Abstract
Host-pathogen interactions are often studied in vitro using primary or immortal cell lines. This set-up avoids ethical problems of animal testing and has the additional advantage of lower costs. However, the influence of cell culture media on bacterial growth and metabolism is not considered or investigated in most cases. To address this question growth and proteome adaptation of Corynebacterium diphtheriae strain ISS3319 were investigated in this study. Bacteria were cultured in standard growth medium, cell culture medium, and fetal calf serum. Mass spectrometric analyses and label-free protein quantification hint at an increased bacterial pathogenicity when grown in cell culture medium as well as an influence of the growth medium on the cell envelope.
1. Introduction
Corynebacterium diphtheriae is a gram-positive, non-motile, facultative anaerobe bacterium [1] which may lead to severe infections in humans. Symptoms of the classical upper respiratory tract diphtheria caused by diphtheria toxin (DT)-producing C. diphtheriae strains may range from mild pharyngitis with low fever to suffocation and death [2]. Due to effective vaccination strategies, diphtheria is generally well-controlled [3,4]. Nevertheless, 22,986 cases were reported by the World Health Organization with outbreaks in India (9622), Ethiopia (7184), Nigeria (2289), and Madagascar (1815) [4].
Besides infections by toxigenic strains, non-toxigenic strains can also cause severe symptoms in humans such as skin lesions [5], myocarditis, [6] and septic arthritis [7,8]. The latest data available showed an increase in infections caused by non-toxigenic strains at least in Germany [9]. Non-toxigenic strain ISS3319, used in this study, was isolated from a patient with severe pharyngitis/tonsillitis and was described as a strain with arthritogenic potential [10]. When mice were challenged with C. diphtheriae ISS3319, the bacteria were able to infiltrate deeper tissues, and joints of infected mice developed mild-to-moderate arthritis. Interestingly, the bacteria were also able to persist in kidneys and spleens for more than two weeks after infection [10].
To unravel mechanisms of host-pathogen interaction, a number of in vitro experiments with non-toxigenic corynebacterial strains and human and animal cell lines were carried out (e.g., see [10,11,12,13,14,15,16,17]). However, until now, the influence of cell culture medium and serum were completely neglected. To address this problem, we started a proteomics approach to characterize putative influences of different cell culture media on the proteome of C. diphtheriae.
Proteomics have already been used successfully in studying different pathogenic corynebacteria [18,19,20,21,22]. Nevertheless, most of the currently existing proteomic studies were carried out using bacteria grown in complex media; however, these do not represent the conditions the bacteria are facing during the infection process in vitro. For this purpose, the influence of two different cell culture media on viability, cell growth, and proteome of a non-toxigenic C. diphtheriae strain was analyzed. In total, 781 proteins were identified, 41 differentially expressed proteins and 115 proteins exclusively expressed in cell culture medium were found (49 proteins were exclusively observed for RPMI 1640, 35 proteins exclusive for FCS, and 31 proteins present in both conditions). The data suggest an increased bacterial pathogenicity when grown under stress conditions as well as an influence of the growth medium on the cell envelope. An increased occurrence of two-component systems indicates an adaption and stress response of the bacteria.
2. Materials and Methods
2.1. Bacteria and Growth Conditions
C. diphtheriae strain ISS3319 [10] (kindly provided by Christina von Hunolstein, Istituto Superiore di Sanità, Rome, Italy) was cultured under constant shaking in brain heart infusion (BHI) broth (Oxoid), RPMI 1640 (Capricorn, Epsdorfergrund, Germany), or fetal calf serum (FCS) (Capricorn, Epsdorfergrund, Germany) in bluffed flasks at 37 °C. For growth experiments an over-day culture was inoculated to an optical density at 600 nm (OD600) of 0.15 and grown at 37 °C under constant shaking. Colony forming units (CFU) and OD600 were determined every hour for 6 h and after 24 h. Experiments were carried out in three independent biological replicates, mean and standard deviation were calculated for every time point.
2.2. Sample Preparation of Proteomic Analysis
Protein samples were prepared as described [18]. Bacteria grown in BHI medium, RPMI 1640, and FCS were harvested for whole proteome analysis and lysed with a homogenizer using glass beads (5.5 m s−1, 30 s, 5 cycles, 4 °C). 40 µg of extracted proteins were alkylated (40 mM CAA final concentration), precipitated with acetone (80% final concentration, overnight, 4 °C), resuspended in 100 mM TEAB buffer with 2 µg trypsin, and digested overnight at 37 °C. Twenty-five micrograms of the resulting peptides (three biological replicates of secreted proteins and whole proteome from bacteria grown in BHI, RPMI 1640, and FCS) were purified using C18 stage tips, vacuum dried, and resuspended in 0.1% trifluoroacetic acid (TFA) before being supplied for LC-MS/MS analysis [18].
2.3. Mass Spectrometry
Mass spectrometric analysis was carried out as described previously [18,19,23]. For separation of peptides, 10 µg were supplied to a nanoflow Ultimate 3000 HPLC (Dionex, Sunnydale, CA, USA) and loaded onto an EASY-Spray column (Thermo Fisher Scientific; C18 with 2 µm particle size, 50 cm × 75 µm) with a flow rate of 200 nL min−1 and an increasing acetonitrile concentration over 120 min. An Orbitap Fusion mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was used for analysis of all samples using the following settings: spray voltage 2000 V, transfer tube temperature 275 °C, scan range for MS 1 detection in the Orbitrap 300–2000 (m/z), 50 ms maximum injection time, automatic gain control (AGC) target of 400,000, and Orbitrap resolution of 120.000. For collision-induced dissociation the most intense ions were selected with collision energy of 35%. For ion trap detection a maximum injection time of 250 ms and an AGC target of 100 were set [18,19,23]. The Proteome Discoverer program package 1.4 (Thermo Fisher Scientific, Bremen, Germany) and C. diphtheriae NCTC13129 database (Proteome Id: UP000002198) in UniProt (www.uniprot/proteomes) (accessed on 12 March 2021) were used to analyze the resulting raw data files with following settings: (I) theoretical masses of peptides were generated with a maximum of two missed cleavages, as described in [24], (II) fixed modification: carbamidomethyl on cysteine, (II) dynamic: oxidation of methionine, (III) mass tolerance for survey scans: 10 ppm and 0.6 Da for fragment mass measurements, (IV) false discovery rate (FDR): 1%. FDR was determined based on a target-decoy search and q-value the implemented percolator workflow from the Proteome Discoverer 1.4 software package (Thermo Fisher Scientific, Bremen, Germany) [19].
2.4. Data Analysis
Only proteins, which are present in all three independent biological replicates, were considered as identified. The peak areas of the identified proteins were normalized via the molecular weight. For label-free quantification, the relative abundance of each protein was calculated based on the total protein approach (TPA) method [25]. A multiple sample test (ANOVA) was applied to identify significant expression levels of the common proteins. A Z-score was calculated before hierarchical cluster analysis using the Euclidian algorithm. Information in respect to prediction location of identified proteins were extracted from published data [26]. Data on pathway analysis were extracted from Sangal et al., 2015 [26] and the KEGG consortium [27,28,29]. Collected data (see supplementary Table S1) were visualized using the proteomaps program (https://bionic-vis.biologie.uni-greifswald.de/) (accessed on 12 March 2021) [30,31,32].
