Abstract
The precise molecular mechanisms of diabetic retinopathy (DR) pathogenesis are unclear, and treatment options are limited. There is an urgent need to discover and develop novel therapeutic targets for the treatment of this disease. Glycosylation is a post-translational modification that plays a critical role in determining protein structure, function, and stability. Recent studies have found that serum glycoproteomic changes are associated with the presence or progression of several inflammatory diseases. However, very little is known about the glycoproteomic changes associated with DR. In this study, glycoproteomic profiling of the serum of diabetic patients with and without DR was performed. A total of 15 glycopeptides from 11 glycoproteins were found to be significantly altered (5 upregulated and 10 downregulated) within the serum glycoproteome of DR patients. These glycoproteins are known to be involved in the maintenance of the extracellular matrix and complement system through peptidolytic activity or regulation.
1. Introduction
Diabetic retinopathy (DR) is a microvascular complication of diabetes and remains the primary cause of permanent blindness in the working age population [1]. The prevalence of DR is expected to increase due to the global uptrend of both diabetes mellitus and life expectancy [2]. The precise molecular mechanisms of DR pathogenesis are not clear, and treatment options are limited [3]. Additionally, detection of this disease at an early stage is very difficult mainly due to a late onset of irreversible symptoms [4]. There is an urgent need to discover novel therapeutic targets and develop early detection methods for the treatment of DR.
Glycosylation is a common post-translational modification (PTM), in which an oligosaccharide (glycan) is enzymatically attached to a protein at either an asparagine (N) residue (N-linked) or a serine (S)/threonine (T) residue (O-linked). Glycosylation plays a critical role in determining protein structure, function, and stability. Glycoproteins participate in the regulation of a variety of cellular and biological functions, including cell growth, adhesion, signaling, and inflammation [5]. Changes in glycosylation patterns of systemic proteins have been observed in inflammation, immune deficiencies, cancers, and autoimmune diseases [6,7,8,9,10,11]. Additionally, in the case of chronic inflammatory conditions, acute phase proteins, transport proteins, and proteins related to the complement system were found to contain altered glycosylation patterns [12]. However, studies exploring the serum glycoproteomic profiles in the context of DR are limited.
In this study, we performed comprehensive glycoproteomic profiling of serum samples from diabetic patients with and without DR using LC–MS/MS technology. Significant differences were found in the glycosylation patterns in the serum of DR patients. Discovery of altered glycosylation profiles associated with DR opens a new area of research for DR diagnosis and surveillance.
2. Materials and Methods
2.1. Human Subjects and Sample Collection
This study was approved by the Institutional Review Board (IRB) of the Augusta University, Augusta, Georgia (IRB#1458143). Intravenous peripheral blood samples from 23 type-2 diabetic patients, 10 with DR (cases) and 13 without DR (controls), were collected after obtaining informed consent. The patients with other diabetic complications including peripheral neuropathy, diabetic foot, and chronic kidney disease were excluded from this study. The demographics of the patient population used in the study are presented in Table 1. Approximately 10 mL of venous blood was collected from each consenting patient in a serum separator tube (Vacutainer SST; Catalog#BD367985; BD Biosciences, San Jose, CA, USA). Blood samples were allowed to clot for 30 min and subsequently centrifuged at 1300× g in a fixed angle centrifuge for 15 min. The serum (approximately 5 mL) was then isolated and immediately stored at −80 °C until further studies.

Table 1.
Patient demographics for subjects with and without diabetic retinopathy (DR).
2.2. Sample Preparation and Digestion
Analysis of serum proteins is often masked by the presence of high concentrations of albumin and IgG that can make up a large proportion (>70%) of total serum protein. These proteins were depleted, to improve the dynamic range and detection of low-abundant proteins, using a spin depletion cartridge (Multiple Affinity Removal Spin Cartridge Human 14, MARS-14, Agilent, Santa Clara, CA, USA). The depleted serum samples were lyophilized and reconstituted into 40 µL of 8 M urea in 50 mM Tris-HCl (pH 8). Reduction and alkylation of the cysteine residues were then performed with 20 mM DTT and 55 mM iodoacetamide, respectively, followed by adding 360 µL of 50 mM ammonium bicarbonate buffer to reduce the urea concentration to below 1 M. Protein concentration of the samples was measured using a Bradford Assay Kit according to the manufacturer’s instructions (Pierce, Rockford, IL, USA). Trypsin (Pierce, Rockford, IL, USA) was then added to 50 µg protein from each sample at a 1:20 ratio (w/w) to perform protein digestion at 37 °C overnight.
2.3. LC–MS/MS Analysis
Digested serum samples were first cleaned using a C18 spin plate (Nest Group, Southborough, MA, USA) and then analyzed using an Orbitrap Fusion tribrid mass spectrometer (Thermo Scientific, Waltham, MA, USA) coupled with an Ultimate 3000 nano-UPLC system (Thermo Scientific). Two micrograms of reconstituted peptide (1 µg/mL) was first trapped and washed on a Pepmap100 C18 trap (5 μm, 0.3 × 5 mm) at 20 μL/min using 2% acetonitrile in water (with 0.1% formic acid) for 10 min and then separated on a Pepman100 RSLC C18 column (2.0 μm, 75 μm × 150 mm) using a gradient of 2 to 40% acetonitrile with 0.1% formic acid over 120 min at a flow rate of 300 nL/min and a column temperature of 40 °C. Eluted peptides were introduced into an Orbitrap Fusion MS via nano-electrospray ionization source with a temperature of 300 °C and spray voltage of 2000 V. The peptides were analyzed by data-dependent acquisition in positive mode using an Orbitrap MS analyzer for a precursor scan at 120,000 FWHM from 400 to 2000 m/z and an ion-trap MS analyzer for MS/MS scans in top speed mode (3 s cycle time) with dynamic exclusion settings (repeat count 1 and exclusion duration 15 s). Higher-energy collisional dissociation (HCD) was used as a fragmentation method with a normalized collision energy of 32%.
