2020, 325, 126866. [Google Scholar] [CrossRef] [PubMed]
- Ngara, R.; Ndimba, R.; Borch-Jensen, J.; Jensen, O.N.; Ndimba, B. Identification and profiling of salinity stress-responsive proteins in Sorghum bicolor seedlings. J. Proteom. 2012, 75, 4139–4150. [Google Scholar] [CrossRef] [PubMed]
- Vengavasi, K.; Pandey, R.; Abraham, G.; Yadav, R.K. Comparative analysis of soybean root proteome reveals molecular basis of differential carboxylate efflux under low phosphorus stress. Genes 2017, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Chakraborty, D.; Pal, A. Proteomic analysis of salicylic acid induced resistance to Mungbean Yellow Mosaic India Virus in Vigna mungo. J. Proteom. 2011, 74, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F.A. Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Fauquet, C.; Fargette, D. African cassava mosaic virus: Etiology, epidemiology and control. Plant Dis 1990, 74, 404–411. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.F.; Huang, Y.P.; Chen, L.H.; Hsu, Y.H.; Tsai, C.H. Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 2013, 163, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Anitha, V.; Ram, R. Distinct Roles of Alpha/Beta Hydrolase Domain Containing Proteins. Biochem. Mol. Biol. J. 2016, 2, 1–3. [Google Scholar]
- Kim, J.; Jung, J.H.; Lee, S.B.; Go, Y.S.; Kim, H.J.; Cahoon, R.; Markham, J.E.; Cahoon, E.B.; Suh, M.C. Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 2013, 162, 567–580. [Google Scholar] [CrossRef]
- Blake, C.C.; Rice, D.W. Phosphoglycerate kinase. Biol. Sci. 1981, 293, 93–104. [Google Scholar]
- Warner, J.R.; McIntosh, K.B. How Common Are Extraribosomal Functions of Ribosomal Proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef]
- Edmondson, S.P.; Turri, J.; Smith, K.; Clark, A.; Shriver, J.W. Structure, stability, and flexibility of ribosomal protein L14e from Sulfolobus solfataricus. Biochemistry 2009, 48, 5553–5562. [Google Scholar] [CrossRef]
- Solano de la Cruz, M.T.; Adame-Garcia, J.; Gregorio-Jorge, J.; Jimenez-Jacinto, V.; Vega-Alvarado, L.; Iglesias-Andreu, L.; Escobar-Hernández, E.E.; Luna-Rodríguez, M. Increase in ribosomal proteins activity: Translational reprogramming in Vanilla planifolia Jacks., against Fusarium infection. bioRxiv 2019, 1–42. [Google Scholar] [CrossRef]
- Ditt, R.F.; Kerr, K.F.; De Figueiredo, P.; Delrow, J.; Comai, L.; Nester, E.W. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant-Microbe Interact. 2006, 19, 665–681. [Google Scholar] [CrossRef] [PubMed]
- Bureau, M.; Leh, V.; Haas, M.; Geldreich, A.; Ryabova, L.; Keller, M. P6 protein of Cauliflower mosaic virus, a translation reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. J. Gen. Virol. 2004, 85, 3765–3775. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, T.; Matsui, M.; Wei, N. The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol. 2001, 305, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Rosas-Diaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Deng, X.W.; Bejarano, E.R. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signalling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef] [PubMed]
- Inaba, T.; Ito-Inaba, Y. Versatile roles of plastids in plant growth and development. Plant Cell Physiol. 2010, 51, 1847–1853. [Google Scholar] [CrossRef][Green Version]
