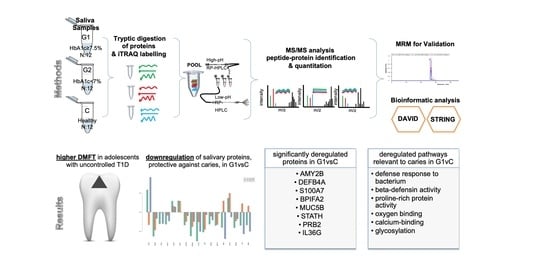
Downregulation of Salivary Proteins, Protective against Dental Caries, in Type 1 Diabetes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Design and Clinical Data
2.3. Standardized Sample Collection
2.4. LC-MS Analysis amd Pathway Analysis
2.5. LC-MRM Validation
2.6. Bioinformatics Analysis
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics of the Study Population and Dental Caries
3.2. Molecular Pathways and Differential Expression of Proteins Involved in Dental Caries
4. Discussion
4.1. Saliva as a Diagnostic Body Fluid
4.2. Salivary Alterations in Diabetes
4.3. Salivary Proteins and Dental Caries
- PRP’s (PRB2, PRR4)
- Phosphopeptides (PRB2, STATH)
- Albumin (ALB)
- Mucins (MUC5B)
- Histatin-1 and BPI (BPIFB1, BPIFA2)
- Keratins (KRT74, KRT19)
- S100A7 is a calcium- and zinc-binding protein with a prominent role in regulating the immune response and antimicrobial humoral response, and has also been associated with dental caries [31]. Accordingly, it was found to be significantly downregulated (fold change = −1.7) in G1–C comparison.
- Apolipoproteins (APOA1, APOA2)Apolipoproteins have recently been suggested to be particularly relevant to the aging process and longevity by playing crucial human immune functions [50]. Both APOA1 and APOA2 were found to be significantly downregulated in G1–C and G1–G2 comparisons. While their role in periodontitis is recently investigated [51], their possible association with caries susceptibility has yet to be explored.
4.4. Gingival Inflammation and Diabetes
4.5. From the Molecular Characteristics to the Clinical Practice
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Approval Statement
References
- Schmoeckel, J.; Gorseta, K.; Splieth, C.H.; Juric, H. How to Intervene in the Caries Process: Early Child-hood Caries—A Systematic Review. Caries Res. 2020, 54, 102–112. [Google Scholar] [CrossRef]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the Oral Microbiota in Health: Mechanisms That Prevent Dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef]
- Gao, X.; Jiang, S.; Koh, D.; Hsu, C.-Y. Salivary biomarkers for dental caries. Periodontology 2000 2015, 70, 128–141. [Google Scholar] [CrossRef]
- Frencken, J.E.; Sharma, P.; Stenhouse, L.; Green, D.; Laverty, D.; Dietrich, T. Global epidemiology of dental caries and severe periodontitis—A comprehensive review. J. Clin. Periodontol. 2017, 44, S94–S105. [Google Scholar] [CrossRef]
- Alves, C.; Menezes, R.; Brandao, M. Salivary flow and dental caries in Brazilian youth with type 1 diabetes mellitus. Indian J. Dent. Res. 2012, 23, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Arheiam, A.; Omar, S. Dental caries experience and periodontal treatment needs of 10- to 15-year old children with type 1 diabetes mellitus. Int. Dent. J. 2014, 64, 150–154. [Google Scholar] [CrossRef]