3. Results
3.1. Growth Experiments under Different Culture Conditions
In order to evaluate the influence of standard cell culture media on bacterial growth, optical density and colony-forming units of ISS3319 cultures were monitored. Bacterial growth in standard BHI medium was comparable with growth in cell culture media, i.e., FCS and RPMI 1640 (Figure 1a). For bacteria grown in BHI a doubling time of 57 ± 2 min was determined, while 74 ± 3 min were measured for RPMI 1640 and 51 ± 2 min for FCS. After 24 h incubation bacteria reached a final OD600 of 5.3 ± 0.4 in BHI, 2.2 ± 0.2 in RMPI 1640, and 7.4 ± 1.4 in FCS. To monitor cell viability, bacteria were plated after two, four, and six h. The number of CFU increased from the beginning of the experiment (zero h) to four h under all conditions. No growth for bacteria in RPMI 1640 medium at six h was noticed, whereas bacteria in BHI and FCS were able to increase biomass (Figure 1b).
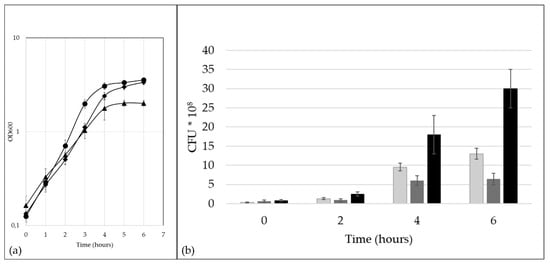
Figure 1.
Growth of C. diphtheriae ISS3319. (a) Optical density in brain heart infusion (BHI) medium (rhombi), RPMI 1640 (triangles), and fetal calf serum (FCS) (circles). (b) Colony forming units (CFU) at different time points for bacteria grown in BHI medium (light grey), RPMI 1640 (dark grey), and FCS (black). Three independent biological replicates were carried out for all experiments. Data show the calculated mean value and the resulting standard deviation.
3.2. Mass Spectrometric Analysis
By mass spectrometry, 666 proteins from C. diphtheriae ISS3319 grown in BHI medium were identified, 602 proteins from bacteria grown in RPMI 1640, and 562 proteins from bacteria grown in FCS. In total, 781 distinct proteins were found comprising 34.5% of the annotated genome. 92 proteins of the BHI sample, 49 proteins from RPMI 1640-grown cells, and 35 proteins from bacteria grown in FCS were found to be exclusive for the respective conditions (Figure 2, supplemental Table S2).
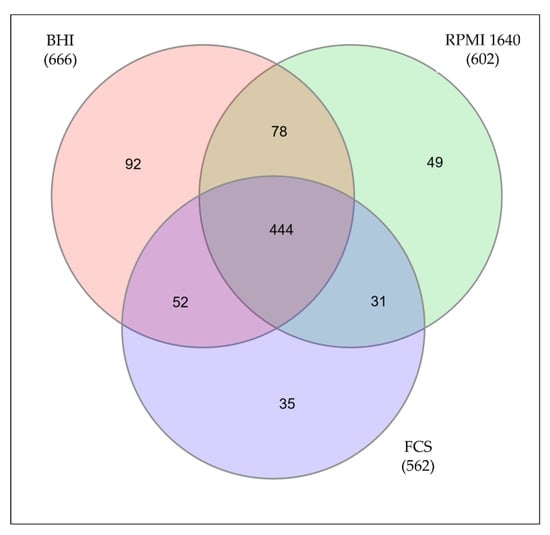
Figure 2.
Proteins identified under different growth conditions. Only proteins identified in all three independent replicates were considered for further analysis. The Venn diagram shows the common proteins as well as the unique proteins for all three growth conditions.
The prediction of protein localization showed a similar distribution for all three media. The main parts of the identified proteins are located to the cytoplasm (BHI: 87%; RMPI 1640: 87%; FCS: 85%) followed by proteins with extracellular localization (BHI: 6%; RPMI 1640: 7%; FCS: 6%), proteins located to the membrane (BHI: 4%; RPMI 1640: 3%; FCS: 4%), and proteins with ambiguous localization (BHI: 3%; RPMI 1640: 3%; FCS: 5%) (Figure 3a–c).
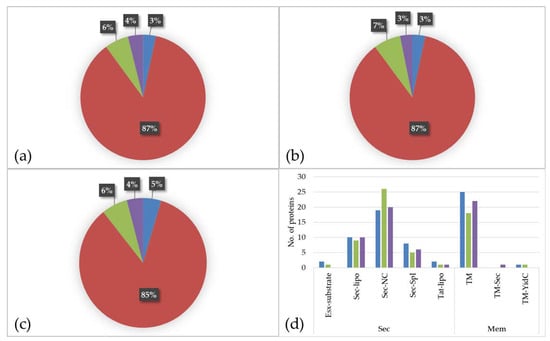
Figure 3.
Proteome of C. diphtheriae strain ISS3319 grown in various culture media ((a): BHI; (b): RPMI 1640; (c): FCS). Percentage of different proteins located to the cytoplasm (red), extracellular (green), located to the membrane (purple), and proteins with ambiguous localization (blue). (d) shows the classification of secreted proteins and proteins with localization to the membrane regarding the secretion system. Blue: BHI, green: RPMI 1640 and purple: FCS.
An analysis of the secreted and transmembrane proteins revealed two ESAT-6-like proteins, EsxA (DIP0559) and EsxB (DIP0558), both present in BHI and the latter one also in RPMI 1640. These proteins belong to WGX100 family, which are type VII secretion substrates and thought to play a role in pathogenicity in taxonomically closely related bacteria such as Mycobacterium tuberculosis [26,33]. In total, 14 lipoproteins (Sec-lipo) (BHI: 10; RPMI 1640: 9; FCS: 10), 29 non-classical secreted (Sec-NC) proteins (BHI: 19; RPMI 1640: 26; FCS: 20), 10 proteins with an SPI signal peptide (Sec-SPI) (BHI: 8; RPMI 1640: 5; FCS: 6), and two lipoproteins secreted by Twin-Arginine Translocation pathway (TAT-lipo) were identified (BHI: 2; RPMI 1640: 1; FCS: 1). Within the Sec-SPI proteins are two putative secreted antigens and predicted mycolyltransferases involved in cell envelope assembly (DIP2193 and DIP2194) [34] and the putative invasion protein DIP1281, which was shown to be involved in virulence [35]. The TAT-lipo protein DIP1389 is a putative Dyp family peroxidase, which is thought to bind heme (Figure 3d).
When proteins with localization to the membrane (TM) were analyzed in total 32 proteins were found (BHI: 25; RPMI 1640: 18; FCS: 22). The multifunctional protein DIP0733, a fibronectin-binding protein involved in adhesion and invasion of host cells [17], was present in all three samples (BHI, RPMI 1640, and FCS). One single YidC protein (TM-YidC), namely DIP2379, which is involved in insertion of integral membrane proteins [36], and one transmembrane protein (DIP0736) with an SPI (TM-sec) was found in the dataset (Figure 3d).