- Yang, F.; Xiao, K.; Pan, H.; Liu, J. Chloroplast: The Emerging Battlefield in Plant-Microbe Interactions. Front. Plant Sci. 2021, 12, 637853. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Hong, Y.; Liu, Y. Chloroplast in plant-virus interaction. Front. Microbiol. 2016, 7, 1565. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Jin, P.; Chen, X.; Zhang, T.-Y.; Zhong, K.-L.; Liu, P.; Chen, J.-P.; Yang, J. Comparative proteomic analysis of Nicotiana benthamiana plants under Chinese wheat mosaic virus infection. BMC Plant Biol. 2021, 21, 51. [Google Scholar] [CrossRef]
- Lehto, K.; Tikkanen, M.; Hiriart, J.B.; Paakkarinen, V.; Aro, E.M. Depletion of the Photosystem II Core Complex in Mature Tobacco Leaves Infected by the Flavum Strain of Tobacco mosaic virus. Mol. Plant-Microbe Interact. 2003, 16, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Li, H.F.; Wong, S.M.; Fan, Z.F. Plastocyanin transit peptide interacts with Potato virus X coat protein, while silencing of plastocyanin reduces coat protein accumulation in chloroplasts and symptom severity in host plants. Mol. Plant-Microbe Interact. 2009, 22, 1523–1534. [Google Scholar] [CrossRef]
- Mazidah, M.; Lau, W.H.; Yusoff, K.; Habibuddin, H.; Tan, Y.H. Ultrastructural Features of Catharanthus roseus Leaves Infected with Cucumber Mosaic Virus. Pertanika J. Trop. Agric. Sci. 2012, 35, 85–92. [Google Scholar]
- Gunasinghe, U.B. Association of Potato Virus Y Gene Products with Chloroplasts in Tobacco. Mol. Plant-Microbe Interact. 1991, 4, 452–457. [Google Scholar] [CrossRef]
- Hodgson, R.A.J.; Beachy, R.N.; Pakrasi, H.B. Selective inhibition of photosystem II in spinach by tobacco mosaic virus: An effect of the viral coat protein. Febs Lett. 1989, 245, 267–270. [Google Scholar] [CrossRef]
- Yang, S.M.; Kim, B.J.; Toro, L.N.; Skoultchi, A.I. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl. Acad. Sci. USA 2013, 110, 1708–1713. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Mateos, M.; Sabarit, B.; Maio, F.; Sanchez-Duran, M.A.; Rosas-Diaz, T.; Prins, M.; Ruiz-Albert, J.; Luna, A.P.; van den Burg, H.A.; Bejarano, E.R. Geminivirus replication protein impairs SUMO conjugation of proliferating cellular nuclear antigen at two acceptor sites. J. Virol. 2018, 92, e00611-18. [Google Scholar] [CrossRef]
- Azevedo, C.; Betsuyaku, S.; Peart, J.; Takahashi, A.; Noël, L.; Sadanandom, A.; Casais, C.; Parker, J.; Shirasu, K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006, 25, 2007–2016. [Google Scholar] [CrossRef]
- Lu, R.; Malcuit, I.; Moffett, P.; Ruiz, M.T.; Wu, A.-J.; Rathjen, J.P.; Bendahmane, A.; Day, L.; Baulcombe, D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003, 22, 5690–5699. [Google Scholar] [CrossRef]
- Moshe, A.; Gorovits, R.; Liu, Y.; Czosnek, H. Tomato plant cell death induced by inhibition of HSP90 is alleviated by Tomato yellow leaf curl virus infection. Mol. Plant Pathol. 2016, 17, 247–260. [Google Scholar] [CrossRef]
- Langin, G.; Gouguet, P.; ÜstünMicrobial, S. Effector Proteins—A Journey through the Proteolytic Landscape. Trends Microbiol. 2020, 28, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Ustun, S.; Hafrén, A. Proteasome homeostasis is essential for a robust cauliflower mosaic virus infection. bioRxiv 2021, 436740, 1–22. [Google Scholar] [CrossRef]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2017, 119, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.; Carvalho, C.M.; Florentino, L.H.; Ramos, H.J.O.; Fontes, E.P.B. Conserved threonine residues within the A-loop of the receptor NIK differentially regulate the kinase function required for antiviral signalling. PLoS ONE 2009, 4, e5781. [Google Scholar] [CrossRef]