- Buysschaert, M.; Jamart, J.; Buysschaert, B. Dental caries and diabetes: A Belgian survey of patients with type 1 and type 2 diabetes. Diabetes Metab. 2020, 46, 248–249. [Google Scholar] [CrossRef] [PubMed]
- El-Tekeya, M.; El Tantawi, M.; Fetouh, H.; Mowafy, E.; Khedr, N.A. Caries risk indicators in children with type 1 diabetes mellitus in relation to metabolic control. Pediatr. Dent. 2012, 34, 510–516. [Google Scholar] [PubMed]
- Ferizi, L.; Dragidella, F.; Spahiu, L.; Begzati, A.; Kotori, V. The Influence of Type 1 Diabetes Mellitus on Dental Caries and Salivary Composition. Int. J. Dent. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Malhotra, S.; Sharma, V.; Hiremath, S.S. The Influence of Insulin Dependent Diabetes Mellitus on Dental Caries and Salivary Flow. Int. J. Chronic Dis. 2014, 2014, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jawed, M.; Shahid, S.M.; Qader, S.A.; Azhar, A. Dental caries in diabetes mellitus: Role of salivary flow rate and minerals. J. Diabetes Complicat. 2011, 25, 183–186. [Google Scholar] [CrossRef]
- Miko, S.; Ambrus, S.J.; Sahafian, S.; Dinya, E.; Tamas, G.; Albrecht, M.G. Dental caries and adolescents with type 1 diabetes. Br. Dent. J. 2010, 208, E12. [Google Scholar] [CrossRef]
- Pappa, E.; Vastardis, H.; Rahiotis, C. Chair-side saliva diagnostic tests: An evaluation tool for xerostomia and caries risk assessment in children with type 1 diabetes. J. Dent. 2020, 93, 103224. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Hegde, A.M.; Kamath, A.; Shetty, S. Dental Caries and Salivary Alterations in Type I Diabetes. J. Clin. Pediatr. Dent. 2011, 36, 181–184. [Google Scholar] [CrossRef]
- Sampaio, N.; Mello, S.; Alves, C. Dental caries-associated risk factors and type 1 diabetes mellitus. Pediatr. Endocrinol. Diabetes Metab. 2011, 17, 152–157. [Google Scholar] [PubMed]
- Siudikiene, J.; Machiulskiene, V.; Nyvad, B.; Tenovuo, J.; Nedzelskiene, I. Dental Caries Increments and Related Factors in Children with Type 1 Diabetes Mellitus. Caries Res. 2008, 42, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Brignardello-Petersen, R. Insufficient evidence about the association between caries and type 1 diabetes and metabolic status in children and adolescents. J. Am. Dent. Assoc. 2018, 149, e88. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.S.; Amaro, I.F.; Caramelo, F.; Paula, A.; Marto, C.M.; Ferreira, M.M.; Botelho, M.F.; Carrilho, E.V. Dental caries, diabetes mellitus, metabolic control and diabetes duration: A systematic review and meta-analysis. J. Esthet. Restor. Dent. 2020, 32, 291–309. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.D. Children with type 1 diabetes and caries—Are they linked? Evid. Based Dent. 2020, 21, 94–95. [Google Scholar] [CrossRef] [PubMed]
- Akpata, E.S.; Alomari, Q.; Mojiminiyi, O.A.; Al-Sanae, H. Caries experience among children with type 1 diabetes in Kuwait. Pediatr. Dent. 2012, 34, 468–472. [Google Scholar] [PubMed]