3.3. Differentially Expressed Proteins in Cell Culture Media and Serum
Proteins found under all three culture conditions were subjected to a pathway analysis (see ProteoTreeMap (Figure 4)). Most proteins were attributed to the categories metabolism, information storage and processing, and cellular processes and signaling as would be expected since proteins were extracted from similarly growing cells.
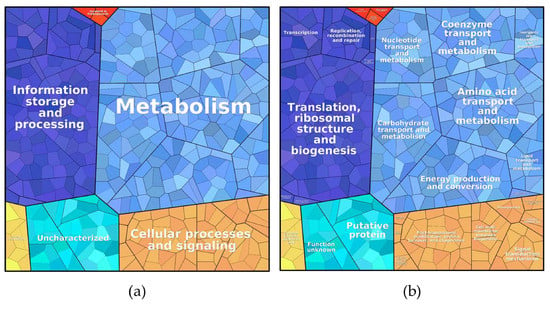
Figure 4.
ProteoTreeMap of the total protein content in all three samples. (a) level 1: metabolic pathway and (b) level 3: function.
Interestingly, three proteins involved in pathogenesis were identified: the multifunctional protein DIP0733, the conserved hypothetical protein DIP1546, and the resuscitation-promoting factor RpfB DIP0874. Based on a label-free quantification method (TPA-method) the relative abundance of each proteins was calculated. The most abundant protein in all three samples was the iron repressible polypeptide (dirA) (DIP1420) and is involved in cellular processes and signaling. This protein has an ortholog protein AhpC in M. tuberculosis involved in tolerance of reactive oxygen and nitric oxide tolerance during macrophage infection [21,37,38]. The DtxR-regulated protein DIP2303 [39], which is involved in DNA protection during starvation, was among the most abundant proteins. This protein is similar to the starvation inducible non-specific DNA-binding protein (Dps) in Escherichia coli [40] and protects DNA from oxidative stress damage [41]. The manganese superoxide dismutase (sodA) DIP2261 also showed a high abundance in all three samples. SodA is involved in cell viability and knock-out mutants of Corynebacterium melassecola showed increased sensitivity to superoxide radicals [42].
Based on the abundance of each protein a multiple sample test using the ANOVA algorithm was applied. Forty-one proteins showed significantly different expression patterns among the three different growth conditions. The hierarchical cluster analysis revealed a separation from bacteria grown in BHI from the bacteria grown in RPMI 1640 medium and FCS. The proteins can be divided into five main clusters (Figure 5; supplemental Table S3). In the first cluster are proteins with a high expression in RPMI 1640 and lower expression rates in BHI and FCS represented. Two proteins of this cluster, DIP0550 and DIP0620, are under the regulation of DtxR [39,42], which indicates iron starvation under these conditions.
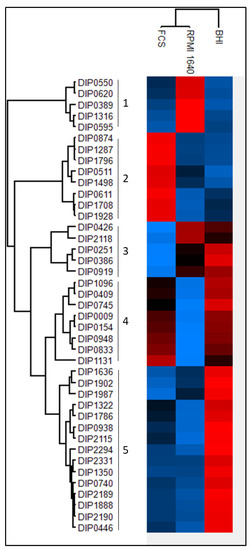
Figure 5.
Differences in expression level of common proteins. A high intensity is shown in red and goes to a low intensity in blue. The relative abundance was used for a multiple sample test (ANOVA). A Z-score was calculated for the resulting 41 proteins and clustered using the Euclidian algorithm.
The second cluster shows the enriched proteins when bacteria were grown in serum, and clusters 3, 4, and 5 show proteins with a lesser abundance in RPMI 1640 and FCS compared to BHI. The resuscitation-promoting factor RpfB (DIP0874) shows a high expression level in serum compared to BHI and RPMI 1640. Cluster four comprises eight proteins (DIP1096, DIP0409, DIP0745, DIP0009, DIP0154, DIP0948, DIP0833, and DIP1131). Significantly increased abundance of proteins from bacteria grown in BHI medium are represented in cluster five. This cluster includes 15 proteins (DIP1636, DIP1902, DIP1987, DIP1322, DIP1786, DIP0938, DIP2115, DIP2294, DIP2331, DIP1350, DIP0740, DIP2189, DIP1888, DIP2190, and DIP0446), three proteins (DIP0740, DIP2189, and DIP2190) are thought to be involved in mycolic acid synthesis [43] and a homologue of DIP1350 is essential in M. tuberculosis [44].
3.4. Newly Synthesised Proteins under Two Different Growth Conditions
The analysis of the proteins exclusively found in bacteria grown under different culture conditions (Figure 6; Table 1 and Table 2) revealed 92 proteins specific to BHI, 35 proteins for FCS, and 49 proteins for RPMI 1640.
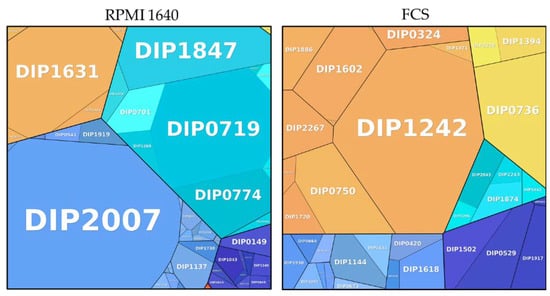
Figure 6.
Metabolic pathways of proteins exclusively found in different growth conditions. Information storage and processing: dark blue; metabolism: light blue; cellular processes and signaling: orange; involved in pathogenesis: red; uncharacterized: turquoise; poorly characterized: yellow. The size of the area is proportional to the abundance calculated based on the total protein approach (TPA) method for label-free quantification.

Table 1.
Distribution of metabolic pathways of exclusive proteins found in the respective medium. The numbers show the abundance in percent [%].

Table 2.
Distribution of proteins exclusively found under two different growth conditions (RPMI 1640 and FCS). Pathway is shown in the first column (C: Cellular processes and signaling; I: Information storage and processing; P: Involved in Pathogenesis; M: Metabolism). The numbers represent the relative amount [%] when the exclusive proteins were set to 100%.
Bacteria grown in RPMI 1640 showed an enrichment of proteins involved in metabolism (40.00%) followed by a similar amount of uncharacterized proteins (39.31%), proteins involved in cellular processes and signaling (15.94%), information storage and processing (4.65%), poorly characterized (0.08%), and one protein involved in pathogenesis (DIP1281).
Proteins involved in cellular processes and signaling were enriched (56.65%) in exclusive proteins from bacteria grown in FCS. Poorly characterized proteins were represented with a ratio of 14.07%, proteins involved in metabolism 12.90%, proteins involved in information storage and processing with 10.12%, and uncharacterized proteins with 6.24%.
A detailed breakdown of the function of the pathways from proteins exclusively found under growth conditions showed a high amount of ribosomal proteins (33.63%) in BHI, whereas an enrichment of proteins for inorganic ion transport and metabolism were noticed in RPMI (33.77%). One protein attributed to the category intracellular trafficking; secretion and vesicular transport contributed to almost one third (30.73%) of the exclusive proteins from FCS. DIP1242 is a Sec-independent protein translocase protein TatA, which is part of the twin-arginine (Tat) pathway. TatA was shown to be essential for protein export via the Tat pathway in Corynebacterium glutamicum [45].