- Zorzatto, C.; Machado, J.P.B.; Lopes, K.V.G.; Nascimento, K.-J.T.; Pereira, W.A.; Brustolini, O.J.B.; Reis, P.A.B.; Calil, L.P.; Deguchi, D.; Sachetto-Martin, S.M.; et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef]
- Brustolini, O.J.B.; Machado, J.P.B.; Condori-Apfata, J.A.; Coco, D.; Deguchi, M.; Loriato, V.A.P.; Pereira, W.A.; Alfenas-Zerbini, P.; Zerbini, F.M.; Inoue-Nagata, A.K.; et al. Sustained NIK-mediated antiviral signalling confers broad-spectrum tolerance to begomoviruses in cultivated plants. Plant Biotechnol. J. 2015, 13, 1300–1311. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.; Lopes, K.V.G.; Apfata, J.A.C.; Fontes, E.P.B. NSP-interacting kinase, NIK: A transducer of plant defence signalling. J. Exp. Bot. 2010, 61, 3839–3845. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Santos, A.A.; Pires, S.R.; Rocha, C.S.; Saraiva, D.I.; Machado, J.P.; Mattos, E.C.; Fietto, L.G.; Fontes, E.P. Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PloS Pathog. 2008, 4, e1000247. [Google Scholar] [CrossRef]
- Rocha, C.S.; Santos, A.A.; Machado, J.P.B.; Fontes, E.P.B. The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signalling. Virology 2008, 380, 165–169. [Google Scholar] [CrossRef]
- Akano, A.O.; Dixon, A.G.O.; Mba, C.; Barrera, E.; Fregene, M. Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor. Appl. Genet. 2002, 105, 521–525. [Google Scholar] [CrossRef]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Rabbi, I.Y.; Egesi, C.; Hamblin, M.; Kawuki, R.; Kulakow, P.; Lozano, R.; Pino Del Carpio, D.; Ramu, P.; Jannink, J.-L. Genome-Wide Association and Prediction Reveals Genetic Architecture of Cassava Mosaic Disease Resistance and Prospects for Rapid Genetic Improvement. Plant Genome 2016, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chatukuta, P.; Rey, M.E.C. A cassava protoplast system for screening genes associated with the response to South African cassava mosaic virus. Virol. J. 2020, 17, 184. [Google Scholar] [CrossRef]
- Shen, Q.; Hu, T.; Bao, M.; Cao, L.; Zhang, H.; Song, F.; Xie, Q.; Zhou, X. Tobacco RING E3 Ligase NtRFP1 Mediates Ubiquitination and Proteasomal Degradation of a Geminivirus-Encoded β C1. Mol. Plant 2016, 9, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cheng, Z.; Liu, M.; Yang, X.; Qiu, D. C3HC4-type RING finger protein NbZFP1 is involved in growth and fruit development in Nicotiana benthamiana. PLoS ONE 2014, 9, e99352. [Google Scholar] [CrossRef]
- Yu, Y.; Xu, W.; Wang, J.; Wang, L.; Yao, W.; Yang, Y.; Xu, Y.; Ma, F.; Du, Y.; Wang, Y. The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 2013, 200, 834–846. [Google Scholar] [CrossRef]
- Louis, B.; Rey, C. Resistance gene analogs involved in tolerant cassava—Geminivirus interaction that shows a recovery phenotype. Virus Genes 2015, 51, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.M.; Ferreira, M.A.; Raimundo, G.A.S.; Fontes, E.P.B. Geminiviral Triggers and Suppressors of Plant Antiviral Immunity. Microorganisms 2021, 9, 775. [Google Scholar] [CrossRef]
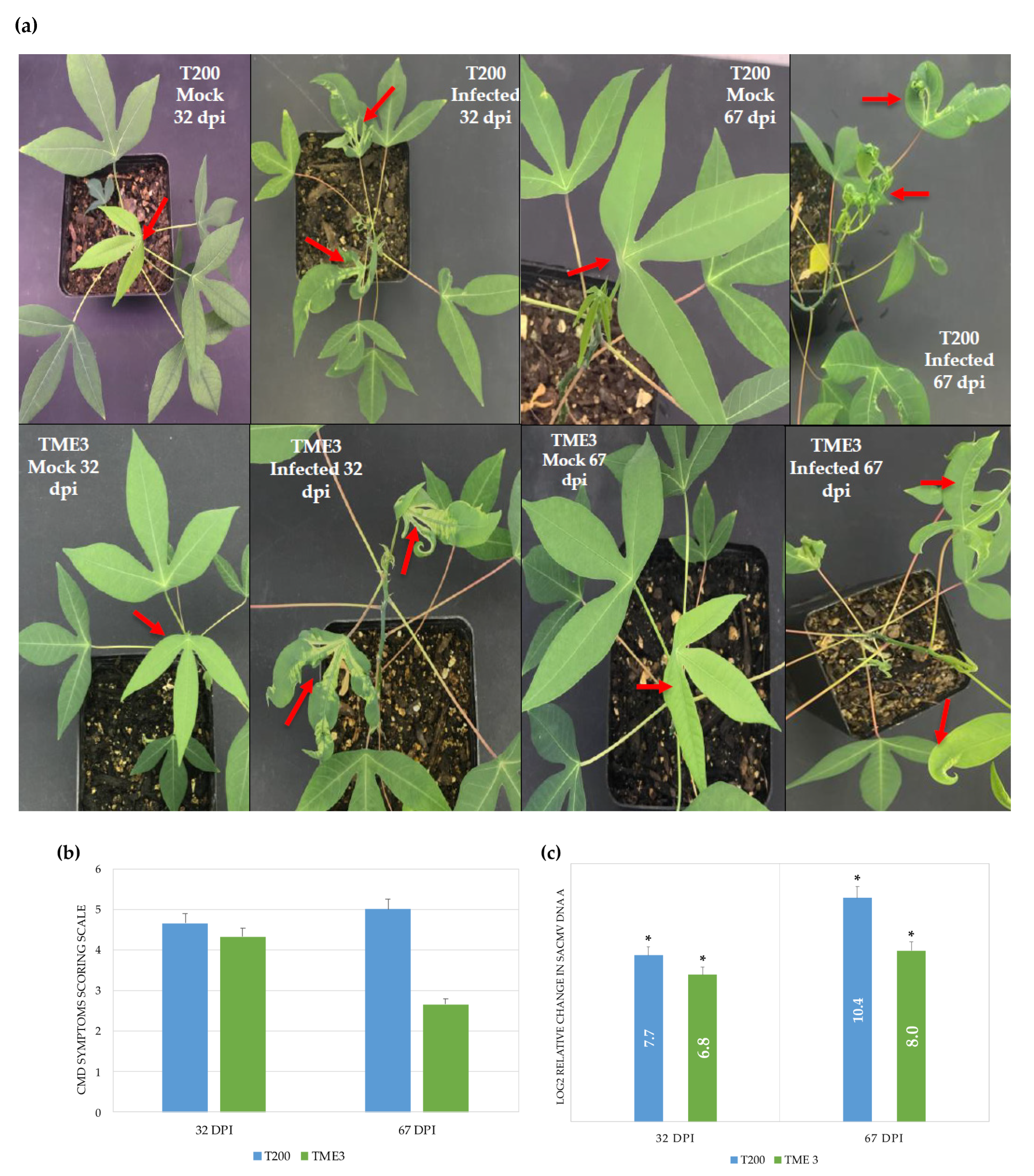