2.4. Protein Identification and Quantification
Identification and quantification of glycopeptides were performed using Byonic software (v3.7.13, Protein Metrics, Cupertino, CA, USA) against the UniProt human protein database (reviewed and annotated UniProt Knowledge Human Database; Release August 2017) with the following settings: fully specific trypsin digestion, one missed cleavage site, precursor mass tolerance of 10 ppm, fragmentation type of QTOF/HCD, fragment mass tolerance of 0.5 Da, fixed carbidomethylation (+57.021 Da) for cysteine, dynamic oxidation (+15.995 Da) for methionine, and 2% FDR. N- and O-glycans were searched against their respective glycan databases. Total modifications per peptide were set to 3 with a maximum of 2 common modifications and 1 rare modification.
2.5. Statistical and Bioinformatics Analysis
All statistical analyses were conducted using the R Project for Statistical Computing (v3.2.5, R Core Team, Vienna, Austria). Peptide-spectrum match (PSM) count data for the identified glycopeptides were log2 transformed, and differential expression analyses were conducted using the “edgeR” (v3.26.8) package [13]. FDR adjusted p-value < 0.05 was considered significant. Gene ontology enrichment analysis was conducted to identify biological processes, cellular components, and molecular functions associated with the identified serum glycoproteins. The “goana” function from “limma” (v3.40.6) package was used to perform over-representation analyses for gene ontology terms [14]. The p-values were adjusted for multiple testing using the FDR method.
3. Results
3.1. Glycosylation Profiling of Human Serum
After protein digestion, samples were analyzed via LC–MS/MS, and Byonic software was used to identify intact glycopeptides and sites of glycosylation on glycoproteins in all 23 serum samples, as shown in Figure 1. Overall, the average number of total glycopeptides detected in serum samples from patients with DR (717.0 ± 5.0) was significantly higher than in patients without DR (648.9 ± 6.8) (Figure 2). The number of unique glycoproteins detected per sample was also significantly higher in patients with DR (116.1 ± 4.1) as compared to patients without DR (105.2 ± 3.2). There was no difference in the number of unique glycopeptides (229.3 ± 9.8 vs. 204.8 ± 7.4), unique sites of glycosylation (264.6 ± 11.5 vs. 239.6 ± 7.8), or unique glycans (71.5 ± 0.9 vs. 69.5 ± 1.4) found in serum of DR samples as compared to those without DR (Figure 2). The most abundant glycopeptides detected in human serum are listed in Table 2. Hemopexin (HPX), alpha-2-HS-glycoprotein (AHSG), inter-alpha-trypsin inhibitor heavy chain H1 (ITIH1), complement factor H (CFH), ceruloplasmin (CP), haptoglobin (HP), inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4), complement C4-B (C4B), prothrombin (F2), plasma protease C1 inhibitor (SERPING1), beta-2-glycoprotein 1 (APOH), heparin cofactor 2 (SERPIND1), and antithrombin-III (SERPINC1) were found to be the most prevalent glycoproteins in the human serum. Among the glycans, HexNAc(1)Hex(1)NeuAc(1) (1.1.0.1) and HexNAc(4)Hex(5)NeuAc(2) (4.5.0.2) were the two most prevalent detected on the human serum glycoproteins (Table 2).
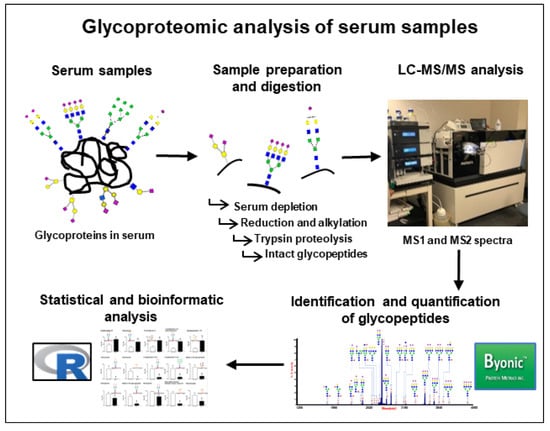
Figure 1.
Workflow of glycoproteomic analysis to identify and quantify glycopeptides in serum of patients with and without diabetic retinopathy (DR). After depletion of the most abundant serum proteins, reduction and alkylation of the cysteine residues were performed with DTT and iodoacetamide, respectively. Protein digestion was performed using trypsin, and digested samples were analyzed using an Orbitrap Fusion tribrid mass spectrometer coupled with an Ultimate 3000 nano-UPLC system. Identification and quantification of glycopeptides was performed using Byonic software, and statistical analyses were performed using R-software.
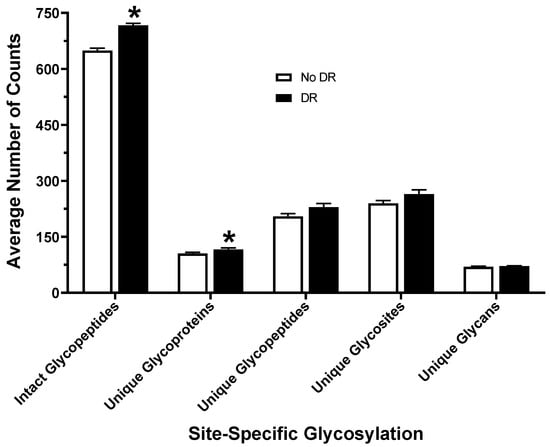
Figure 2.