To examine the influence of cell culture media on bacteria, high abundance proteins exclusively found in cells grown in RPMI 1640 and FCS were analyzed in more detail (Figure 6).
Among the 48 proteins found in RPMI 1640-grown bacteria, two were indicating iron limitation: The gene encoding the putative succinate dehydrogenase iron-sulfur protein DIP0372 (0.002%) was found in a predicted DtxR-regulated operon [39], and the gene of the uncharacterized protein DIP0579 (0.016%) also had has a DtxR binding site and was under the direct transcriptional control [46]. Five proteins were found to be involved in oxidative or nitrosative stress response (DIP0574: 0.003%; DIP1231: 0.004%; DIP1362: 0.044%; DIP1779: 0.002% and DIP1911: 0.011%). DIP0574 is identical to TsaD from mycobacteria and is upregulated under oxidative stress and contributes to pathogenicity of M. tuberculosis and M. leprae [47]. The thiol peroxidase Tpx (DIP1362) and the mycothiol acetyltransferase MshD (DIP1911) have homologues in M. tuberculosis. A study by Hu and Coates [48] showed that Tpx is involved in survival in macrophages and is crucial to establish acute and persistent infections with M. tuberculosis. Mutants of mshD have a limited stress tolerance and were not able to grow and survive in macrophages [49]. Homologous proteins of DIP0541 (0.002%), DIP0655 (0.006%), and DIP1634 (0.027%) play a role in infection and immune recognition in M. tuberculosis [50,51,52]. A Blast search of the sigma factor DIP0577 revealed an 83.6% identity with the RNA polymerase sigma-D factor from Corynebacterium pseudotuberculosis. SigD was found to affect the growth rate of C. glutamicum and regulates genes involved in mycolic acid synthesis [53].
The protein with the highest abundance in FCS is DIP1242 (30.07%), a Sec-independent protein translocase (tatA). A tatA mutant in C. glutamicum showed reduced cell growth and was susceptible to SDS [45]. Five proteins (DIP2043: 0.008%; DIP2005: 0.0001%; DIP1618: 2.74%; DIP1602: 6.82%; DIP0055: 6.8 × 10−5%) are involved in bacterial cell envelope formation. DIP1618 is a putative acyltransferase (PlsC) involved in lipid synthesis as shown for C. glutamicum [54]. Three proteins, the putative D-Ala D-Ala carboxypeptidase DIP2005, the putative secreted penicillin-binding protein PpbpA (DIP0055), and the UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase MurF (DIP1602) are involved in peptidoglycan synthesis and regulation of cell shape. A study by Arora and co-workers [55] showed that PpbpA functions in peptidoglycan biosynthesis in M. tuberculosis are involved in the regulation of bacterial cell length but do not affect growth. The putative secreted protein DIP2043 has a LytR_C domain, which is required for synthesis of anionic cell wall polymers [56] and present in bacterial cell wall assembly proteins. Five proteins are involved in membrane transport: the putative ABC transport system ATP-binding protein DIP1886 (2.70%), the protein-export membrane protein SecF (DIP1371, 0.002%), the putative uptake hydrogenase large subunit DIP0673 (0.002%), the Opt family protein (putative membrane protein) DIP0329 (0.005%), and the putative secreted protein DIP0324 (0.009%). Two proteins are involved in heme homeostasis (DIP2267 and DIP1394). The putative two component system response regulator protein HrrA (DIP2267: 3.23%) has a function for heme homeostasis [57,58] and the uncharacterized protein DIP1394 (0.009%) was identified as coproheme decarboxylase (ChdC) in C. diphtheriae [59]. Two probable drug targets were also exclusively present FCS, the probable nicotinate-nucleotide adenylyltransferase (NadD) DIP1775 and the SsrA-binding protein (SmpB) DIP0750. A study by Rodionova and co-workers [60] showed that NadD in mycobacteria acts as drug-target, and an in silico study by Jamal and co-workers identified DIP0750 as a putative drug target for corynebacteria [61]. A probable pathogenic function was found for two proteins in this data set: A putative serine-like trypsin protease (DIP0736: 10.51%) showed 30.86% identity to venom serine protease 2 (vsp2) from C. ulcerans 809. The putative protein DIP1144 (2.85%) shows 72.4% identity with PknD from C. pseudotuberculosis. PknD plays a role in pathogenesis of M. tuberculosis on the central nervous system. This tissue specific serine-threonine protein kinase triggers the invasion of brain endothelial cells [62].
When the two growth conditions, FCS and RPMI 1640, were compared to BHI, 115 proteins were found to be exclusively expressed (see supplemental Table S2). Of these 115 proteins, 49 proteins are exclusive for RPMI 1640, 35 proteins were exclusive for FCS, and 31 proteins were present under both growth conditions. An analysis of the latter revealed an involvement in bacterial stress response, pathogenicity, or increased antibiotic tolerance (Table 3). Interestingly, two proteins (DIP0600 and DIP2327) are two component response regulator proteins. The protein DIP0600 is part of an pathogenic island (PAI) with proteins involved in siderophore biosynthesis and transport, which includes the proteins DIP0597 and DIP0585, also found in both conditions [63]. Beside the putative iron transport system ATP-binding protein DIP0585, a second protein, the putative iron transport system exported solute-binding component protein DIP1086 was identified. Another protein which is part of a PAI, is the conserved hypothetical protein DIP0823 involved in cellular processes and signaling [64]. The homologue protein of DIP1225, RbpA in M. tuberculosis, was found to increase the resistance to the antibiotic rifampicin [65].

Table 3.
Unique proteins found in bacteria grown in cell culture medium and serum. Data show the UniProt accession number, the gene onthology (GO), the abundance in RPMI and FCS in percent and a short description of the protein.
4. Discussion
In this study, we analyzed the proteome of the non-toxigenic C. diphtheriae strain ISS3319 grown under three different culture conditions. In total, we identified 781 proteins in all three data sets. 444 proteins were found under all three conditions, 92 were only found in BHI-, 49 proteins in RPMI 1640-, and 35 proteins in FCS-grown bacteria.
When the common proteins were analyzed in respect to their annotated function, the majority were involved in metabolism followed by information storage and processing. This may simply reflect the similar growth observed in standard complex medium (BHI), cell culture medium (RPMI 1640), and serum (FCS). In addition, specific proteome adaptation to cell culture medium and serum was observed.
Obviously, iron especially is a critical nutrient under the tested growth conditions. Bacteria grown in cell culture medium and serum expressed two proteins involved in iron acquisition, DIP1086 and DIP2327. DIP1086 is involved in iron acquisition and transport, and DIP2327 (ChrA) activates the gene expression of hmuO and hrtAB by the binding of the upstream regions of their promoters [66]. HmuO is a heme oxygenase and therefore degrades heme to acquire heme-iron sources [67]. Heme is the most abundant iron source of vertebrates to acquire iron, but it is also required as cofactor of many different enzymes, has a role in bacterial metabolism, and provides an advantage for bacteria during infection [68,69]. For this purpose, bacteria have to balance the intracellular heme concentrations. One protein from the novel identified coproporphyrin-dependent heme biosynthesis pathway, DIP1394, was identified in bacteria grown in FCS. It is speculated that heme biosynthesis occurs only in environments where iron is available [69,70,71]. It is probable that, due to the detection limit of the mass spectrometer, this protein was not detected in the data sets of bacteria grown in BHI or RPMI 1640. Despite the function of DIP1420 as putative virulence factor, the homologue protein AhpC was found to serve as heme-binding protein for storage [68,69]. DIP1420 was the most abundant protein under all examined growth conditions, suggesting an essential role in bacterial metabolism. Availability of heme, and therefore heme storage, might play an essential role in survival of bacteria in macrophages, since heme is a crucial component of catalases which protect bacteria against oxidative burst [69].