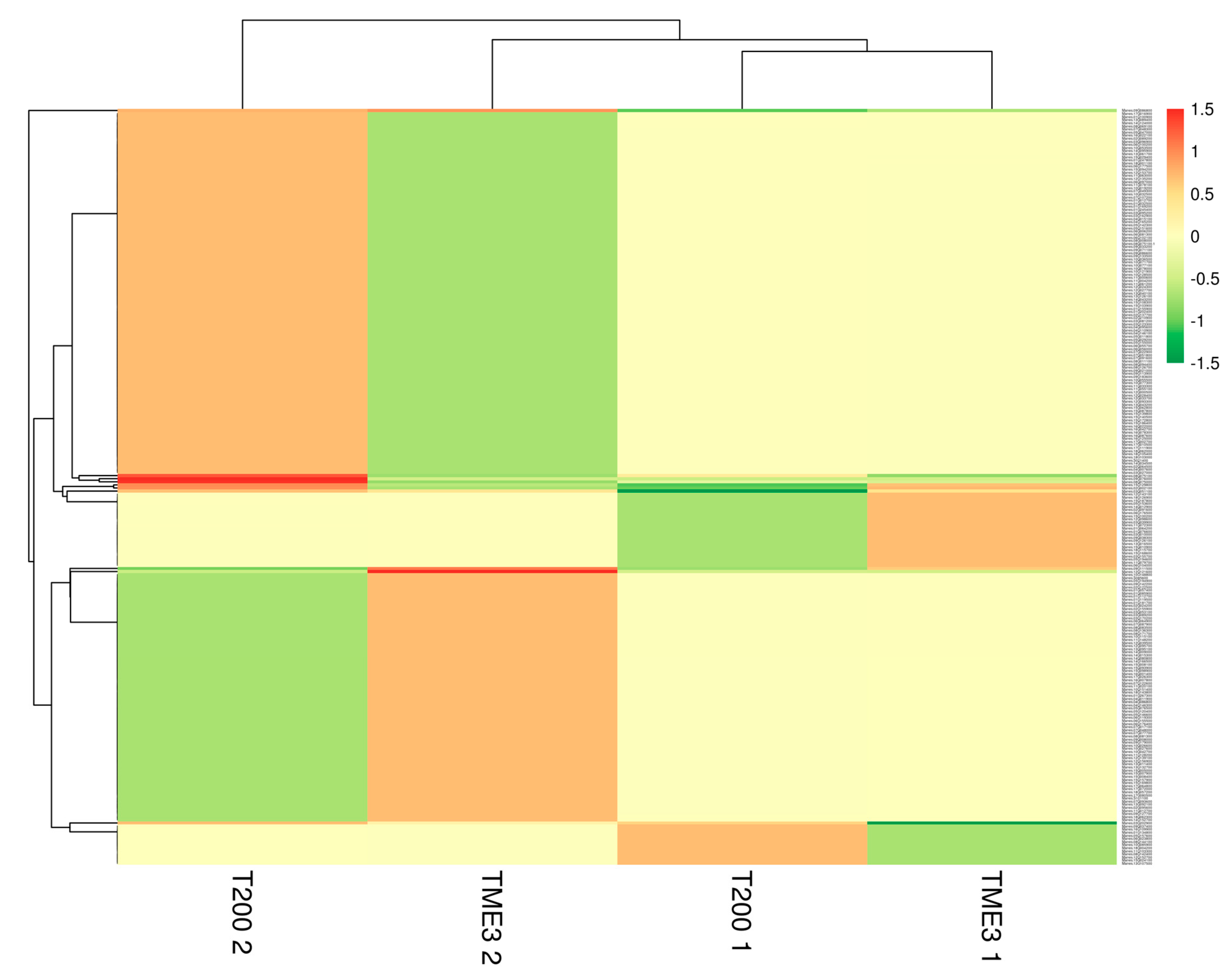
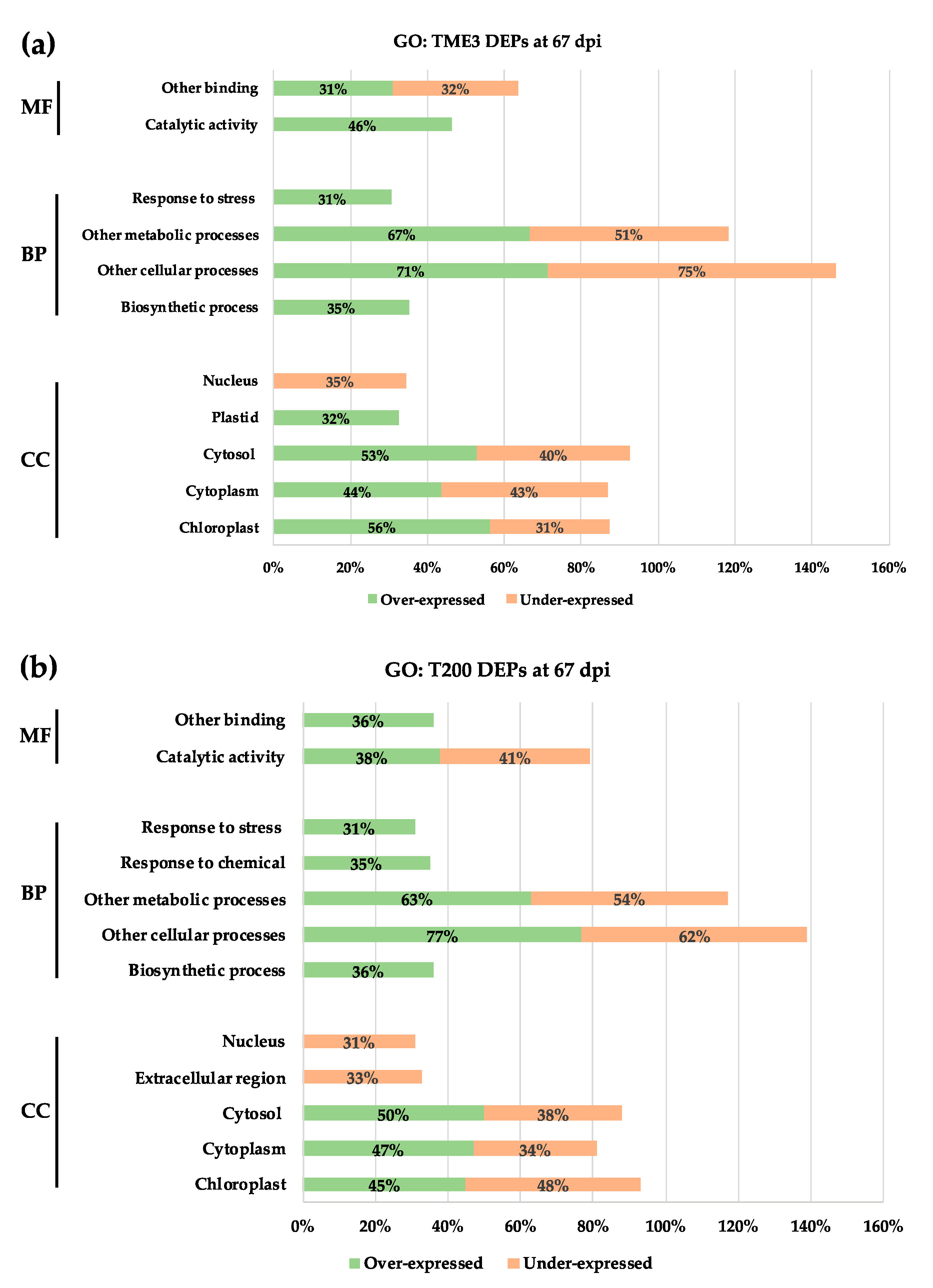
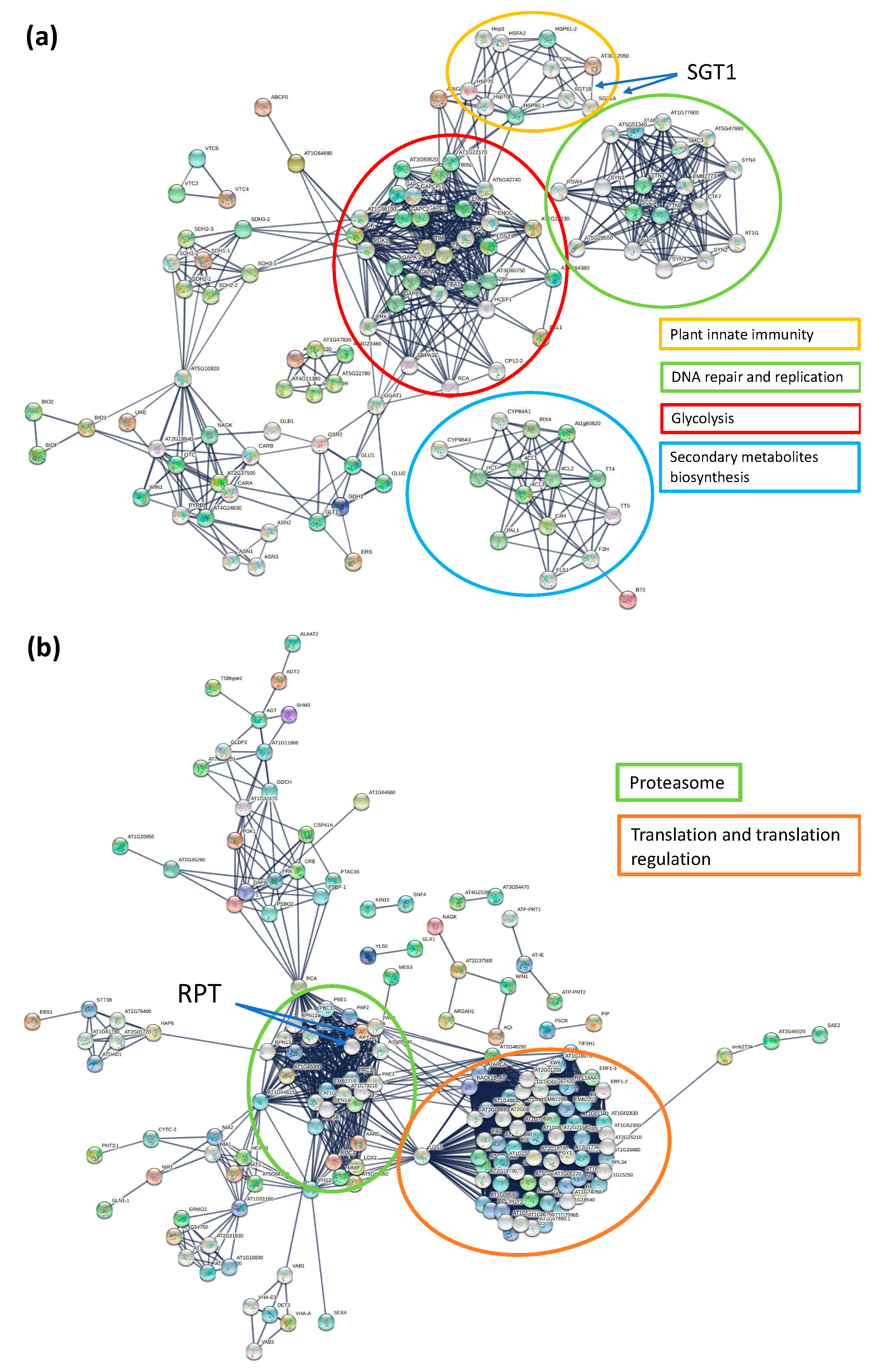

| Timepoint | Regulation | Protein Count | Protein Characterisation | GO Categories a | Functional Groups |
|---|---|---|---|---|---|
| 32 dpi | Under-expressed | 30 T200 | 14 characterised 35 uncharacterised | 49 CC | 19 |
| 27 TME3 | 42 BP | 35 | |||
| Over-expressed | 19 T200 | 36 MF | 13 | ||
| 22 TME3 | 49 KEGG | 11 | |||
| Total=49 | |||||
| 67 dpi | Under-expressed | 63 T200 | 128 characterised 78 uncharacterised | 198 CC | 21 |
| 81 TME3 | 178 BP | 44 | |||
| Over-expressed | 143 T200 | 198 MF | 23 | ||
| 125 TME3 | 199 KEGG | 22 | |||
| Total=206 |
| Timepoint | Pathway | Protein Count | FDR a |
|---|---|---|---|
| 32 dpi | Metabolic pathways | 13 | 2.30 × 10−4 |
| Biosynthesis of secondary metabolites | 8 | 2.30 × 10−3 | |
| Carbon metabolism | 5 | 7.30 × 10−4 | |
| Arginine biosynthesis | 3 | 5.40 × 10−4 | |
| Biosynthesis of amino acids | 3 | 3.86 × 10−2 | |
| Carbon fixation in photosynthetic organisms | 2 | 3.64 × 10−2 | |
| 67 dpi | Metabolic pathways | 56 | 1.46 × 10−18 |
| Biosynthesis of secondary metabolites | 36 | 2.88 × 10−13 | |
| Biosynthesis of amino acids | 14 | 1.04 × 10−7 | |
| Glycine, serine and threonine metabolism | 9 | 1.04 × 10−7 | |
| Carbon metabolism | 9 | 1.30 × 10−3 | |
| Arginine biosynthesis | 7 | 2.83 × 10−7 | |
| Ribosome | 7 | 3.26 × 10−2 | |
| Amino sugar and nucleotide sugar metabolism | 6 | 2.80 × 10−3 | |
| Alanine, aspartate and glutamate metabolism | 5 | 4.80 × 10−4 | |
| Carbon fixation in photosynthetic organisms | 5 | 1.60 × 10−3 | |
| 2-Oxocarboxylic acid metabolism | 5 | 1.80 × 10−3 | |
| Glyoxylate and dicarboxylate metabolism | 5 | 1.90 × 10−3 | |
| Glycolysis/Gluconeogenesis | 5 | 9.10 × 10−3 | |
| Arginine and proline metabolism | 4 | 4.60 × 10−3 | |
| Pyruvate metabolism | 4 | 1.92 × 10−2 | |
| Nitrogen metabolism | 3 | 2.07 × 10−2 | |
| Ascorbate and aldarate metabolism | 3 | 2.32 × 10−2 | |
| Valine, leucine and isoleucine degradation | 3 | 2.32 × 10−2 | |
| Pentose phosphate pathway | 3 | 3.41 × 10−2 | |
| Proteasome | 3 | 3.41 × 10−2 | |
| Histidine metabolism | 2 | 3.26 × 10−2 | |
| Other glycan degradation | 2 | 3.41 × 10−2 |
| Accession | Manes ID | Protein Name | Unique Peptides | q Value | T200 Ratio a | TME3 Ratio a |
|---|---|---|---|---|---|---|
| tr|A0A2C9U824 | Manes.16G022000 | PKS_ER domain-containing protein | 3.00 | 2.83 × 10−4 | 2.38 | −1.15 |
| tr|A0A2C9VEA2 | Manes.08G075000 | Uncharacterized protein | 4.00 | 7.56 × 10−4 | 2.37 | −2.04 |
| tr|A0A2C9U4J9 | Manes.17G026300 | Lactoylglutathione lyase | 2.00 | 2.59 × 10−3 | −1.35 | 2.82 |
| tr|A0A2C9VIL5 | Manes.07G049300 | Proline iminopeptidase | 7.00 | 3.91 × 10−3 | 2.26 | 1.05 |
| tr|A0A0A1E5H6 | Manes.11G078100 | Phosphate transporter | 2.00 | 4.63 × 10−3 | 5.62 | 1.01 |
| tr|A0A251JR98 | Manes.11G020100 | Germin-like protein | 6.00 | 5.29 × 10−3 | −3.26 | −1.03 |
| tr|A0A2C9VJP2 | Manes.07G087900 | Glycine cleavage system H protein | 2.00 | 6.82 × 10−3 | −3.32 | 1.43 |
| tr|A0A2C9WL85 | Manes.01G155900 | Uncharacterized protein | 2.00 | 6.82 × 10−3 | 2.46 | −2.07 |
| tr|A0A2C9V1F6 | Manes.11G148200 | Uncharacterized protein | 3.00 | 7.50 × 10−3 | −2.79 | 5.63 |
| tr|A0A2C9UCJ8 | Manes.15G029400 | Cysteine synthase | 10.00 | 7.85 × 10−3 | 2.94 | 1.25 |