- Busato, I.M.S.; Ignacio, S.A.; Brancher, J.A.; GregioHardy, A.M.; Machado, M.A.N.; Azevedo-Alanis, L.R. Impact of xerostomia on the quality of life of adolescents with type 1 diabetes mellitus. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2009, 108, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Babatzia, A.; Papaioannou, W.; Stavropoulou, A.; Pandis, N.; Kanaka-Gantenbein, C.; Papagiannoulis, L.; Gizani, S. Clinical and microbial oral health status in children and adolescents with type 1 diabetes mellitus. Int. Dent. J. 2020, 70, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xing, L.; Yu, H.; Zhao, L. Prevalence of dental caries in children and adolescents with type 1 diabetes: A systematic review and meta-analysis. BMC Oral Health 2019, 19, 1–9. [Google Scholar] [CrossRef]
- Liu, T.; Wei, Y.; Zhu, Y.; Yang, W. Caries Status and Salivary Alterations of Type-1 Diabetes Mellitus in Children and Adolescents: A Systematic Review and Meta-analysis. J. Évid. Based Dent. Pract. 2021, 21, 101496. [Google Scholar] [CrossRef]
- Pappa, E.; Kousvelari, E.; Vastardis, H. Saliva in the “Omics” era: A promising tool in paediatrics. Oral Dis. 2019, 25, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegde, M.N.; Hegde, N.; Ashok, A.; Shetty, S. Biochemical Indicators of Dental Caries in Saliva: An in vivo Study. Caries Res. 2014, 48, 170–173. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Y.; Wang, X.; Ren, Q.; Han, S.; Ding, L.; Li, Z.; Zhou, X.; Li, W.; Zhang, L. Comparative salivary proteomics analysis of children with and without dental caries using the iTRAQ/MRM approach. J. Transl. Med. 2018, 16, 11. [Google Scholar] [CrossRef] [Green Version]
- Pachoński, M.; Jarosz-Chobot, P.; Koczor-Rozmus, A.; Łanowy, P.; Mocny-Pachońska, K. Dental caries and periodontal status in children with type 1 diabetes mellitus. Pediatr. Endocrinol. Diabetes Metab. 2020, 26, 39–44. [Google Scholar] [CrossRef]
- Angwaravong, O.; Pitiphat, W.; Bolscher, J.G.M.; Chaiyarit, P. Evaluation of salivary mucins in children with deciduous and mixed dentition: Comparative analysis between high and low caries-risk groups. Clin. Oral Investig. 2015, 19, 1931–1937. [Google Scholar] [CrossRef]
- Wang, K.; Zhou, X.; Li, W.; Zhang, L. Human salivary proteins and their peptidomimetics: Values of function, early diagnosis, and therapeutic potential in combating dental caries. Arch. Oral Biol. 2019, 99, 31–42. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, Q.; Yan, G.; Yang, D. The oral microbiome and salivary proteins influence caries in children aged 6 to 8 years. BMC Oral. Health 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Sun, X.; Huang, X.; Tan, X.; Si, Y.; Wang, X.; Chen, F.; Zheng, S. Salivary peptidome profiling for diagnosis of severe early childhood caries. J. Transl. Med. 2016, 14, 240. [Google Scholar] [CrossRef] [Green Version]