During bacterial infection of the host, macrophages are part of the first line defense against invading pathogens. A study by Weerasekera and co-workers [11] revealed high intracellular survival rates of C. diphtheriae strain ISS3319 in human and murine macrophages compared to other C. diphtheriae isolates. After bacteria are phagocytosed by macrophages, phagosomes are fused with lysosome to form phagolysosomes. Within the phagolysosome bacteria are killed by production of reactive oxygen species (ROS) [11]. Many bacteria evolved strategies to resist to this specific stress from macrophages [38,72,73]. For example, SodA protects Salmonella Typhimurium from killing by macrophages within the early infection stage [74]. Two proteins involved in survival under oxidative stress, SodA and Dps, were present in all three datasets, which hints at a high tolerance of the bacteria against oxidative stress under these growth conditions.
Virulence factors play a crucial role in bacterial pathogenicity. Three proteins involved in pathogenesis were present in all three data sets. The multifunctional protein DIP0733, the hypothetical protein DIP1546 and the resuscitation-promoting factor RpfB (DIP0874). DIP0733 is involved in adherence to erythrocytes [75] and human epithelial cells [76], in colonization and killing of Caenorhabditis elegans, binding to collagen and to fibrinogen. The ability of C. diphtheriae to bind to fibrinogen by DIP0733 is thought to play a role in evading the recognition by the host immune system [17]. The hypothetical protein DIP1546 was shown in a previous study from Ott and co-workers [77] to act as colonization factor in a C. elegans model system. Resuscitation-promoting factors (Rpf) are peptidoglycan-hydrolyzing enzymes and known to function in resuscitation and virulence of M. tuberculosis [78,79]. C. diphtheriae RpfB (DIP0874) showed significantly higher abundance in bacteria grown in FCS compared to bacteria grown in BHI or RPMI 1640 medium. Pathogenic islands (PAI) in pathogenic bacteria include disease- related factors which serves for antibiotic resistance, or proteins which are helpful for the current growth environment [80]. Twenty-four PAIs are reported in the genome of C. diphtheriae NCTC13129 [81]. Three proteins of a PAI, including one protein of a putative two component system response regulator (DIP0600) and two proteins involved in siderophore biosynthesis and transport (DIP0585 and DIP0597), were found in bacteria grown in RPMI 1640 and serum and not in bacteria grown in BHI.
A considerable number of known C. diphtheriae virulence factors was present under the three growth conditions tested, indicating a preadaptation of this species to host conditions. The putative invasion protein DIP1281 was exclusively found in RPMI 1640, while a Vsp2 homologous protein DIP0736 and a PknD homologous protein DIP1144, both involved in virulence of pathogenic corynebacteria [62,82], were found in the data set of bacteria grown in FCS. This suggests an influence of growth media on the virulence potential of C. diphtheriae.
5. Conclusions
In this study, the influence of cell culture media on bacterial growth and the bacterial proteome was investigated for C. diphtheriae strain ISS3319. The data suggest an increased virulence potential when bacteria are grown in cell culture medium and serum even without host cell contact. This result supports the idea that an influence of cell culture conditions on bacterial pathogenicity has to be considered in host-pathogen interactions studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/2227-7382/9/1/14/s1, in a single excel file entitled Supplementary material_Cd_ISS3319_proteomics. Table S1: ProteoTreeMap, Table S2: Identified proteins, Table S3: Analysis of differentially expressed proteins in all three data sets. The Supplementary material_Cd_ISS3319_proteomics_output_file includes the score, coverage, number of peptides, PSM and the area of each identified protein, of all three independent biological replicates (R1, R2 and R3) and under all three conditions (BHI, RPMI and FCS).
Author Contributions
Protein preparation, data analysis, data storage, visualization of results and writing of the manuscript was carried out by J.M.; F.N. carried out growth experiments; L.M. was involved in the data visualization; J.H. was responsible for mass spectrometry analyses; project administration, supervision of experiments and writing of the draft was carried out by A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw data files and .msf files of mass spectrometric analysis were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) (accessed on 12 March 2021) via the PRIDE partner repository [83]. Data are available via ProteomeXchange ID PXD024381. The excel output files are provided as a single supplementary file (supplementary material_Cd_ISS3319_proteomics_output_file). The proteotreemaps are accessible via following links: Figure 4: http://bionic-vis.biologie.uni-greifswald.de/result.php?jobID=16098528745332&version=UserSpec; Figure 6 (RPMI): http://bionic-vis.biologie.uni-greifswald.de/result.php?jobID=16093262330360&version=UserSpec; Figure 6 (FCS): http://bionic-vis.biologie.uni-greifswald.de/result.php?job_ID=16093270429281&-version=UserSpec.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AGC | automatic gain control |
| BHI | brain heart infusion |
| CAA | chloroacetamide |
| CFU | colony forming units |
| DT | diphtheria toxin |
| DtxR | diphtheria toxin repressor |
| FCS | fetal calf serum |
| FDR | false discovery rate |
| GO | gene onthology |
| HPLC | High-performance liquid chromatography |
| LC-MS/MS | liquid chromatography mass spectrometry/mass spectrometry |
| OD | optical density |
| PAI | pathogenic island |
| ROS | reactive oxygen species |
| RPMI | Roswell Park Memorial Institute |
| TEAB | triethylammonium bicarbonate |
| TFA | trifluoroacetic acid |
References
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Burkovski, A. Diphtheria and its etiological agents. In Corynebacterium Diphtheriae and Related Toxigenic Species; Burkovski, A., Ed.; Springer International Publishing: Dordrecht, The Netherlands, 2014; pp. 1–14. [Google Scholar]
- Sharma, N.C.; Efstratiou, A.; Mokrousov, I.; Mutreja, A.; Das, B.; Ramamurthy, T. Diphtheria. Nat. Rev. Dis. Prim. 2019, 5. [Google Scholar] [CrossRef]
- World Health Organization Diphtheria Reported Cases. Available online: https://apps.who.int/immunization_monitoring/globalsummary/timeseries/tsincidencediphtheria.html (accessed on 16 March 2020).