| tr|A0A2C9VK05 | Manes.07G093600 | GLOBIN domain-containing protein | 2.00 | 1.11 × 10−2 | 1.14 | 3.05 |
| tr|A0A2C9UGM3 | Manes.15G103900 | Peroxidase | 9.00 | 1.16 × 10−2 | 2.09 | −1.03 |
| tr|A0A2C9UYJ6 | Manes.11G012700 | Aminotran_1_2 domain-containing protein | 10.00 | 1.16 × 10−2 | 1.01 | 2.01 |
| tr|A0A2C9VAT8 | Manes.09G071100 | Uncharacterized protein | 2.00 | 1.22 × 10−2 | 2.23 | 1.27 |
| tr|A0A2C9VSW1 | Manes.05G029200 | AB hydrolase-1 domain-containing protein | 2.00 | 1.26 × 10−2 | 2.09 | 1.25 |
| tr|A0A2C9WBE7 | Manes.02G064500 | PALP domain-containing protein | 3.00 | 1.30 × 10−2 | 3.27 | 1.14 |
| tr|A0A2C9URN3 | Manes.13G132700 | Uncharacterized protein | 4.00 | 1.32 × 10−2 | −1.40 | 3.51 |
| tr|A0A2C9VLP8 | Manes.06G006200 | Uncharacterized protein | 3.00 | 1.34 × 10−2 | 3.25 | −1.85 |
| tr|A0A2C9UGX0 | Manes.15G172800 | Phosphoacetylglucosamine mutase | 9.00 | 1.49 × 10−2 | 2.46 | −1.12 |
| tr|A0A2C9VP29 | Manes.06G081300 | Uncharacterized protein | 3.00 | 1.51 × 10−2 | 18.46 | −1.91 |
| tr|A0A2C9UBX9 | Manes.15G008100 | Bet_v_1 domain-containing protein | 9.00 | 1.76 × 10−2 | −1.11 | 3.20 |
| tr|A0A2C9V4E1 | Manes.10G042700 | Uncharacterized protein | 2.00 | 1.76 × 10−2 | −1.20 | 2.12 |
| tr|A0A2C9WLB8 | Manes.01G085900 | Uncharacterized protein | 2.00 | 1.76 × 10−2 | 1.17 | 2.43 |
| tr|A0A251K3C9 | Manes.09G021000 | HP domain-containing protein | 18.00 | 1.81 × 10−2 | 2.18 | −1.71 |
| tr|A0A2C9UTJ3 | Manes.12G039500 | Uncharacterized protein | 6.00 | 1.81 × 10−2 | −1.18 | 2.10 |
| tr|A0A2C9W314 | Manes.04G086800 | SUMO-activating enzyme subunit | 2.00 | 1.81 × 10−2 | −5.15 | 4.65 |
| tr|A0A251K980 | Manes.08G011100 | Uncharacterized protein | 3.00 | 1.81 × 10−2 | 11.82 | −2.66 |
| tr|A0A251K915 | Manes.08G008000 | Uncharacterized protein | 4.00 | 1.81 × 10−2 | 1.82 | −2.13 |
| tr|A0A2C9VGL2 | Manes.08G075100 | VWFA domain-containing protein | 11.00 | 2.00 × 10−2 | 3.13 | −1.29 |
| tr|A0A2C9WEH9 | Manes.02G089200 | Uncharacterized protein | 6.00 | 2.07 × 10−2 | 2.41 | 1.99 |
| tr|A0A2C9UID8 | Manes.15G186400 | Uncharacterized protein | 6.00 | 2.07 × 10−2 | 4.47 | −5.30 |
| tr|A0A2C9UV72 | Manes.12G093300 | Glycosyltransferase | 2.00 | 2.24 × 10−2 | 9.06 | −1.92 |
| tr|A0A2C9VFU9 | Manes.08G126700 | Protein kinase domain-containing protein | 2.00 | 2.66 × 10−2 | 23.94 | −1.43 |
| tr|A0A2C9W2J9 | Manes.04G146100 | Uncharacterized protein | 7.00 | 2.66 × 10−2 | 2.21 | −1.44 |
| tr|A0A2C9UPJ8 | Manes.13G061700 | FAS1 domain-containing protein | 10.00 | 2.66 × 10−2 | −1.45 | −2.01 |
| tr|A0A2C9U6Y4 | Manes.17G080500 | Uncharacterized protein | 5.00 | 2.67 × 10−2 | −2.30 | 1.59 |
| tr|A0A2C9U1Y7 | Manes.18G103000 | Uncharacterized protein | 3.00 | 2.68 × 10−2 | 3.18 | −1.22 |
| tr|A0A2C9UQH1 | Manes.13G092100 | AB hydrolase-1 domain-containing protein | 18.00 | 2.70 × 10−2 | 1.30 | 2.76 |
| tr|A0A2C9UYF6 | Manes.11G055100 | Phospholipase D | 34.00 | 2.73 × 10−2 | 2.05 | −1.17 |
| tr|A0A251K1M8 | Manes.10G151400 | Uncharacterized protein | 2.00 | 2.73 × 10−2 | −3.90 | −1.14 |
| tr|A0A2C9WAR9 | Manes.03G202900 | Uncharacterized protein | 9.00 | 2.73 × 10−2 | 2.28 | 1.45 |
| tr|A0A2C9UXW4 | Manes.12G139100 | Ribosomal_L7Ae domain-containing protein | 3.00 | 2.73 × 10−2 | −5.24 | 2.18 |
| tr|A0A2C9UYT0 | Manes.12G153700 | Uncharacterized protein | 9.00 | 2.79 × 10−2 | 2.40 | 1.20 |