- Busato, I.M.; Ignacio, S.A.; Brancher, J.A.; Moyses, S.T.; Azevedo-Alanis, L.R. Impact of clinical status and salivary conditions on xerostomia and oral health-related quality of life of adolescents with type 1 diabetes mellitus. Community Dent. Oral Epidemiol. 2012, 40, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Guglielmo Campus; Cocco, F.; Ottolenghi, L.; Cagetti, M.G. Comparison of ICDAS, CAST, Nyvad’s Criteria, and WHO-DMFT for Caries Detection in a Sample of Italian Schoolchildren. Int. J. Environ. Res. Public Health 2019, 16, 4120. [Google Scholar] [CrossRef] [Green Version]
- Loe, H. The Gingival Index, the Plaque Index and the Retention Index Systems. J. Periodontol. 1967, 38, 610–616. [Google Scholar] [CrossRef]
- Castagnola, M.; Cabras, T.; Iavarone, F.; Fanali, C.; Nemolato, S.; Peluso, G.; Bosello, S.L.; Faa, G.; Ferraccioli, G.; Messana, I. The human salivary proteome: A critical overview of the results obtained by different proteomic platforms. Expert Rev. Proteom. 2012, 9, 33–46. [Google Scholar] [CrossRef]
- Craig, R.; Beavis, R.C. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 2004, 20, 1466–1467. [Google Scholar] [CrossRef] [PubMed]
- Vempati, U.D.; Chung, C.; Mader, C.; Koleti, A.; Datar, N.; Vidovic, D.; Wrobel, D.; Erickson, S.; Muhlich, J.L.; Berriz, G.; et al. Metadata Standard and Data Exchange Specifications to Describe, Model, and Integrate Complex and Diverse High-Throughput Screening Data from the Library of Integrated Network-based Cellular Signatures (LINCS). J. Biomol. Screen 2014, 19, 803–816. [Google Scholar] [CrossRef] [Green Version]
- Pappa, E.; Vastardis, H.; Mermelekas, G.; Gerasimidi-Vazeou, A.; Zoidakis, J.; Vougas, K. Saliva Proteomics Analysis Offers Insights on Type 1 Diabetes Pathology in a Pediatric Population. Front. Physiol. 2018, 9, 444. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W. The Peptide Atlas Project. Methods Mol. Biol. 2010, 604, 285–296. [Google Scholar] [PubMed]
- Hicks, J.; Garcia-Godoy, F.; Flaitz, C. Biological factors in dental caries: Role of saliva and dental plaque in the dynamic process of demineralization and remineralization (part 1). J. Clin. Pediatr. Dent. 2004, 28, 47–52. [Google Scholar] [CrossRef]
- Nobbs, A.H.; Jenkinson, H.F.; Jakubovics, N.S. Stick to your gums: Mechanisms of oral microbial adherence. J. Dent. Res. 2011, 90, 1271–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegde, M.N.; Attavar, S.; Shetty, N.; Hegde, N.D.; Hegde, N.N. Saliva as a biomarker for dental caries: A systematic review. J. Conserv. Dent. 2019, 22, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Duncan, S.E.; Dietrich, A.M.; O’Keefe, S.F.; Eigel, W.N.; Mallikarjunan, K. Interaction of Copper and Human Salivary Proteins. J. Agric. Food Chem. 2009, 57, 6967–6975. [Google Scholar] [CrossRef]
- Huo, L.; Zhang, K.; Ling, J.; Peng, Z.; Huang, X.; Liu, H.; Gu, L. Antimicrobial and DNA-binding activities of the peptide fragments of human lactoferrin and histatin 5 against Streptococcus mutans. Arch. Oral Biol. 2011, 56, 869–876. [Google Scholar] [CrossRef]