- Hadfield, T.L.; McEvoy, P.; Polotsky, Y.; Tzinserling, V.A.; Yakovlev, A.A. The pathology of diphtheria. J. Infect. Dis. 2000, 181, S116–S120. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Hacker, E.; Antunes, C.A.; Weerasekera, D.; Alves, A.; Oliveira, D.S.; Martins, C.A.; Júnior, R.H.; Burkovski, A.; Mattos-guaraldi, A.L. Pathogenic properties of a Corynebacterium diphtheriae strain isolated from a case of osteomyelitis. J. Med. Microbiol. 2016, 65, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Tiley, S.M.; Kociuba, K.R.; Heron, L.G.; Munro, R. Infective endocarditis due to nontoxigenic Corynebacterium diphtheriae: Report of seven cases and review. Clin. Infect. Dis. 1993, 16, 271–275. [Google Scholar] [CrossRef]
- Patey, O.; Bimet, F.; Riegel, P.; Halioua, B.; Emond, J.P.; Estrangin, E.; Dellion, S.; Alonso, J.M.; Kiredjian, M.; Dublanchet, A.; et al. Clinical and molecular study of Corynebacterium diphtheriae systemic infections in France. J. Clin. Microbiol. 1997, 35, 441–445. [Google Scholar] [CrossRef]
- Berger, A.; Teusch, B.; Heinzinger, S.; Sing, A. Corynebacterium ulcerans—ein Emerging Pathogen? Daten des Konsiliarlabors für Diphtherie 2011–2016. Epidemiol. Bull. 2018. [Google Scholar] [CrossRef]
- Puliti, M.; Von Hunolstein, C.; Marangi, M.; Bistoni, F.; Tissi, L. Experimental model of infection with non-toxigenic strains of Corynebacterium diphtheriae and development of septic arthritis. J. Med. Microbiol. 2006, 55, 229–235. [Google Scholar] [CrossRef]
- Weerasekera, D.; Fastner, T.; Lang, R.; Burkovski, A.; Ott, L. Of mice and men: Interaction of Corynebacterium diphtheriae strains with murine and human phagocytes. Virulence 2019, 10, 414–428. [Google Scholar] [CrossRef]
- Sabbadini, P.S.; Assis, M.C.; Trost, E.; Gomes, D.L.R.; Moreira, L.O.; dos Santos, C.S.; Pereira, G.A.; Nagao, P.E.; de Azevedo, V.A.; Hirata Júnior, R.; et al. Corynebacterium diphtheriae 67-72p hemagglutinin, characterized as the protein DIP0733, contributes to invasion and induction of apoptosis in HEp-2 cells. Microb. Pathog. 2012, 52, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.S.; Antunes, C.A.; Lourêdo, L.S.; Viana, V.G.; dos Santos, C.S.; da Silva, J.F.R.; Hirata, R.; Hacker, E.; Mattos-Guaraldi, A.L.; Burkovski, A. Functional characterization of the collagen-binding protein DIP2093 and its influence on host–pathogen interaction and arthritogenic potential of Corynebacterium diphtheriae. Microbiology 2017, 163, 692–701. [Google Scholar] [CrossRef]
- Kolodkina, V.; Denisevich, T.; Titov, L. Identification of Corynebacterium diphtheriae gene involved in adherence to epithelial cells. Infect. Genet. Evol. 2011, 11, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, L.S.; Antunes, C.A.; dos Santos, C.S.; Pereira, J.A.A.; Sabbadini, P.S.; de Luna, M.; Azevedo, V.; Hirata Junior, R.; Burkovski, A.; Asad, L.M.B.; et al. Corynebacterium diphtheriae putative tellurite-resistance protein (CDCE8392_0813) contributes to the intracellular survival in human epithelial cells and lethality of Caenorhabditis elegans. Mem. Inst. Oswaldo Cruz 2015, 110, 662–668. [Google Scholar] [CrossRef]
- Weerasekera, D.; Hahn, J.; Herrmann, M.; Burkovski, A. Live cell imaging of macrophage/bacterium interaction demonstrates cell lysis induced by Corynebacterium diphtheriae and Corynebacterium ulcerans. BMC Res. Notes 2019, 12, 19–21. [Google Scholar] [CrossRef]
- Antunes, C.A.; dos Santos, L.S.; Hacker, E.; Köhler, S.; Bösl, K.; Ott, L.; de Luna, M.G.; Hirata, R.; Azevedo, V.A.; Mattos-Guaraldi, A.L.; et al. Characterization of DIP0733, a multi-functional virulence factor of Corynebacterium diphtheriae. Microbiology 2015, 161, 639–647. [Google Scholar] [CrossRef]
- Bittel, M.; Gastiger, S.; Amin, B.; Hofmann, J.; Burkovski, A. Surface and extracellular proteome of the emerging pathogen Corynebacterium ulcerans. Proteomes 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Möller, J.; Schorlemmer, S.; Hofmann, J.; Burkovski, A. Cellular and extracellular proteome of the animal pathogen Corynebacterium silvaticum, a close relative of zoonotic Corynebacterium ulcerans and Corynebacterium pseudotuberculosis. Proteomes 2020, 8, 19. [Google Scholar] [CrossRef]
- Goring, A.K.; Chen, Y.; Clubb, R.T.; Loo, J.A. Secreted and cell surface proteome analysis of pathogenic Corynebacterium diphtheriae reveals proteins relevant to virulence. bioRxiv 2020, 1–11. [Google Scholar] [CrossRef]
- Hansmeier, N.; Chao, T.C.; Kalinowski, J.; Pühler, A.; Tauch, A. Mapping and comprehensive analysis of the extracellular and cell surface proteome of the human pathogen Corynebacterium diphtheriae. Proteomics 2006, 6, 2465–2476. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.M.; Folador, E.L.; Soares, S.C.; Souza, G.H.M.F.; Santos, A.V.; Sousa, C.S.; Figueiredo, H.; Miyoshi, A.; Le Loir, Y.; Silva, A.; et al. Label-free quantitative proteomics of Corynebacterium pseudotuberculosis isolates reveals differences between Biovars ovis and equi strains. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Kraner, M.E.; Müller, C.; Sonnewald, U. Comparative proteomic profiling of the choline transporter-like1 (CHER1) mutant provides insights into plasmodesmata composition of fully developed Arabidopsis thaliana leaves. Plant. J. 2017, 92, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, W.; Eckart, R.A.; Schmid, B.; Cagköylü, H.; Hof, K.; Muller, Y.A.; Amin, B.; Lührmann, A. Nuclear trafficking of the anti-apoptotic Coxiella burnetii effector protein AnkG requires binding to p32 and Importin-α1. Cell. Microbiol. 2016, 19, 43–48. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Hein, M.Y.; Cox, J.; Mann, M. A “Proteomic Ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteom. 2014, 13, 3497–3506. [Google Scholar] [CrossRef] [PubMed]
- Sangal, V.; Blom, J.; Sutcliffe, I.C.; von Hunolstein, C.; Burkovski, A.; Hoskisson, P.A. Adherence and invasive properties of Corynebacterium diphtheriae strains correlates with the predicted membrane-associated and secreted proteome. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Yi, Y.; Fang, Y.; Wu, K.; Liu, Y.; Zhang, W. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, J.; Funke, S.; Hecker, M.; Siebourg, J. Visualizng gene expression data via Voroni treemaps. Int. Symp. Vor. Diagrams 2009, 233–241. [Google Scholar] [CrossRef]