| tr|A0A2C9UNI9 | Manes.14G166500 | Non-specific lipid-transfer protein | 6.00 | 2.81 × 10−2 | −5.88 | 10.33 |
| tr|A0A2C9V3Z4 | Manes.10G027600 | ATPase_AAA_core domain-containing protein | 3.00 | 2.89 × 10−2 | −52.29 | 6.89 |
| tr|A0A251JSV5 | Manes.11G063000 | Uncharacterized protein | 3.00 | 2.94 × 10−2 | 2.16 | 1.11 |
| tr|A0A2C9WF62 | Manes.02G137700 | Alpha-mannosidase | 13.00 | 3.05 × 10−2 | 2.32 | 1.63 |
| tr|A0A2C9W858 | Manes.03G096900 | M20_dimer domain-containing protein | 3.00 | 3.05 × 10−2 | 2.47 | 1.62 |
| tr|A0A2C9VPI9 | Manes.06G102100 | Uncharacterized protein | 2.00 | 3.05 × 10−2 | 2.09 | −2.96 |
| tr|A0A2C9VT96 | Manes.06G155500 | Glyco_hydro_18 domain-containing protein | 4.00 | 3.05 × 10−2 | 2.67 | 2.71 |
| tr|A0A2C9VIN8 | Manes.07G051800 | Uncharacterized protein | 3.00 | 3.07 × 10−2 | 2.44 | 1.35 |
| tr|A0A2C9UXQ1 | Manes.11G033300 | TauD domain-containing protein | 4.00 | 3.14 × 10−2 | 2.87 | 1.67 |
| tr|A0A0C4ZQZ2 | Manes.12G135200 | Annexin | 13.00 | 3.14 × 10−2 | 2.78 | 1.41 |
| tr|A0A2C9UCR5 | Manes.15G007900 | Bet_v_1 domain-containing protein | 2.00 | 3.14 × 10−2 | −1.08 | 3.69 |
| tr|A0A2C9U3Q4 | Manes.17G010500 | RRM domain-containing protein | 4.00 | 3.14 × 10−2 | 2.41 | −1.45 |
| tr|A0A2C9WMF8 | Manes.01G202400 | Uncharacterized protein | 3.00 | 3.21 × 10−2 | 6.53 | −1.13 |
| tr|A0A2C9VRV6 | Manes.06G177500 | Uncharacterized protein | 2.00 | 3.26 × 10−2 | 3.26 | 1.27 |
| tr|A0A2C9UUG2 | Manes.12G027700 | Uncharacterized protein | 14.00 | 3.32 × 10−2 | 2.28 | 1.56 |
| tr|A0A2C9U185 | Manes.18G062000 | NAD(P)H-hydrate epimerase | 7.00 | 3.42 × 10−2 | 4.79 | −2.12 |
| tr|A0A2C9V3P1 | Manes.10G055500 | WD_REPEATS_REGION domain-containing protein | 2.00 | 3.43 × 10−2 | 2.24 | −1.84 |
| tr|A0A2C9VBZ8 | Manes.09G142200 | Uncharacterized protein | 2.00 | 3.59 × 10−2 | −1.63 | 67.74 |
| tr|A0A2C9UV10 | Manes.12G095700 | AB hydrolase-1 domain-containing protein | 3.00 | 3.59 × 10−2 | 1.42 | 2.21 |
| tr|A0A2C9UJE4 | Manes.14G034500 | UMP-CMP kinase | 2.00 | 3.59 × 10−2 | 3.10 | 1.14 |
| tr|A0A2C9WRD2 | Manes.01G245400 | Importin N-terminal domain-containing protein | 9.00 | 3.59 × 10−2 | 3.16 | −1.16 |
| tr|A0A251KS16 | Manes.05G011800 | Uncharacterized protein | 12.00 | 3.61 × 10−2 | 2.05 | −1.21 |
| tr|A0A2C9UGV6 | Manes.15G140500 | Eukaryotic translation initiation factor 3 subunit I | 10.00 | 3.61 × 10−2 | 2.10 | −1.40 |
| tr|A0A2C9UA86 | Manes.16G087600 | 3-ketoacyl-CoA synthase | 2.00 | 3.61 × 10−2 | 8.18 | −6.12 |
| tr|A0A2C9UX42 | Manes.12G156900 | Hist_deacetyl domain-containing protein | 5.00 | 3.74 × 10−2 | −4.08 | 1.33 |
| tr|A0A2C9W735 | Manes.03G081200 | FAD-binding PCMH-type domain-containing protein | 8.00 | 3.77 × 10−2 | 5.66 | −1.54 |
| tr|A0A2C9VMD1 | Manes.07G122600 | AAI domain-containing protein | 4.00 | 3.77 × 10−2 | −10.20 | −1.40 |
| tr|A0A2C9VAA7 | Manes.09G127700 | Bet_v_1 domain-containing protein | 7.00 | 3.77 × 10−2 | 1.32 | 2.22 |
| tr|A0A2C9URF0 | Manes.13G126100 | Uncharacterized protein | 2.00 | 3.77 × 10−2 | 2.16 | −1.04 |
| tr|A0A2C9U7U7 | Manes.16G007800 | Uncharacterized protein | 7.00 | 3.77 × 10−2 | −2.85 | 1.88 |
| tr|A0A2C9W3J5 | Manes.04G095600 | eRF1_1 domain-containing protein | 4.00 | 3.77 × 10−2 | 2.26 | −1.02 |
| tr|A0A199UC10 | Manes.s021400 | ZnMc domain-containing protein | 2.00 | 3.77 × 10−2 | 3.57 | 1.09 |