Number of total and unique glycopeptides, unique glycoproteins, glycosites, and glycans identified in the serum from diabetic patients with and without DR. * p-value < 0.05.

Table 2.
Most abundant glycopeptides detected in human serum.
3.2. N-Glycans versus O-Glycans
N-linked glycans have a core containing two GlcNAc residues and three mannose residues attached to the asparagine (N) at the consensus sequence N-X-S/T (where X denotes any amino acid except proline). O-glycans have the linkage of an N-acetylgalactosamine (GalNAc) moiety to the hydroxyl of serine (S) or threonine (T). Among total N-glycans detected in the serum, the N-X-T binding motif was more common than the N-X-S motif (59% vs. 41%) (Figure 3A), whereas among the O-glycans, 63% were linked to a serine residue, and 37% were linked to a threonine residue (Figure 3B). Glycoproteins have one or more sites of glycosylation. In our study, a total of 51.9% of glycoproteins were found to be glycosylated at one site, while 23.1%, 8.2%, 5.4%, and 11.4% of glycoproteins were glycosylated at two, three, four, and more than four glycosites, respectively (Figure 3C).

Figure 3.
Relative abundance of typical N- and O- glycan sequence motifs and glycosites per protein detected in serum samples. (A) N-linked glycans have a consensus sequence N-X-S or N-X-T (where X denotes any amino acid except proline). In analysis of serum samples, the prevalence of N-glycan sequence motifs, N-X-T, was 59%, whereas the prevalence of N-X-S was 41%. (B) Among O-glycans, 63% were linked to a serine (S) residue, and 37% were linked to a threonine (T) residue. (C) Glycoproteins can have more than one site of glycosylation. The percentages of glycoproteins with 1, 2, 3, 4, >4 glycosite(s) are 51.9%, 23.1%, 8.2%, 5.4%, and 11.4%, respectively.
A summary of the number of glycosites and the number of unique glycans for each N-glycoprotein found in all serum samples is shown in Figure 4A. The glycoproteins with the most unique N-glycosites were apolipoprotein B-100 (APOB, 7 glycosites, 9 glycans), titin (TTN, 7 glycosites, 6 glycans), complement factor H (CFH, 6 glycosites, 4 glycans), fibronectin (FN1, 5 glycosites, 3 glycans), hemopexin (HPX, 5 glycosites, 11 glycans), attractin (ATRN, 4 glycosites, 3 glycans), ceruloplasmin (CP, 4 glycosites, 4 glycans), complement factor 4A (C4A, 4 glycosites, 4 glycans,) complement C4-B (C4B, 4 glycosites, 4 glycans), antithrombin-III (SERPINC1, 3 glycosites, 2 glycans), alpha-1-antichymotrypsin (SERPINA3, 3 glycosites, 2 glycans), and haptoglobin (HP, 2 glycosites, 7 glycans). Glycoproteins found to have the most O-glycosites were titin (TTN, 185 glycosites, 64 glycans), hornerin (HRNR, 43 glycosites, 28 glycans), apolipoprotein (APOB, (39 glycosites, 35 glycans), fibronectin (FN1, 37 glycosites, 16 glycans), mucin-19 (MUC19, 37 glycosites, 27 glycans), alstrom syndrome protein (ALMS1, 31 glycosites, 25 glycans), proteoglycan 4 (PRG4, 27 glycosites, 19 glycans), kinonogen-1 (KNG1, 26 glycosites, 9 glycans), inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4, 22 glycosites, 14 glycans), inter-alpha-trypsin inhibitor heavy chain H1 (ITIH1, 17 glycosites, 13 glycans), plasminogen (PLG, 16 glycosites, 12 glycans), and alpha-2-HS glycoprotein (AHSG, 16 glycosites, 30 glycans) (Figure 4B). These findings are in agreement with current literature, as the O-glycoproteome features more heterogeneity than the N-glycoproteome. This may be due to O-glycans having a simpler structure compared to N-glycans. Additionally, there are two amino acid residues (S and T) where O-glycans are linked versus the one amino acid (N) in the case of N-glycans.
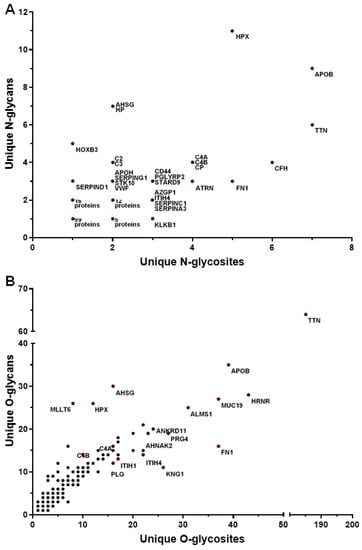
Figure 4.
The distribution of unique glycans and glycosites on the serum glycoproteins. For each glycoprotein, the x-axis represents the number of glycosites, and the y-axis represents the number of unique glycans. The scatterplots depicting the distribution of unique N-linked glycans and glycosites and unique O-linked glycans and glycosites detected in serum glycoproteins are shown in panels (A) and (B), respectively.