- Cabras, T.; Pisano, E.; Mastinu, A.; Denotti, G.; Pusceddu, P.P.; Inzitari, R.; Fanali, C.; Nemolato, S.; Castagnola, M.; Messana, I. Alterations of the Salivary Secretory Peptidome Profile in Children Affected by Type 1 Diabetes. Mol. Cell. Proteom. 2010, 9, 2099–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amerongen, A.V.N.; Bolscher, J.; Veerman, E. Salivary Proteins: Protective and Diagnostic Value in Cariology? Caries Res. 2004, 38, 247–253. [Google Scholar] [CrossRef]
- Wang, K.; Wang, X.; Zheng, S.; Niu, Y.; Zheng, W.; Qin, X.; Li, Z.; Luo, J.; Jiang, W.; Zhou, X.; et al. iTRAQ-based quantitative analysis of age-specific variations in salivary proteome of caries-susceptible individuals. J. Transl. Med. 2018, 16, 293. [Google Scholar] [CrossRef]
- Duverger, O.; Beniash, E.; Morasso, M.I. Keratins as components of the enamel organic matrix. Matrix Biol. 2016, 52–54, 260–265. [Google Scholar] [CrossRef] [Green Version]
- Dominiczak, M.H.; Caslake, M.J. Apolipoproteins: Metabolic role and clinical biochemistry applications. Ann. Clin. Biochem. 2011, 48, 498–515. [Google Scholar] [CrossRef]
- Ljunggren, S.; Bengtsson, T.; Karlsson, H.; Johansson, C.S.; Palm, E.; Nayeri, F.; Ghafouri, B.; Davies, J.; Svensäter, G.; Lönn, J. Modified lipoproteins in periodontitis: A link to cardiovascular disease? Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Haukka, A.; Heikkinen, A.M.; Haukka, J.; Kaila, M. Oral health indices predict individualised recall interval. Clin. Exp. Dent. Res. 2020, 6, 585–595. [Google Scholar] [CrossRef]
- Novotna, M.; Podzimek, S.; Broukal, Z.; Lencova, E.; Duškova, J. Periodontal Diseases and Dental Caries in Children with Type 1 Diabetes Mellitus. Mediat. Inflamm. 2015, 2015, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belibasakis, G.N.; Bostanci, N.; Marsh, P.D.; Zaura, E. Applications of the oral microbiome in personalized dentistry. Arch. Oral Biol. 2019, 104, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Featherstone, J.D.B.; Chaffee, B.W. The Evidence for Caries Management by Risk Assessment (CAMBRA(R)). Adv. Dent. Res. 2018, 29, 9–14. [Google Scholar] [CrossRef] [PubMed]
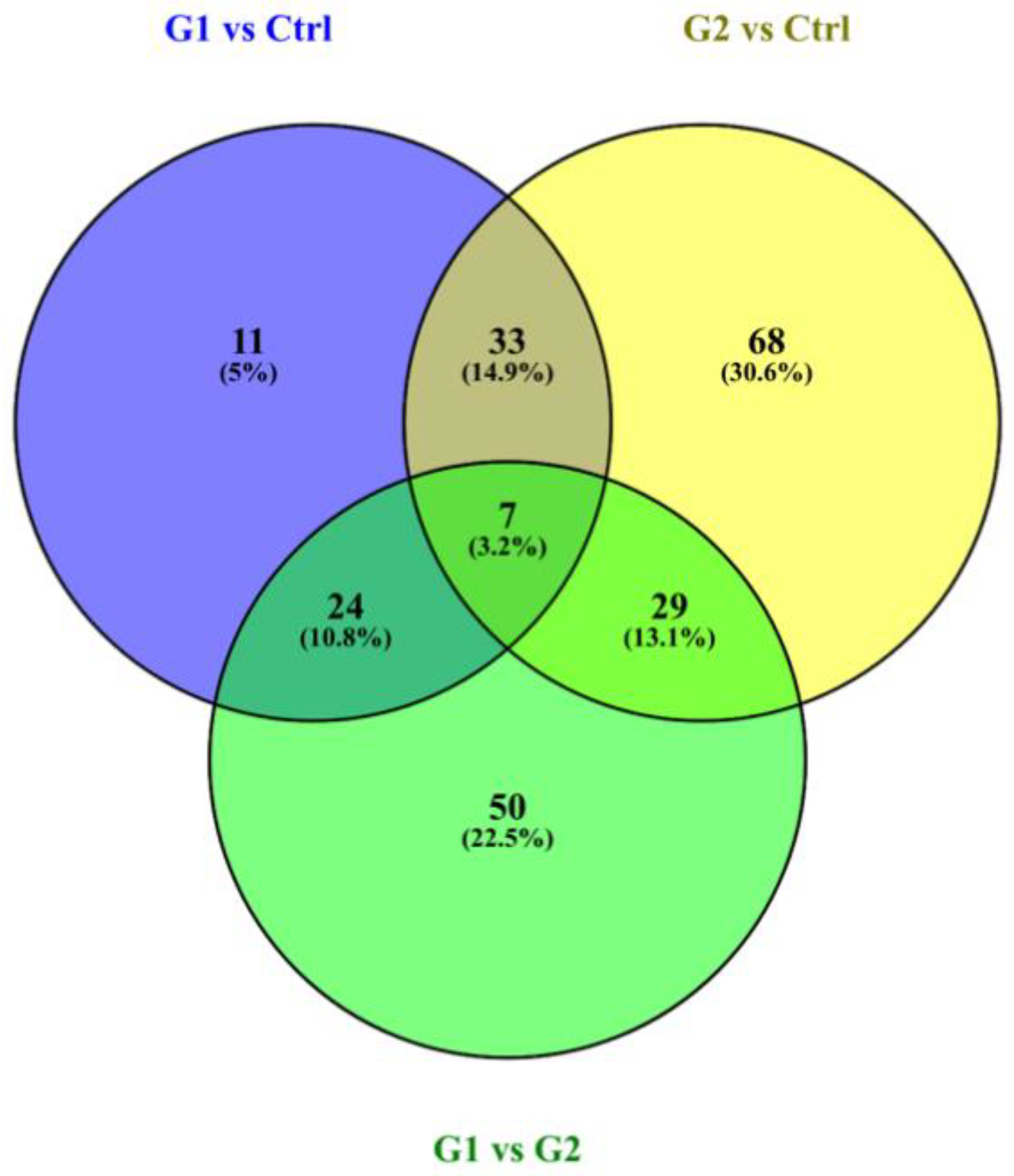
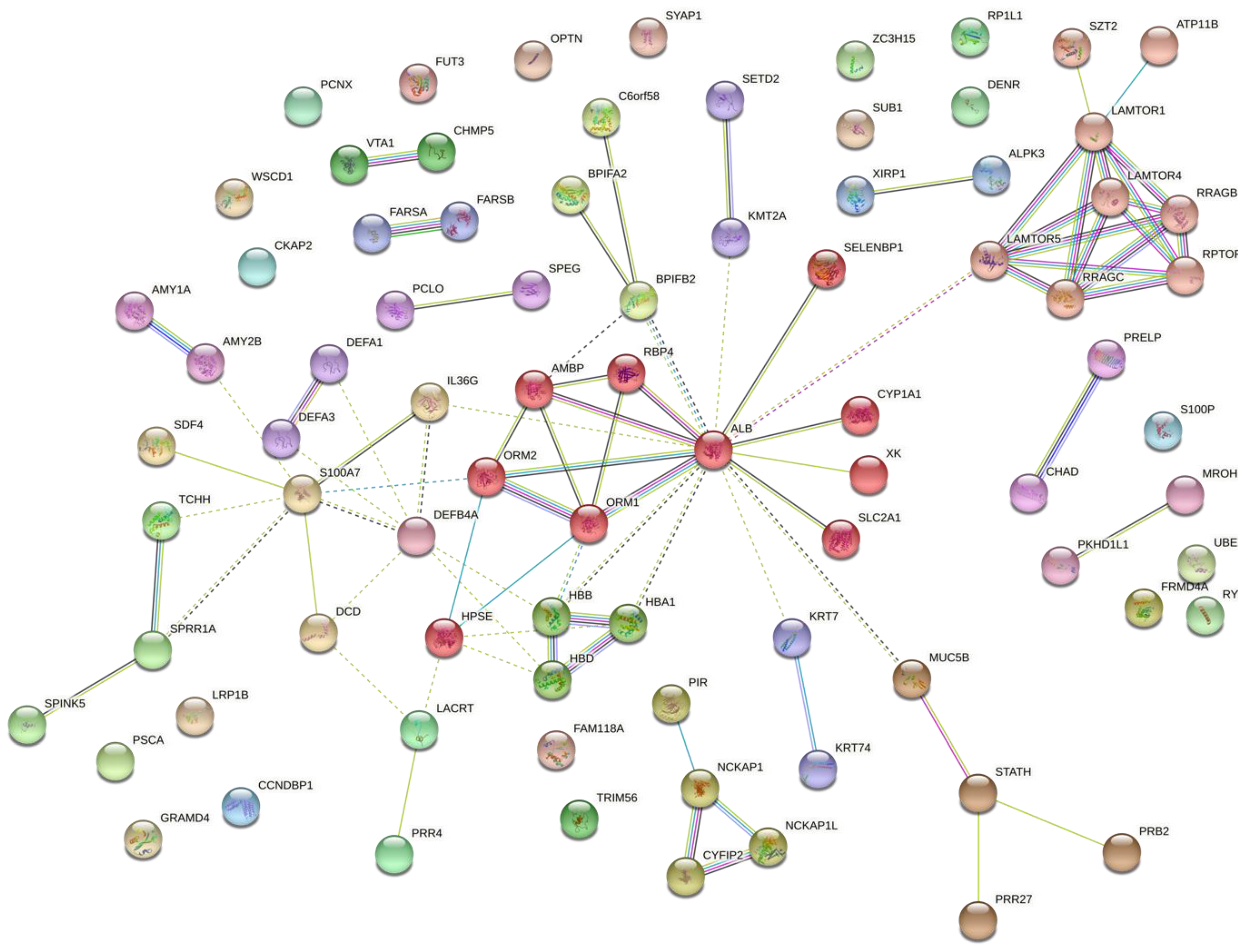
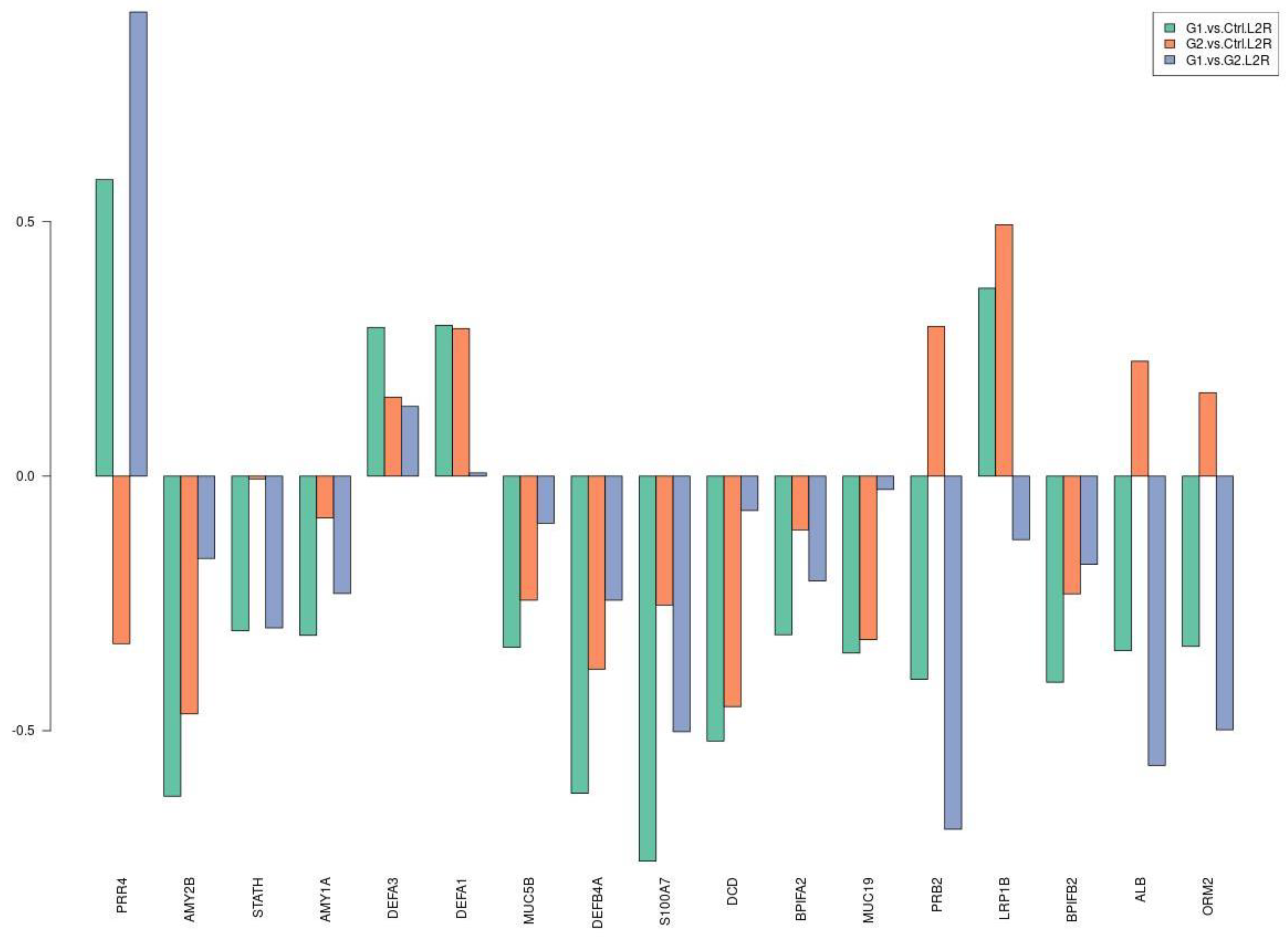
| Participants’ Characteristics | G1 | G2 | C |
|---|---|---|---|
| Age (yrs), mean (SD) | 14.5 ± 1.7 | 14.1 ± 1.3 | 14.9 ± 1.8 |
| Gender, n (Male/Female) | 5/7 | 5/7 | 5/7 |
| Time with DM1 (yrs) | 5.8 ± 1.9 | 6.4 ± 2.8 | - |
| HbA1c% (mmol/mol) | 9.7 ± 0.7 * (83) | 6.2 ± 0.4 * (44) | 4.2 ± 0.4 * (22) |
| BMI (kg/m2) | 22.9 ± 4.0 | 20.7 ± 5.0 | 24.3 ± 3.0 |
| Blood Pressure (mmHg) | 82 ± 5 | 79 ± 4 | 85 ± 5 |
| Diastolic Blood Pressure (mmHg) | 67 ± 3 | 63 ± 3 | 70 ± 4 |
| Systolic Blood Pressure (mmHg) | 113 ± 4 | 109 ± 3 | 114 ± 3 |