| tr|A0A2C9WH55 | Manes.01G012700 | Vac14_Fab1_bd domain-containing protein | 3.00 | 3.80 × 10−2 | 2.00 | 1.10 |
| tr|A0A2C9U3Q3 | Manes.18G143800 | Uncharacterized protein | 3.00 | 3.80 × 10−2 | −5.26 | 1.18 |
| tr|A0A2C9V4F7 | Manes.10G077300 | Uncharacterized protein | 2.00 | 3.80 × 10−2 | 2.46 | 1.12 |
| tr|A0A251LVH3 | Manes.01G267300 | Uncharacterized protein | 3.00 | 3.87 × 10−2 | 1.54 | 2.11 |
| tr|A0A2C9UJT8 | Manes.14G080800 | Peptidyl-prolyl cis-trans isomerase | 2.00 | 3.90 × 10−2 | −3.45 | 4.03 |
| tr|A0A2C9V4B1 | Manes.10G077100 | Lipase_3 domain-containing protein | 5.00 | 3.92 × 10−2 | 2.03 | −1.24 |
| tr|A0A2C9VG07 | Manes.08G081300 | Uncharacterized protein | 2.00 | 3.93 × 10−2 | −5.02 | 6.17 |
| tr|A0A2C9WEB2 | Manes.02G155900 | Uncharacterized protein | 4.00 | 3.93 × 10−2 | 1.17 | 4.80 |
| tr|A0A2C9V5Y8 | Manes.10G128500 | Uncharacterized protein | 5.00 | 3.93 × 10−2 | 4.19 | −1.96 |
| tr|A0A2C9WJN9 | Manes.01G112700 | AAI domain-containing protein | 3.00 | 4.06 × 10−2 | −2.73 | 6.45 |
| tr|A0A2C9VN74 | Manes.06G056000 | Uncharacterized protein | 2.00 | 4.06 × 10−2 | 3.26 | −1.66 |
| tr|A0A2C9WNU1 | Manes.01G247800 | Glycosyltransferase | 2.00 | 4.06 × 10−2 | 2.17 | 1.39 |
| tr|A0A2C9VYT5 | Manes.05G155000 | AAA domain-containing protein | 9.00 | 4.06 × 10−2 | 2.82 | −1.07 |
| tr|A0A2C9W393 | Manes.04G165200 | Uncharacterized protein | 4.00 | 4.06 × 10−2 | 4.42 | −1.42 |
| tr|A0A2C9VKB0 | Manes.07G107200 | Chalcone-flavonone isomerase family protein | 10.00 | 4.06 × 10−2 | 2.89 | 1.20 |
| tr|A0A2C9UBT3 | Manes.15G008400 | Bet_v_1 domain-containing protein | 7.00 | 4.06 × 10−2 | 2.85 | 13.10 |
| tr|A0A2C9V9W8 | Manes.09G113900 | Uncharacterized protein | 4.00 | 4.06 × 10−2 | 2.31 | 1.48 |
| tr|A0A2C9WD78 | Manes.02G122500 | Alpha-1,4 glucan phosphorylase | 5.00 | 4.06 × 10−2 | 2.32 | 4.83 |
| tr|A0A2C9WCC9 | Manes.02G095600 | CTP_transf_like domain-containing protein | 3.00 | 4.18 × 10−2 | 1.16 | 2.35 |
| tr|A0A2C9UNS6 | Manes.13G043200 | AAA domain-containing protein | 9.00 | 4.22 × 10−2 | 4.75 | −1.32 |
| tr|A0A2C9U6K5 | Manes.17G064800 | Uncharacterized protein | 2.00 | 4.23 × 10−2 | −4.61 | 1.83 |
| tr|A0A2C9VE34 | Manes.08G069100 | Uncharacterized protein | 15.00 | 4.30 × 10−2 | 2.06 | 1.34 |
| tr|A0A2C9V5M6 | Manes.10G121900 | Uncharacterized protein | 13.00 | 4.30 × 10−2 | 2.52 | −1.27 |
| tr|A0A2C9U8P4 | Manes.17G111900 | Uncharacterized protein | 3.00 | 4.30 × 10−2 | 2.33 | 1.50 |
| tr|A0A2C9USP9 | Manes.12G024300 | Uncharacterized protein | 2.00 | 4.30 × 10−2 | 2.64 | −1.54 |
| tr|A0A2C9V2I1 | Manes.10G019200 | Uncharacterized protein | 2.00 | 4.34 × 10−2 | 3.30 | −1.16 |
| tr|A0A2C9VWB9 | Manes.05G146600 | Peptidyl-prolyl cis-trans isomeras | 3.00 | 4.34 × 10−2 | −1.52 | 2.06 |
| tr|A0A2C9UGS7 | Manes.15G108300 | Uncharacterized protein | 6.00 | 4.45 × 10−2 | 3.10 | −1.11 |
| tr|A0A2C9VJ85 | Manes.07G017100 | 60S ribosomal protein L13 | 2.00 | 4.45 × 10−2 | −3.86 | 2.11 |
| tr|A0A2C9UTD1 | Manes.12G033700 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 | 4.00 | 4.49 × 10−2 | 5.39 | −1.86 |
| tr|A0A2C9WHE0 | Manes.01G032500 | PCI domain-containing protein | 2.00 | 4.52 × 10−2 | 7.14 | −1.40 |
| tr|A0A2C9UFY6 | Manes.15G139800 | Transmembrane 9 superfamily member | 4.00 | 4.55 × 10−2 | 4.51 | −2.11 |
| tr|A0A2C9VXS5 | Manes.05G194900 | Protein kinase domain-containing protein | 2.00 | 4.55 × 10−2 | −8.12 | −2.18 |