3.3. Gene Ontology Enrichment Analyses of Glycoproteins Detected in Serum
Gene ontology (GO) enrichment analyses were performed to annotate the detected serum glycoproteins with biological processes, molecular functions, and cellular compartments (Figure 5). The biological functions enriched in the serum glycoproteins were found to include protein activation cascade, regulation of protein maturation, complement activation, inflammatory response, leukocyte mediated immunity, vesicle-mediated transport, response to stress, exocytosis, blood coagulation, wound healing, secretion, endopeptidase activity, and establishment of localization (Figure 5A). As expected, the cellular components enriched in the serum glycoproteins were found to include blood microparticle, extracellular vesicle, extracellular matrix, secretory vesicle, cytoplasmic vesicle, cell surface, endoplasmic reticulum, Golgi apparatus, and cytoplasm (Figure 5B). The molecular functions enriched in the serum glycoproteome included endopeptidase inhibitor activity, glycosaminoglycan binding, heparin binding, serine-type peptidase activity, sulfur compound binding, signaling receptor binding, lipid binding, structural molecule activity, and carbohydrate derivative binding (Figure 5C).
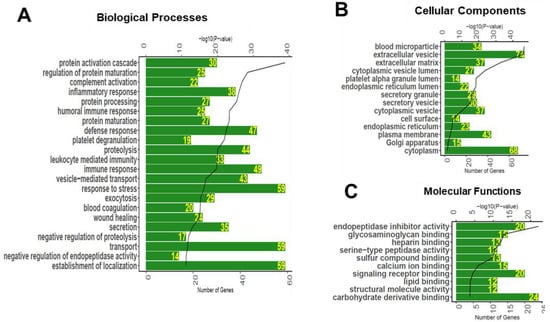
Figure 5.
Biological processes (A), cellular compartments (B), and molecular functions (C) associated with the glycoproteins detected in the human serum. Bioinformatics analysis was performed to associate significantly enriched Gene Ontology (GO) terms to the serum glycoproteins. The green bar represents the number of proteins annotated to each GO term, and black line represents the p-value of enrichment.
3.4. Serum Glycoproteomic Alterations Associated with Diabetic Retinopathy
The quantification of glycopeptides was performed by counting peptide-spectrum match (PSM) values. PSM values for each peptide indicate the total number of mass spectra associated with each peptide. Statistical analysis revealed 15 glycopeptides significantly altered in the serum of patients with DR as compared to those without DR (Table 3). These 15 glycopeptides belong to 11 glycoproteins including apolipoprotein C-III (APOC3), complement C1s subcomponent (C1S), proteoglycan 4 (PRG4), vitronectin (VTN), antithrombin-III (SERPINC1), fibronectin (FN1), complement C4-B (C4B), alpha-2-HS-glycoprotein (AHSG), hemopexin (HPX), inter-alpha-trypsin inhibitor heavy chain H1 (ITIH1), and plasminogen (PLG). The significantly upregulated glycopeptides within DR serum belong to the glycoproteins APOC3 (T-94; 3.21-fold), C1S (N-174; 2.86-fold), PRG4 (T-1004; 2.21-fold), VTN (N-86; 2.10-fold), and SERPINC1 (N-128; 1.79-fold). Glycopeptides that are downregulated in DR belong to the following glycoproteins: FN1 (N-542; 0.069-fold and T-2156; 0.107-fold), C4B (N-226; 0.207-fold and N-1328; 0.185-fold), AHSG (N-156; 0.641-fold and S-297; 0.328-fold), HPX (N-240; 0.256-fold and N-246; 0.639-fold), ITIH1 (S-656; 0.193-fold), and PLG (S-384; 0.468-fold). The bar plots depicting the glycopeptide levels detected in serum of patients with and without DR are shown in Figure 6.

Table 3.
Significant alterations in the serum glycoproteome of patients with diabetic retinopathy.
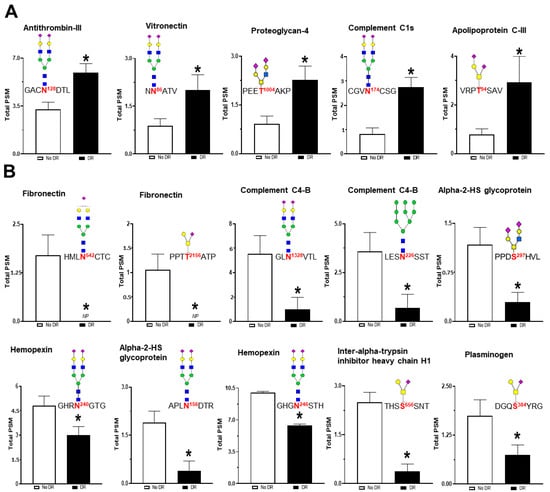
Figure 6.
The difference in the glycopeptide levels in the serum of patients with and without DR. Each bar represents the PSM (peptide spectrum match) values of the glycopeptides detected in the serum samples. The site of glycosylation and the structure of the glycan attached are also shown for each peptide. Statistical analysis revealed that levels of five glycopeptides were significantly upregulated (A), whereas the levels of 10 glycopeptides were significantly downregulated (B). * p-value < 0.05.
3.5. Trends in Sialylation and Fucosylation of Glycans
The terminal sugar residues of glycans such as sialic acid and fucose participate in cell–cell interactions and play an important role in the function of glycoproteins. Changes in the sialylation and fucosylation of glycopeptides in the serum were also investigated. There was an 8.3% increase in mono-sialylated glycopeptides and a 13.2% increase in di-sialylated glycopeptides in patients with DR as compared to patients without DR, whereas no significant differences were observed in non-sialylated or tri-sialylated glycopeptides (Figure 7A). Analysis of fucosylation revealed that non-fucosylated glycopeptides were significantly increased (12% increase) in patients with DR, and there was no significant difference in mono-, di-, tri-, and tetra-fucosylated glycopeptides (Figure 7B).
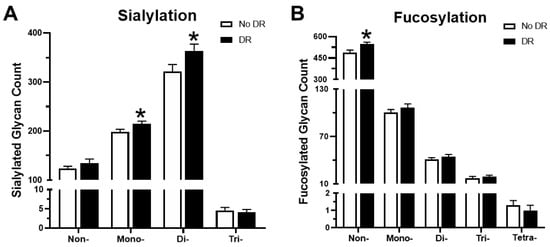
Figure 7.