| Total cholesterol (mg/dL) | 165 ± 10 | 160 ± 12 | 168 ± 15 |
| LDL cholesterol (mg/dL) | 92 ± 6 | 88 ± 5 | 94 ± 8 |
| Plaque Index (PI) | 0.80 ± 0.05 | 0.60 ± 0.03 | 0.70 ± 0.02 |
| Gingival Index (GI) | 0.7 ± 0.2 | 0.5 ± 0.2 | 0.8 ± 0.1 |
| DMFT | 3.9 ± 0.7 * | 0.9 ± 0.2 * | 1.1 ± 0.3 * |
| G1 vs. C L2R-p-DAVID | ||
| Term | Proteins | p value |
| Secreted | PRR4, AMY2B, PRELP, STATH, ORM2, BPIFA2, C6ORF58, CHAD, PRB2, AMBP, DCD, AMY1A, MUC19, SPINK5, DEFA3, IL36G, DEFA1, MUC5B, BPIFB2, RBP4, ALB, HPSE, DEFB4A, LACRT, S100A7 | 1.79 × 10−8 |
| GO:0042742~defense response to bacterium | IGLC7, DCD, BPIFA2, DEFA3, MUC5B, DEFB4A, STATH, LACRT | 1.42 × 10−6 |
| GO:0005796~Golgi lumen | MUC19, DEFA3, SDF4, PRELP, DEFA1, MUC5B, DEFB4A | 1.98 × 10−6 |
| GO:0019825~oxygen binding | ALB, CYP1A1, HBB, HBD, HBA1 | 2.00 × 10−5 |
| signal peptide | PRR4, PSCA, AMY2B, PRELP, STATH, ORM2, BPIFA2, C6ORF58, CHAD, PRB2, SDF4, AMBP, DCD, AMY1A, MUC19, SPINK5, DEFA3, DEFA1, MUC5B, LRP1B, BPIFB2, RBP4, ALB, HPSE, PKHD1L1, DEFB4A, LACRT | 3.07 × 10−5 |
| IPR006080:Beta defensin-Neutrophil defensin | DEFA3, DEFA1, DEFB4A | 2.49 × 10−4 |
| Signal | PRR4, PSCA, AMY2B, PRELP, STATH, ORM2, BPIFA2, C6ORF58, CHAD, PRB2, SDF4, AMBP, DCD, AMY1A, MUC19, SPINK5, DEFA3, DEFA1, MUC5B, LRP1B, BPIFB2, RBP4, ALB, CYP1A1, HPSE, PKHD1L1, DEFB4A, LACRT | 3.27 × 10−4 |
| glycosylation site:N-linked (Glc) (glycation) | ALB, HBB, HBA1 | 3.54 × 10−4 |
| Antimicrobial | DCD, BPIFA2, DEFA3, DEFA1, DEFB4A | 3.84 × 10−4 |
| Glycation | ALB, HBB, HBA1 | 4.04 × 10−4 |
| hsa04970:Salivary secretion | PRB2, MUC5B, STATH, RYR3 | 0.001844339 |
| calcium-binding region:2; high affinity | TCHH, S100P, S100A7 | 0.002649627 |
| Calcium | PCLO, AMY1A, AMY2B, TCHH, SDF4, S100P, HPSE, RYR3, LRP1B, S100A7 | 0.003100924 |
| Pyrrolidone carboxylic acid | AMY1A, AMY2B, PRB2, ORM2 | 0.004088861 |
| GO:0050832~defense response to fungus | DCD, DEFA3, DEFA1 | 0.004847206 |
| IPR000566:Lipocalin-cytosolic fatty-acid binding protein domain | RBP4, AMBP, ORM2 | 0.006685494 |
| Defensin | DEFA3, DEFA1, DEFB4A | 0.013921585 |
| GO:0004556~alpha-amylase activity | AMY1A, AMY2B | 0.017063442 |
| IPR006046:Alpha amylase | AMY1A, AMY2B | 0.0179226 |
| GO:0050829~defense response to Gram-negative bacterium | DEFB4A, OPTN, S100A7 | 0.019204342 |
| IPR016327:Alpha-defensin | DEFA3, DEFA1 | 0.021468961 |
| IPR026086:Proline-rich protein | PRR4, PRB2 | 0.025002707 |
| IPR002345:Lipocalin | RBP4, AMBP | 0.028523881 |
| Glycoprotein | PSCA, AMBP, WSCD1, AMY1A, SLC2A1, HBB, PRELP, HBA1, MUC5B, ORM2, LRP1B, FUT3, BPIFB2, C6ORF58, BPIFA2, CHAD, ALB, PRB2, CYP1A1, SDF4, HPSE, PKHD1L1, VN1R5, LACRT | 0.029468179 |
| GO:0050830~defense response to Gram-positive bacterium | DEFA3, DEFA1, DEFB4A | 0.042886244 |