- Otto, A.; Bernhardt, J.; Meyer, H.; Schaffer, M.; Herbst, F.A.; Siebourg, J.; Mäder, U.; Lalk, M.; Hecker, M.; Becher, D. Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat. Commun. 2010, 1. [Google Scholar] [CrossRef]
- Liebermeister, W.; Noor, E.; Flamholz, A.; Davidi, D.; Bernhardt, J.; Milo, R. Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. USA 2014, 111, 8488–8493. [Google Scholar] [CrossRef]
- Houben, E.N.G.; Korotkov, K.V.; Bitter, W. Take five—Type VII secretion systems of Mycobacteria. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 1707–1716. [Google Scholar] [CrossRef]
- Dover, L.G.; Cerde, A.M.; Pallen, M.J.; Parkhill, J.; Besra, G.S. Comparative cell wall core biosynthesis in the mycolated pathogens, Mycobacterium tuberculosis and Corynebacterium diphtheriae. FEMS Microbiol Rev. 2004, 28, 225–250. [Google Scholar] [CrossRef] [PubMed]
- Ott, L.; Höller, M.; Gerlach, R.G.; Hensel, M.; Rheinlaender, J.; Schäffer, T.E.; Burkovski, A. Corynebacterium diphtheriae invasion-associated protein (DIP1281) is involved in cell surface organization, adhesion and internalization in epithelial cells. BMC Microbiol. 2010, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Dalbey, R.E.; Wang, P.; Kuhn, A. Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 2011, 80, 161–187. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C.; Hazlitt, W. Antimicrobial reactive oxygen and nitrogen species: Concepts and controverses. Nat. Rev. Microbiol. 2004, 2, 820–832. [Google Scholar] [CrossRef]
- Zahrt, T.C.; Deretic, V. Reactive nitrogen and oxygen intermediates and bacterial defenses: Unusual adaptations in Mycobacterium tuberculosis. Antioxid. Redox Signal. 2002, 4, 141–159. [Google Scholar] [CrossRef]
- Yellaboina, S.; Ranjan, S.; Chakhaiyar, P.; Hasnain, S.E.; Ranjan, A. Prediction of DtxR regulon: Identification of binding sites and operons controlled by diphtheria toxin repressor in Corynebacterium diphtheriae. BMC Microbiol. 2004, 8, 1–8. [Google Scholar] [CrossRef]
- Martinez, A.; Kolter, R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1997, 179, 5188–5194. [Google Scholar] [CrossRef]
- Merkamm, M.; Guyonvarch, A. Cloning of the sodA Gene from Corynebacterium melassecola and role of superoxide dismutase in cellular viability. J. Bacteriol. 2001, 183, 1284–1295. [Google Scholar] [CrossRef]
- Spinler, J.K.; Zajdowicz, S.L.W.; Haller, J.C.; Oram, D.M.; Ronald, E.; Holmes, R.K. Development and use of a selectable, broad-host-range reporter transposon for identifying environmentally regulated promoters in bacteria. FEMS Microbiol. Lett. 2010, 291, 143–150. [Google Scholar] [CrossRef]
- Ott, L.; Hacker, E.; Kunert, T.; Karrington, I.; Etschel, P.; Lang, R.; Wiesmann, V.; Wittenberg, T.; Singh, A.; Varela, C.; et al. Analysis of Corynebacterium diphtheriae macrophage interaction: Dispensability of corynomycolic acids for inhibition of phagolysosome maturation and identification of a new gene involved in synthesis of the corynomycolic acid layer. PLoS ONE 2017, 12, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, B.P.; Nelson, K.M.; Aldrich, C.C. Adenylating enzymes in Mycobacterium tuberculosis as drug targets. Curr. Top. Med. Chem. 2012, 12, 766–796. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Date, M.; Itaya, H.; Matsui, K.; Wu, L. Functional analysis of the Twin-Arginine translocation pathway in Corynebacterium glutamicum ATCC 13869. Appl. Environ. Microbiol. 2006, 72, 7183–7192. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brune, I.; Werner, H.; Hüser, A.T.; Kalinowski, J.; Pühler, A.; Tauch, A. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genom. 2006, 7, 1–19. [Google Scholar] [CrossRef]
- Bhat, A.H.; Pathak, D.; Rao, A. The alr-groEL1 operon in Mycobacterium tuberculosis: An interplay of multiple regulatory elements. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Hu, Y.; Coates, A.R.M. Acute and persistent Mycobacterium tuberculosis infections depend on the thiol peroxidase TPX. PLoS ONE 2009, 4, e5150. [Google Scholar] [CrossRef] [PubMed]
- Buchmeier, N.A.; Newton, G.L.; Fahey, R.C. A mycothiol synthase mutant of Mycobacterium tuberculosis has an altered thiol-disulfide content and limited tolerance to stress. J. Bacteriol. 2006, 188, 6245–6252. [Google Scholar] [CrossRef]
- Meena, L.S.; Chopra, P.; Bedwal, R.S.; Singh, Y. Nucleoside diphosphate kinase-like activity in adenylate kinase of Mycobacterium tuberculosis. Biotechnol. Appl. Biochem. 2003, 38, 169. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; De Sessions, P.F.; Teoh, G.H.K.; Mohamed, A.N.N.; Zhu, Y.O.; Koh, V.H.Q.; Ang, M.L.T.; Dedon, P.C.; Hibberd, M.L.; Alonsoa, S. Transcriptional profiling of Mycobacterium tuberculosis exposed to in vitro lysosomal stress. Infect. Immun. 2016, 84, 2505–2523. [Google Scholar] [CrossRef]
- Byun, E.H.; Kim, W.S.; Kim, J.S.; Won, C.J.; Choi, H.G.; Kim, H.J.; Cho, S.N.; Lee, K.; Zhang, T.; Hur, G.M.; et al. Mycobacterium paratuberculosis CobT activates dendritic cells via engagement of toll-like receptor 4 resulting in Th1 cell expansion. J. Biol. Chem. 2012, 287, 38609–38624. [Google Scholar] [CrossRef]
- Taniguchi, H.; Busche, T.; Patschkowski, T.; Niehaus, K.; Pátek, M.; Kalinowski, J.; Wendisch, V.F. Physiological roles of sigma factor SigD in Corynebacterium glutamicum. BMC Microbiol. 2017, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nampoothiri, K.M.; Hoischen, C.; Bathe, B.; Möckel, B.; Pfefferle, W.; Krumbach, K.; Sahm, H.; Eggeling, C. Expression of genes of lipid synthesis and altered lipid composition modulates L -glutamate efflux of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2002, 58, 89–96. [Google Scholar] [CrossRef]
- Arora, D.; Chawla, Y.; Malakar, B.; Singh, A.; Nandicoori, V.K. The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths. J. Biol. Chem. 2018, 293, 6497–6516. [Google Scholar] [CrossRef]
- Kawai, Y.; Marles-Wright, J.; Cleverley, R.M.; Emmins, R.; Ishikawa, S.; Kuwano, M.; Heinz, N.; Bui, N.K.; Hoyland, C.N.; Ogasawara, N.; et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011, 30, 4931–4941. [Google Scholar] [CrossRef] [PubMed]