Variations in sialylated and fucosylated glycopeptides in the serum. (A) The number of non- sialylated, and mono-, di-, and tri-sialylated glycan structures in patients with and without DR. (B) Fucosylation trends between the two groups. * p-value < 0.05.
4. Discussion
Recent studies have shown the significance of serum glycoproteomic changes in several diseases [15,16,17]. However, little information is available regarding the relevance of the serum glycoproteome in the pathogenesis of DR. In this study, using LC–MS/MS, both N- and O-linked intact glycopeptides were identified and quantified in the serum from patients with and without DR. Therefore, this study provides a comprehensive site-specific glycosylation map of serum glycoproteins and glycosylation changes associated with DR. On average, 229 unique glycopeptides from 116 glycoproteins were detected in each serum sample. A total of 15 distinct glycopeptides from 11 glycoproteins displayed altered glycosylation patterns in the DR patients. These glycopeptides belong to proteins involved in a host of functions, including protease regulation, inflammatory effects, cell adhesion, maintenance of the extracellular matrix (ECM), and the complement activation pathway. These pathways are known to play important roles in the pathogenesis of DR.
Two glycopeptides of fibronectin (sites N-542 and T-2156) showed changes in glycosylation in patients with DR. The expression of fibronectin in the retina, plasma, and kidneys has been shown to be increased in diabetes and has been associated with inflammation and retinal dysfunction [18,19,20]. Fibronectin is involved in ECM remodeling via cell-adhesion, migration, and thrombosis regulation [18]. In our study, we found decreased glycosylation of this protein at both sites. Previous studies have shown that the N-linked oligosaccharides of fibronectin act as modulators of biological functions of the glycoprotein, and the lack of carbohydrates significantly increases its ability to promote adhesion and spreading of fibroblasts [21]. N-glycans on fibronectin regulate cell adhesive capability by modulating cell-fibronectin interactions [22].
Proteoglycan 4 (PRG4), also known as lubricin, is a lubricating mucin-like glycoprotein that helps in reducing shear at the cartilage surface. Although the main function of this glycoprotein is lubrication for joints, it has been found to play a protective role at the ocular surface with functions as a boundary lubricant [23]. We found increased glycosylation of PRG4 at site T-1004 in patients with DR. Recent studies have shown inhibitory effects of PRG4 on the migration and proliferation of human venous cells as well as its protective role in preventing damage to the corneal epithelium [24,25]. Additionally, it has been shown that glycosylation of this protein is responsible for recruitment of leucocytes to inflammatory sites [26].
Two proteins within the complement system (C4B, C1S) displayed altered glycosylation patterns in DR. The levels of glycosylation of C4B were decreased at sites N-226 and N-1328, while C1S showed increased glycosylation at site N-174. C4B is a non-enzymatic molecule of complement C4 that is produced when C4 is proteolytically cleaved. C4B acts similar to an antibody, which binds to the surface of pathogens to mark them for phagocytosis [27]. Additionally, C4B has been linked to DR pathology in previous studies [28]. C1S is a serine protease that cleaves and activates the C1 complex, triggering the complement cascade involved in immune responses [29]. Emerging evidence has shown activation of the complement system to be involved in vascular abnormalities associated with DR [30].
Our analysis revealed that several altered glycoproteins, including fibronectin, antithrombin-III, alpha-2-HS-glycoprotein, inter-alpha-trypsin inhibitor heavy chain H1, hemopexin, and vitronectin are involved in peptidolytic processes through regulatory effects [18,31,32,33,34,35]. The altered glycosylation of serum peptidase regulators may offer novel insights into the pathogenesis of DR. ITIH1, a peptidase inhibitor, has previously been identified to be a potential biomarker of DR [36]. In our study, we found that glycosylation of ITIH1 was attenuated in the serum of patients with DR at site S-656. Glycosyl modification of ITIH1 has been shown to produce a physical barrier between insulin receptors and circulating insulin, exacerbating insulin resistance and subsequently increasing inflammation [37]. AHSG is another glycoprotein that exhibits peripheral insulin resistance via protease regulation [32]. Elevated serum AHSG levels have been found in patients with DR and associated with angiogenesis and inflammation [38]. SERPINC1 is a serine protease inhibitor involved in dampening the coagulation cascade that has been observed to provide anti-inflammatory effects to the vascular endothelium by interacting with heparin-like substances [39]. It has been documented that SERPINC1 concentration is normal in the serum of diabetics but shows an inverse correlation between non-enzymatic glycosylation (i.e., glycation) levels and function [40]. Our study found that SERPINC1 was more glycosylated in serum of patients with DR, likely leading to unchecked inflammatory effects on vascular walls.
The interactions of vitronectin and fibronectin with heparin-like substances play a large part in the coagulation cascade and contain novel regulatory functions. Fibronectin has been shown to be involved in the regulation of vascular endothelial-growth factor (VEGF) through a heparin-induced model [41]. Vitronectin also interacts with heparin in circulation and, after interaction, has been shown to oligomerize and transition from freely circulating to a fixture within the local ECM [42]. Vitronectin has been found to be upregulated in the serum of patients with early-stage DR and involved in diabetes-induced angiogenesis that occurs in late-stage DR [43,44]. Our study showed higher levels of glycosylation of vitronectin at site N-86 in DR patients. Hemopexin is an acute phase protein that binds to free heme to prevent its aberrant oxidative function in circulation [45]. However, hemopexin has been shown to have toxic protease activity that can lead to kidney damage reflected in diabetic nephropathy [34].