| Phosphoprotein | SETD2, KMT2A, SYAP1, SLC2A1, HBB, HBD, CCNDBP1, STATH, GRAMD4, SELENBP1, PCLO, PRB2, XIRP1, SDF4, SPEG, DENR, ZC3H15, SZT2, CKAP2, FAM118A, KRT7, FRMD4A, ATP11B, UBE4B, DEFA1, HBA1, KRT74, LRP1B, BPIFB2, RRAGC, SUB1, ALB, ALPK3, FARSA, OPTN, TRIM56, CHMP5 | 0.044573527 |
| G2 vs. C L2R-p-DAVID | ||
| Term | Proteins | p value |
| glycosylation site:N-linked (Glc) (glycation) | ALB, HBB, APOA1, HBA1, CFB | 2.20 × 10−7 |
| Glycation | ALB, HBB, APOA1, HBA1, CFB | 7.69 × 10−7 |
| Pyrrolidone carboxylic acid | ORM1, AMY2B, PRB2, APOA2, PRH1, KNG1 | 3.33 × 10−4 |
| IPR026086:Proline-rich protein | PRR4, PRB2, PRH1 | 9.39 × 10−4 |
| GO:0042742~defense response to bacterium | IGHG1, IGHG2, DCD, MUC5B, DEFB4A, LACRT | 0.004284705 |
| IPR002957:Keratin, type I | KRT33B, KRT12, KRT9 | 0.021047193 |
| GO:0034190~apolipoprotein receptor binding | APOA2, APOA1 | 0.021526132 |
| Antimicrobial | DCD, DEFA1, LYZ, DEFB4A | 0.032341013 |
| G1 vs. G2 L2R-p-DAVID | ||
| Term | Proteins | p value |
| Protease inhibitor | ITIH4, APP, CST1, SERPINA1, ITIH2, AMBP, SERPINC1, SPINK5, SERPING1, A2M, CST5, CST4 | 5.10 × 10−11 |
| glycosylation site:O-linked (GalNAc...) | ITIH4, HPX, TF, ITIH2, AMBP, AHSG, SERPING1, PLG | 9.29 × 10−7 |
| Glycoprotein | APP, ITIH4, ORM1, SERPINA1, ITIH2, CFH, SERPINC1, PON1, LAMA3, SLC2A1, PLG, FURIN, A1BG, ORM2, PRH1, PKHD1, C3, IGHG3, C1QTNF3, IGHG4, HPX, C6, TTR, IGHG2, C9, PRB2, A2M, GC, CPA4, FGB, FGA, AMBP, AHSG, FGG, APOA2, APOA1, CP, TF, BPIFB1, SCGB2A1, ALB, CYP1A1, SERPING1, COL4A5, CFB | 1.01 × 10−5 |
| GO:0006956~complement activation | IGHG3, C3, IGHG4, C6, IGHG2, CFH, CFB | 1.64 × 10−5 |
| GO:0002020~protease binding | CST1, SERPINA1, SERPINC1, FURIN, A2M, CST5, CST4 | 2.37 × 10−5 |
| Complement alternate pathway | C3, CFH, C9, CFB | 2.99 × 10−5 |
| GO:0006953~acute-phase response | ITIH4, ORM1, SERPINA1, AHSG, ORM2 | 9.67 × 10−5 |
| GO:0045087~innate immune response | FGB, IGHG3, FGA, APP, IGHG4, C6, IGHG2, HMGB2, SERPING1, IL36G, S100A7 | 3.34 × 10−4 |
| Complement pathway | C3, C6, C9, SERPING1 | 5.68 × 10−4 |
| IPR026086:Proline-rich protein | PRR4, PRB2, PRH1 | 6.79 × 10−4 |
| region of interest:Coil 1B | KRT19, KRT12, KRT7, KRT75, KRT74 | 6.84 × 10−4 |
| GO:0044267~cellular protein metabolic process | FGA, APP, TTR, APOA1, FURIN, PLG | 8.32 × 10−4 |
| GO:0030674~protein binding, bridging | FGB, FGA, FGG, SPRR1A, OPTN | 9.68 × 10−4 |
| GO:1900026~positive regulation of substrate adhesion-dependent cell spreading | FGB, FGA, FGG, APOA1 | 0.001004558 |
| glycosylation site:N-linked (Glc) (glycation) | ALB, APOA1, CFB | 0.001008382 |
| IPR001664:Intermediate filament protein | KRT19, KRT12, KRT7, KRT75, KRT74 | 0.001073524 |
| Phosphoprotein | APP, SERPINA1, HMGB2, SLC2A1, PTPN23, UNC80, PRB2, DENR, DLAT, KCMF1, FGA, PCYT2, AHSG, CAD, DHPS, FGG, APOA2, CKAP2, KRT7, APOA1, ATP11B, TERF1, KRT74, CCDC80, ESRP2, SUB1, SPECC1L, ALPK3, OPTN, ITIH2, KMT2A, SERPINC1, FURIN, PLG, CCNDBP1, STATH, GRAMD4, PRH1, CST4, C3, RNF213, TTR, C9, TPR, XIRP1, HIVEP2, PCNT, EPS15, SASH1, PEX19, RAB27A, CP, MTOR, KRT19, TF, FUBP1, ALB, TACC2, TARDBP, FARSA | 0.001509071 |