- Frunzke, J.; Ga, C.; Brocker, M.; Bott, M. Control of heme homeostasis in Corynebacterium glutamicum by the two-component system HrrSA. J. Bacteriol. 2011, 193, 1212–1221. [Google Scholar] [CrossRef]
- Bott, M.; Brocker, M. Two-component signal transduction in Corynebacterium glutamicum and other corynebacteria: On the way towards stimuli and targets. Appl. Microbiol. Biotechnol. 2012, 94, 1131–1150. [Google Scholar] [CrossRef]
- Pfanzagl, V.; Holcik, L.; Maresch, D.; Gorgone, G.; Michlits, H.; Paul, G. Coproheme decarboxylases Phylogenetic prediction versus biochemical experiments. Arch. Biochem. Biophys. 2018, 640, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Rodionova, I.A.; Schuster, B.M.; Guinn, K.M.; Sorci, L.; Scott, D.A.; Li, X.; Kheterpal, I.; Shoen, C.; Cynamon, M.; Locher, C.; et al. Metabolic and bactericidal effects of targeted suppression of NadD and NadE enzymes in mycobacteria. MBio 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Jamal, S.B.; Hassan, S.S.; Tiwari, S.; Viana, M.V.; De Jesus Benevides, L.; Ullah, A.; Turjanski, A.G.; Barh, D.; Ghosh, P.; Costa, D.A.; et al. An integrative in-silico approach for therapeutic target identification in the human pathogen Corynebacterium diphtheriae. PLoS ONE 2017, 12, 1–25. [Google Scholar] [CrossRef]
- Be, N.A.; Bishai, W.R.; Jain, S.K. Role of Mycobacterium tuberculosis pknD in the pathogenesis of central nervous system tuberculosis. BMC Microbiol. 2012, 12, 7. [Google Scholar] [CrossRef]
- Yoon, S.H.; Park, Y.K.; Kim, J.F. PAIDB v2.0: Exploration and analysis of pathogenicity and resistance islands. Nucleic Acids Res. 2015, 43, D624–D630. [Google Scholar] [CrossRef]
- Soares, S.C.; Ramos, R.T.J.; Silva, W.M.; Oliveira, L.C.; Amorim, L.G.; Hirata, R.; Mattos-Guaraldi, A.L.; Miyoshi, A.; Silva, A.; Azevedo, V. Corynebacterium pathogenic species in next-generation genomic era: The use of EDGAR and PIPS software and the importance of pathogenicity islands identification in pan-genomic analyses of pathogenic species. Microb. Pathog. Strateg. Combat. Them Sci. Technol. Educ. 2013. [Google Scholar] [CrossRef]
- Hu, Y.; Morichaud, Z.; Chen, S.; Leonetti, J.P.; Brodolin, K. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the σ a-containing RNA polymerase holoenzyme. Nucleic Acids Res. 2012, 40, 6547–6557. [Google Scholar] [CrossRef] [PubMed]
- Burgos, J.; Schmitt, M. The ChrA response regulator in Corynebacterium diphtheriae controls hemin-regulated gene expression through binding to the hmuO and hrtAB promoter regions. J. Bacteriol. 2012, 194, 1717–1729. [Google Scholar] [CrossRef][Green Version]
- Schmitt, M. Iron acquisition and iron-dependent gene expression in Corynebacterium diphtheriae. In Corynebacterium diphtheriae and Related Toxigenic Species: Genomics, Pathogenicity and Applications; Springer: Berlin/Heidelberg, Germany, 2014; pp. 95–121. ISBN 978-94-007-7623-4. [Google Scholar]
- Lechardeur, D.; Fernandez, A.; Robert, B.; Gaudu, P.; Trieu-Cuot, P.; Lamberet, G.; Gruss, A. The 2-Cys peroxiredoxin alkyl hydroperoxide reductase C binds heme and participates in its intracellular availability in Streptococcus agalactiae. J. Biol. Chem. 2010, 285, 16032–16041. [Google Scholar] [CrossRef] [PubMed]
- Anzaldi, L.L.; Skaar, E.P. Overcoming the heme paradox: Heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 2010, 78, 4977–4989. [Google Scholar] [CrossRef]
- Qi, Z.; Hamza, I.; O’Brian, M.R. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. USA 1999, 96, 13056–13061. [Google Scholar] [CrossRef]
- Choby, J.E.; Skaar, E.P. Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 2016, 428, 3408–3428. [Google Scholar] [CrossRef] [PubMed]
- Ott, L.; Burkovski, A. Toxigenic corynebacteria: Adhesion, invasion and host response. In Corynebacterium diphtheriae and Related Toxigenic Species: Genomics, Pathogenicity and Applications; Springer: Berlin/Heidelberg, Germany, 2014; Volume 9789400776, pp. 1–293. ISBN 9789400776241. [Google Scholar]
- Nathan, C.; Shiloh, M.U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 2000, 97, 8841–8848. [Google Scholar] [CrossRef]
- Tsolis, R.M.; Baumler, A.J.; Heffron, F. Role of Salmonella typhimurium mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect. Immun. 1995, 63, 1739–1744. [Google Scholar] [CrossRef]
- Colombo, A.V.; Hirata, R.; De Souza, C.M.R.; Monteiro-Leal, L.H.; Previato, J.O.; Formiga, L.C.D.; Andrade, A.F.B.; Mattos-Guaraldi, A.L. Corynebacterium diphtheriae surface proteins as adhesins to human erythrocytes. FEMS Microbiol. Lett. 2001, 197, 235–239. [Google Scholar] [CrossRef][Green Version]
- Hirata, R.; Souza, S.M.S.; Rocha-de-Souza, C.M.; Andrade, A.F.B.; Monteiro-Leal, L.H.; Formiga, L.C.D.; Mattos-Guaraldi, A.L. Patterns of adherence to HEp-2 cells and actin polymerisation by toxigenic Corynebacterium diphtheriae strains. Microb. Pathog. 2004, 36, 125–130. [Google Scholar] [CrossRef]
- Ott, L.; Mckenzie, A.; Baltazar, M.T.; Britting, S.; Bischof, A.; Burkovski, A.; Hoskisson, P.A. Evaluation of invertebrate infection models for pathogenic corynebacteria. FEMS Immunol. Med. Microbiol. 2012, 65, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Kana, B.D.; Mizrahi, V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol. Med. Microbiol. 2010, 58, 39–50. [Google Scholar] [CrossRef]
- Rosser, A.; Stover, C.; Pareek, M.; Mukamolova, G.V. Resuscitation-promoting factors are important determinants of the pathophysiology in Mycobacterium tuberculosis infection. Crit. Rev. Microbiol. 2017, 43, 621–630. [Google Scholar] [CrossRef]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Trost, E.; Tauch, A. Comparative genomics and pathogenicity islands of Corynebacterium diphtheriae, Corynebacterium ulcerans, and Corynebacterium pseudotuberculosis. In Corynebacterium diphtheriae and Related Toxigenic Species: Genomics, Pathogenicity and Applications; Burkovski, A., Ed.; Springer: Dordrecht, The Netherlands, 2014; pp. 39–65. ISBN 978-94-007-7624-1. [Google Scholar]
- Hacker, E.; Antunes, C.A.; Mattos-guaraldi, A.L.; Burkovski, A.; Tauch, A. Corynebacterium ulcerans, an emerging human pathogen. Future Microbiol. 2016, 11, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Perez-riverol, Y.; Csordas, A.; Bai, J.; Bernal-llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, 442–450. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).