While the main function of plasminogen is to cleave fibrin to prevent blood coagulation, it also possesses anti-angiogenic properties that are enhanced by glycosylation [46,47]. Our study revealed that plasminogen is less glycosylated at site S-384 in the serum of DR patients. This glycosite is within the Kringle 4 fragment of plasminogen, which is the most potent angiostatin of this protein [46].
A glycopeptide of apolipoprotein C-III (APOC3), a known pro-inflammatory mediator [48], showed the most enriched glycosylation in DR. Increased levels of APOC3 in serum, plasma, and vitreous fluids have been associated with the occurrence and the severity of many metabolic disorders, including diabetes and DR [49,50,51]. Elevated serum APOC3 in diabetes has been shown to be a risk factor for DR due to its atherogenic properties [51]. APOC3 is known to be involved in a variety of inflammatory responses, including activation of endothelial cells and monocyte recruitment [52,53]. Di-sialylated APOC3 is associated with improved lipids in diabetes [54]. We also found that mono and di-sialylated glycoproteins were increased in the DR patients. This is in agreement with current literature that demonstrates the direct relationship of sialylation to both the onset of diabetes and the severity and degree of DR [55].
5. Conclusions
In conclusion, this study identified significant alterations in 15 serum glycopeptides belonging to 11 glycoproteins in DR patients. Many of these glycoproteins have been previously tied to vascular complications of diabetes, such as diabetic retinopathy or nephropathy. Considering the significant role these circulating glycoproteins have in chronic low-grade inflammation, these molecules are ideal candidates for future studies to discover precise molecular mechanisms and glycoproteomic-based therapeutics for DR. While the small sample size is the main limitation of our study, this in-depth analysis opens the door to study glycoproteomic alterations in DR using a larger cohort.
Author Contributions
Conceptualization, A.S., S.S., J.C.; methodology, S.K.K., W.Z.; formal analysis, A.S., J.C., T.J.L.; investigation, A.S., J.C., J.G., S.S.; resources, S.S., Z.L., L.U.; data curation, T.J.L., S.K.K.; writing—original draft preparation, J.C., J.G.; writing—review and editing, A.S., S.S.; project administration, A.S., S.S.; funding acquisition, S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study is supported by the National Institutes of Health, National Eye Institute (Bethesda, MD, USA) grant no. R01-EY026936 awarded to Shruti Sharma.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kempen, J.H.; O’Colmain, B.J.; Leske, M.C.; Haffner, S.M.; Klein, R.; Moss, S.E.; Taylor, H.R.; Hamman, R.F.; West, S.K.; Wang, J.J. The prevalence of diabetic retinopathy among adults in the United States. Arch. Ophthalmol. 2004, 122, 552–563. [Google Scholar]
- Guariguata, L.; Whiting, D.R.; Hambleton, I.; Beagley, J.; Linnenkamp, U.; Shaw, J.E. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res. Clin. Pract. 2014, 103, 137–149. [Google Scholar] [CrossRef] [PubMed]
- El-Asrar, A.M.A. Role of inflammation in the pathogenesis of diabetic retinopathy. Middle East Afr. J. Ophthalmol. 2012, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.W.; Sundstrom, J.M. A proposal for early and personalized treatment of diabetic retinopathy based on clinical pathophysiology and molecular phenotyping. Vis. Res. 2017, 139, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef]
- Saldova, R.; Wormald, M.R.; Dwek, R.A.; Rudd, P.M. Glycosylation changes on serum glycoproteins in ovarian cancer may contribute to disease pathogenesis. Dis. Markers 2008, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, L.; Li, X.; Zhu, H.; Gashash, E.A.; Li, Z.; Wang, P.G.; Ma, C. An integrated proteomic and glycoproteomic study for differences on glycosylation occupancy in rheumatoid arthritis. Anal. Bioanal. Chem. 2019, 411, 1331–1338. [Google Scholar] [CrossRef]
- Shah, P.; Wang, X.; Yang, W.; Eshghi, S.T.; Sun, S.; Hoti, N.; Chen, L.; Yang, S.; Pasay, J.; Rubin, A. Integrated proteomic and glycoproteomic analyses of prostate cancer cells reveal glycoprotein alteration in protein abundance and glycosylation. Mol. Cell. Proteom. 2015, 14, 2753–2763. [Google Scholar] [CrossRef]
- Ang, I.L.; Poon, T.C.; Lai, P.B.; Chan, A.T.; Ngai, S.-M.; Hui, A.Y.; Johnson, P.J.; Sung, J.J. Study of serum haptoglobin and its glycoforms in the diagnosis of hepatocellular carcinoma: A glycoproteomic approach. J. Proteome Res. 2006, 5, 2691–2700. [Google Scholar] [CrossRef]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef]
- Mariño, K.; Saldova, R.; Adamczyk, B.; Rudd, P.M.; Rauter, A. Changes in serum N-glycosylation profiles: Functional significance and potential for diagnostics. Carbohydr. Chem. 2012, 37, 57–93. [Google Scholar]