| Glycation | ALB, APOA1, CFB | 0.001739912 |
| IPR027214:Cystatin | CST1, CST5, CST4 | 0.002464241 |
| GO:0050829~defense response to Gram-negative bacterium | APP, HMGB2, OPTN, S100A7 | 0.004793789 |
| IPR003054:Type II keratin | KRT7, KRT75, KRT74 | 0.011288975 |
| IPR001500:Alpha-1-acid glycoprotein | ORM1, ORM2 | 0.011497863 |
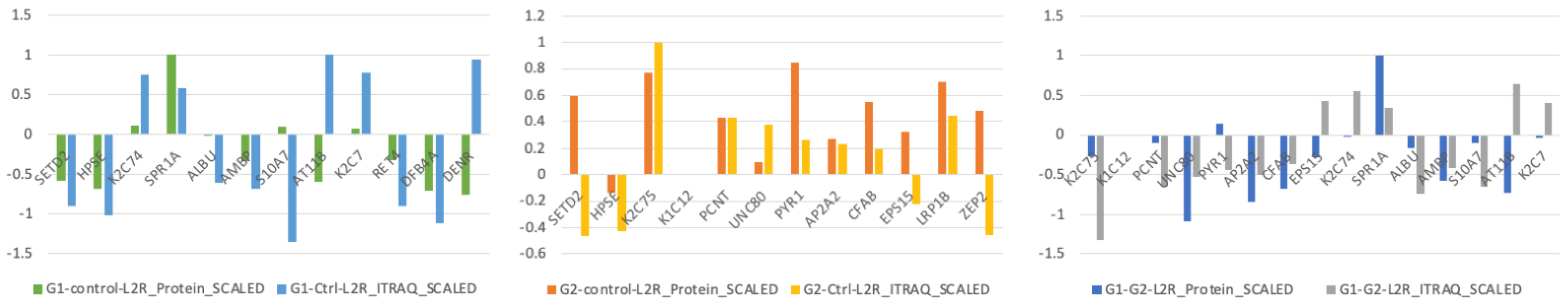
| Proteins | G1. vs. C. Fold Change | G2. vs. C. Fold Change | G1. vs. G. Fold Change |
|---|---|---|---|
| S100A7 | −1.688 | −1.192 | −1.416 |
| AMY2B | −1.546 | −1.382 | −1.119 |
| DEFB4A | −1.540 | −1.300 | −1.184 |
| DCD | −1.434 | −1.369 | −1.048 |
| RBP4 | −1.418 | 1.003 | −1.423 |
| BPIFB2 | −1.324 | −1.174 | −1.127 |
| PRB2 | −1.319 | 1.226 | −1.617 |
| MUC19 | −1.272 | −1.249 | −1.018 |
| MUC5B | −1.262 | −1.184 | −1.067 |
| IL36G | −1.247 | 1.081 | −1.349 |
| AMY1A | −1.242 | −1.058 | −1.173 |
| STATH | −1.235 | −1.004 | −1.229 |
| CST3 | −1.222 | −1.130 | −1.081 |
| APOA2 | −1.222 | 1.174 | −1.435 |
| CST5 | −1.209 | −1.008 | −1.199 |
| KRT9 | −1.204 | −1.165 | −1.034 |
| BPIFB1 | −1.201 | 1.057 | −1.269 |
| APOA1 | −1.198 | 1.247 | −1.496 |
| MUC7 | −1.188 | −1.236 | 1.040 |
| SERPINC1 | −1.166 | 1.058 | −1.235 |
| CST4 | −1.162 | 1.046 | −1.216 |
| CST1 | −1.149 | 1.053 | −1.211 |
| C6 | −1.135 | 1.111 | −1.261 |
| C3 | −1.120 | 1.074 | −1.202 |
| C9 | −1.118 | 1.104 | −1.235 |
| BPI | −1.087 | 1.099 | −1.196 |
| KRT77 | 1.147 | −1.025 | 1.175 |
| KRT19 | 1.223 | 1.010 | 1.211 |
| DEFA3 | 1.224 | 1.113 | 1.100 |
| DEFA1 | 1.228 | 1.223 | 1.004 |
| SPRR1A | 1.255 | 1.049 | 1.197 |
| KRT74 | 1.337 | 1.000 | 1.337 |
| ATP11B | 1.471 | 1.046 | 1.406 |
| PRR4 | 1.497 | −1.256 | 1.880 |
| ORM2 | −1.261 | 1.120 | −1.412 |
| ALB | −1.268 | 1.169 | −1.482 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pappa, E.; Vougas, K.; Zoidakis, J.; Papaioannou, W.; Rahiotis, C.; Vastardis, H. Downregulation of Salivary Proteins, Protective against Dental Caries, in Type 1 Diabetes. Proteomes 2021, 9, 33. https://doi.org/10.3390/proteomes9030033
Pappa E, Vougas K, Zoidakis J, Papaioannou W, Rahiotis C, Vastardis H. Downregulation of Salivary Proteins, Protective against Dental Caries, in Type 1 Diabetes. Proteomes. 2021; 9(3):33. https://doi.org/10.3390/proteomes9030033
Chicago/Turabian StylePappa, Eftychia, Konstantinos Vougas, Jerome Zoidakis, William Papaioannou, Christos Rahiotis, and Heleni Vastardis. 2021. "Downregulation of Salivary Proteins, Protective against Dental Caries, in Type 1 Diabetes" Proteomes 9, no. 3: 33. https://doi.org/10.3390/proteomes9030033
APA StylePappa, E., Vougas, K., Zoidakis, J., Papaioannou, W., Rahiotis, C., & Vastardis, H. (2021). Downregulation of Salivary Proteins, Protective against Dental Caries, in Type 1 Diabetes. Proteomes, 9(3), 33. https://doi.org/10.3390/proteomes9030033