| tr|A0A2C9VQH2 | Manes.06G055700 | Uncharacterized protein | 7.00 | 4.57 × 10−2 | 2.10 | −2.39 |
| tr|A0A2C9VYM5 | Manes.05G142300 | Methyltransferase | 2.00 | 4.62 × 10−2 | 3.32 | −6.67 |
| tr|A0A2C9TZI5 | Manes.18G001100 | Pyrophosphate--fructose 6-phosphate 1-phosphotransferase subunit alpha | 3.00 | 4.62 × 10−2 | 2.13 | 1.34 |
| tr|A0A2C9V233 | Manes.11G128200 | Uncharacterized protein | 3.00 | 4.65 × 10−2 | −6.76 | 9.16 |
| tr|A0A2C9V6N1 | Manes.09G008000 | F420_oxidored domain-containing protein | 4.00 | 4.68 × 10−2 | 1.08 | 2.91 |
| tr|A0A251JLF7 | Manes.12G028400 | Carbonic anhydrase | 2.00 | 4.68 × 10−2 | 2.95 | −1.11 |
| tr|A0A2C9W4R1 | Manes.03G053100 | Uncharacterized protein | 5.00 | 4.68 × 10−2 | −5.50 | 1.68 |
| tr|A0A2C9V313 | Manes.10G032500 | NAD(P)-bd_dom domain-containing protein | 3.00 | 4.68 × 10−2 | 3.11 | 1.28 |
| tr|A0A2C9VBY7 | Manes.09G183600 | Uncharacterized protein | 3.00 | 4.68 × 10−2 | 3.66 | −1.04 |
| tr|A0A2C9UMZ5 | Manes.13G011400 | AAI domain-containing protein | 5.00 | 4.68 × 10−2 | −9.13 | 6.96 |
| tr|A0A2C9VRT4 | Manes.06G097000 | Plasma membrane ATPase | 6.00 | 4.69 × 10−2 | 5.75 | −1.45 |
| tr|A0A2C9WA59 | Manes.03G162900 | Ribosomal_L14e domain-containing protein | 2.00 | 4.69 × 10−2 | 4.36 | −1.86 |
| tr|A0A2C9VYM8 | Manes.04G011900 | Carboxypeptidase | 3.00 | 4.69 × 10−2 | −1.36 | 3.72 |
| tr|A0A251K5T0 | Manes.09G086600 | Uncharacterized protein | 6.00 | 4.69 × 10−2 | 3.00 | −1.22 |
| tr|A0A2C9V462 | Manes.10G078000 | Glycosyltransferase | 5.00 | 4.71 × 10−2 | 3.12 | −1.63 |
| tr|A0A2C9VLD9 | Manes.07G091600 | Uncharacterized protein | 4.00 | 4.81 × 10−2 | 4.30 | −2.64 |
| tr|A0A2C9W8C0 | Manes.03G123300 | Trafficking protein particle complex subunit | 3.00 | 4.81 × 10−2 | 2.54 | −1.12 |
| tr|A0A2C9W8D3 | Manes.03G089200 | Uncharacterized protein | 2.00 | 4.87 × 10−2 | −14.91 | 2.30 |
| tr|A0A199U962 | Manes.S101100 | Lipase_GDSL domain-containing protein | 3.00 | 4.87 × 10−2 | −2.51 | 3.28 |
| tr|A0A2C9UL26 | Manes.14G124000 | Protein kinase domain-containing protein | 3.00 | 4.89E × 10−2 | −1.70 | −2.19 |
| tr|A0A2C9U8J5 | Manes.16G001400 | Abhydrolase_2 domain-containing protein | 2.00 | 4.89 × 10−2 | −1.41 | 8.09 |
| tr|A0A2C9VH42 | Manes.08G171700 | Non-specific lipid-transfer protein | 3.00 | 4.97 × 10−2 | −13.04 | 6.81 |
| tr|A0A2C9VWH1 | Manes.05G151600 | Uncharacterized protein | 10.00 | 4.97 × 10−2 | 3.19 | −3.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramulifho, E.; Rey, M.E.C. Proteome Mapping of South African Cassava Mosaic Virus-Infected Susceptible and Tolerant Landraces of Cassava. Proteomes 2021, 9, 41. https://doi.org/10.3390/proteomes9040041
Ramulifho E, Rey MEC. Proteome Mapping of South African Cassava Mosaic Virus-Infected Susceptible and Tolerant Landraces of Cassava. Proteomes. 2021; 9(4):41. https://doi.org/10.3390/proteomes9040041
Chicago/Turabian StyleRamulifho, Elelwani, and Marie Emma Christine Rey. 2021. "Proteome Mapping of South African Cassava Mosaic Virus-Infected Susceptible and Tolerant Landraces of Cassava" Proteomes 9, no. 4: 41. https://doi.org/10.3390/proteomes9040041
APA StyleRamulifho, E., & Rey, M. E. C. (2021). Proteome Mapping of South African Cassava Mosaic Virus-Infected Susceptible and Tolerant Landraces of Cassava. Proteomes, 9(4), 41. https://doi.org/10.3390/proteomes9040041