- Scott, D.W.; Patel, R.P. Endothelial heterogeneity and adhesion molecules N-glycosylation: Implications in leukocyte trafficking in inflammation. Glycobiology 2013, 23, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Qu, Y.; Zhang, Z.; Wang, Z.; Prytkova, I.; Wu, S. Intact glycopeptide characterization using mass spectrometry. Expert. Rev. Proteom. 2016, 13, 513–522. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, J.; Li, L. Recent advances in mass spectrometry (MS)-based glycoproteomics in complex biological samples. Trac. Trends. Anal. Chem. 2019, 118, 880–892. [Google Scholar] [CrossRef]
- Schumacher, K.N.; Dodds, E.D. A case for protein-level and site-level specificity in glycoproteomic studies of disease. Glycoconj. J. 2016, 33, 377–385. [Google Scholar] [CrossRef]
- Lemańska-Perek, A.; Adamik, B. Fibronectin and its soluble EDA-FN isoform as biomarkers for inflammation and sepsis. Adv. Clin. Exp. Med. 2019. [Google Scholar] [CrossRef]
- Miller, C.G.; Budoff, G.; Prenner, J.L.; Schwarzbauer, J.E. Minireview: Fibronectin in retinal disease. Exp. Biol. Med. 2017, 242, 1–7. [Google Scholar] [CrossRef]
- Kanters, S.D.; Banga, J.-D.; Algra, A.; Frijns, R.C.; Beutler, J.J.; Fijnheer, R. Plasma levels of cellular fibronectin in diabetes. Diabetes Care 2001, 24, 323–327. [Google Scholar] [CrossRef]
- Jones, G.E.; Arumugham, R.G.; Tanzer, M.L. Fibronectin glycosylation modulates fibroblast adhesion and spreading. J. Cell Biol. 1986, 103, 1663–1670. [Google Scholar] [CrossRef]
- Hsiao, C.-T.; Cheng, H.-W.; Huang, C.-M.; Li, H.-R.; Ou, M.-H.; Huang, J.-R.; Khoo, K.-H.; Yu, H.W.; Chen, Y.-Q.; Wang, Y.-K. Fibronectin in cell adhesion and migration via N-glycosylation. Oncotarget 2017, 8, 70653. [Google Scholar] [CrossRef]
- Samsom, M.L.; Morrison, S.; Masala, N.; Sullivan, B.D.; Sullivan, D.A.; Sheardown, H.; Schmidt, T.A. Characterization of full-length recombinant human Proteoglycan 4 as an ocular surface boundary lubricant. Exp. Eye Res. 2014, 127, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.A.; Sullivan, D.A.; Knop, E.; Richards, S.M.; Knop, N.; Liu, S.; Sahin, A.; Darabad, R.R.; Morrison, S.; Kam, W.R. Transcription, translation, and function of lubricin, a boundary lubricant, at the ocular surface. Jama Ophthalmol. 2013, 131, 766–776. [Google Scholar] [CrossRef]
- Wang, L.; Kikuchi, S.; Schmidt, T.A.; Hoofnagle, M.; Wight, T.N.; Azuma, N.; Tang, G.L.; Sobel, M.; Velamoor, G.R.; Mokadam, N.A. Inhibitory Effects of PRG4 on Migration and Proliferation of Human Venous Cells. J. Surg. Res. 2020, 253, 53–62. [Google Scholar] [CrossRef]
- Steele, B.; Alvarez-Veronesi, M.; Schmidt, T. Molecular weight characterization of PRG4 proteins using multi-angle laser light scattering (MALLS). Osteoarthr. Cartil. 2013, 21, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, S.; Kidmose, R.T.; Petersen, S.V.; Szilágyi, Á.; Prohászka, Z.; Andersen, G.R. Structural Basis for the Function of Complement Component C4 within the Classical and Lectin Pathways of Complement. J. Immunol. 2015, 194, 5488–5496. [Google Scholar] [CrossRef]
- Garcia-Ramirez, M.; Canals, F.; Hernández, C.; Colome, N.; Ferrer, C.; Carrasco, E.; Garcia-Arumi, J.; Simo, R. Proteomic analysis of human vitreous fluid by fluorescence-based difference gel electrophoresis (DIGE): A new strategy for identifying potential candidates in the pathogenesis of proliferative diabetic retinopathy. Diabetologia 2007, 50, 1294–1303. [Google Scholar] [CrossRef]
- Gaboriaud, C.; Rossi, V.; Bally, I.; Arlaud, G.J.; Fontecilla-Camps, J.C. Crystal structure of the catalytic domain of human complement C1s: A serine protease with a handle. Embo J. 2000, 19, 1755–1765. [Google Scholar] [CrossRef]
- Østergaard, J.; Hansen, T.K.; Thiel, S.; Flyvbjerg, A. Complement activation and diabetic vascular complications. Clin. Chim. Acta 2005, 361, 10–19. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, F.; Liang, M. SerpinC1/Antithrombin III in kidney-related diseases. Clin. Sci. 2017, 131, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Emoto, M.; Inaba, M. Fetuin-A: A multifunctional protein. Recent Pat. Endocr. Metab. Immune Drug Discov. 2011, 5, 124–146. [Google Scholar] [CrossRef]
- Zhuo, L.; Hascall, V.C.; Kimata, K. Inter-α-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J. Biol. Chem. 2004, 279, 38079–38082. [Google Scholar] [CrossRef] [PubMed]
- Bakker, W.W.; Borghuis, T.; Harmsen, M.C.; Van den Berg, A.; Kema, I.; Niezen, K.E.; Kapojos, J.J. Protease activity of plasma hemopexin. Kidney Int. 2005, 68, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Preissner, K.T.; de Boer, H.; Pannekoek, H.; de Groot, P.G. Thrombin regulation by physiological inhibitors: The role of vitronectin. In Seminars in Thrombosis and Hemostasis; Thieme Medical Publishers Inc.: Leipzig, Germany, 1996; pp. 165–172. [Google Scholar]
- Mcauley, A. Biomarkers in Diabetic Retinopathy: Genetic and Proteomic Profiling. Ph.D. Thesis, The University of Melbourne, Melbourne, Australia, 2014. [Google Scholar]
- Kim, T.H.; Koo, J.H.; Heo, M.J.; Han, C.Y.; Kim, Y.-I.; Park, S.-Y.; Cho, I.J.; Lee, C.H.; Choi, C.S.; Lee, J.W. Overproduction of inter-α-trypsin inhibitor heavy chain 1 after loss of Gα13 in liver exacerbates systemic insulin resistance in mice. Sci. Transl. Med. 2019, 11, eaan4735. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-W.; Ju, H.-X.; Sun, M.-Z.; Fu, Q.-P.; Chen, H.-M.; Ji, H.-B.; Ji, F.; Jiang, D.-M. Serum fetuin-A levels are independently correlated with vascular endothelial growth factor and C-reactive protein concentrations in type 2 diabetic patients with diabetic retinopathy. Clin. Chim. Acta 2016, 455, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.H.; Sniecinski, R.M.; Welsby, I.J.; Levi, M. Antithrombin: Anti-inflammatory properties and clinical applications. Thromb. Haemost. 2016, 115, 712–728. [Google Scholar]
- Ceriello, A.; Giugliano, D.; Russo, P.D.; Tirelli, A.; Passariello, N.; Sgambato, S. Metabolic control may alter antithrombin III activity but not its plasma concentration in diabetes: A possible role for nonenzymatic glycosylation. Diabetes Care 1986, 9, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Mitsi, M.; Forsten-Williams, K.; Gopalakrishnan, M.; Nugent, M.A. A catalytic role of heparin within the extracellular matrix. J. Biol. Chem. 2008, 283, 34796–34807. [Google Scholar] [CrossRef]
- Chillakuri, C.R.; Jones, C.; Mardon, H.J. Heparin binding domain in vitronectin is required for oligomerization and thus enhances integrin mediated cell adhesion and spreading. Febs. Lett. 2010, 584, 3287–3291. [Google Scholar] [CrossRef]
- Preissner, K.; Kanse, S.; Hammes, H.-P. Integrin chatter and vascular function in diabetic retinopathy. Horm. Metab. Res. 1997, 29, 643–645. [Google Scholar] [CrossRef]
- Esser, P.; Bresgen, M.; Weller, M.; Heimann, K.; Wiedemann, P. The significance of vitronectin in proliferative diabetic retinopathy. Graefe’s Arch. Clin. Exp. Ophthalmol. 1994, 232, 477–481. [Google Scholar] [CrossRef]
- Taketani, S.; Immenschuh, S.; Go, S.; Sinclair, P.R.; Stockert, R.J.; Liem, H.H.; Eberhard, U.M. Hemopexin from four species inhibits the association of heme with cultured hepatoma cells or primary rat hepatocytes exhibiting a small number of species specific hemopexin receptors. Hepatology 1998, 27, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Santos, I.C.; Silbiger, V.N.; Higuchi, D.A.; Gomes, M.A.; Barcelos, L.S.; Teixeira, M.M.; Lopes, M.T.P.; Cardoso, V.N.; Lima, M.P.; Araujo, R.C.; et al. Angiostatic activity of human plasminogen fragments is highly dependent on glycosylation. Cancer Sci. 2010, 101, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Collen, D. The plasminogen (fibrinolytic) system. Thromb. Haemost. 1999, 82, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, A.; Aikawa, M.; Libby, P.; Alcaide, P.; Luscinskas, F.W.; Sacks, F.M. Apolipoprotein CIII in apolipoprotein B lipoproteins enhances the adhesion of human monocytic cells to endothelial cells. Circulation 2006, 113, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.L.; McHenry, M.B.; Lok, K.H.; Hunter, S.J.; Le, N.-A.; Jenkins, A.J.; Zheng, D.; Semler, A.; Page, G.; Brown, W.V. Apolipoprotein C-III protein concentrations and gene polymorphisms in Type 1 diabetes: Associations with microvascular disease complications in the DCCT/EDIC cohort. J. Diabetes Complicat. 2005, 19, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Gao, J.; Pu, C.; Zhang, Y. Apolipoprotein status in type 2 diabetes mellitus and its complications. Mol. Med. Rep. 2017, 16, 9279–9286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, J.; Hu, Y.; Ding, Y.; Zhu, J.; Zhuang, C. Relationship between serum apolipoproteins levels and retinopathy risk in subjects with type 2 diabetes mellitus. Acta. Diabetol. 2018, 55, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, A.; Aikawa, M.; Alcaide, P.; Luscinskas, F.W.; Libby, P.; Sacks, F.M. Apolipoprotein CIII induces expression of vascular cell adhesion molecule-1 (VCAM-1) in vascular endothelial cells and increases adhesion of monocytic cells. Am. Heart Assoc. 2006. [Google Scholar] [CrossRef]
- Kawakami, A.; Aikawa, M.; Nitta, N.; Yoshida, M.; Libby, P.; Sacks, F.M. Apolipoprotein CIII-induced THP-1 cell adhesion to endothelial cells involves pertussis toxin-sensitive G protein-and protein kinase Cα-mediated nuclear factor-κB activation. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Koska, J.; Yassine, H.; Trenchevska, O.; Sinari, S.; Schwenke, D.C.; Yen, F.T.; Billheimer, D.; Nelson, R.W.; Nedelkov, D.; Reaven, P.D. Disialylated apolipoprotein C-III proteoform is associated with improved lipids in prediabetes and type 2 diabetes. J. Lipid Res. 2016, 57, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Akbri, M.Z.A.; Sheikh, A.S.; Bhatti, M.S.; Hussnain, M.; Chaudhry, Z.A. Serum sialic acid level in diabetic retinopathy. J. Ayub Med. Coll. Abbottabad 2001, 13, 29–30. [Google Scholar] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).