Abstract
The mass spectrometric analysis of skeletal muscle proteins has used both peptide-centric and protein-focused approaches. The term ‘top-down proteomics’ is often used in relation to studying purified proteoforms and their post-translational modifications. Two-dimensional gel electrophoresis, in combination with peptide generation for the identification and characterization of intact proteoforms being present in two-dimensional spots, plays a critical role in specific applications of top-down proteomics. A decisive bioanalytical advantage of gel-based and top-down approaches is the initial bioanalytical focus on intact proteins, which usually enables the swift identification and detailed characterisation of specific proteoforms. In this review, we describe the usage of two-dimensional gel electrophoretic top-down proteomics and related approaches for the systematic analysis of key components of the contractile apparatus, with a special focus on myosin heavy and light chains and their associated regulatory proteins. The detailed biochemical analysis of proteins belonging to the thick and thin skeletal muscle filaments has decisively improved our biochemical understanding of structure-function relationships within the contractile apparatus. Gel-based and top-down proteomics has clearly established a variety of slow and fast isoforms of myosin, troponin and tropomyosin as excellent markers of fibre type specification and dynamic muscle transition processes.
1. Introduction
Mass spectrometry (MS) based proteomics is concerned with the systematic analysis of the protein components of a particular biological fluid, cell type, tissue, organ or organism [1], and plays a key role in protein biochemistry and molecular cell biology [2]. Modern proteomics is based on high-performance mass analysers [3,4,5] and is routinely used for cell-specific protein expression profiling, large-scale comparative protein studies, the biochemical characterization of the composition and structure of individual subcellular proteomes, and the determination of the temporal and spatial dynamics within specific protein constituents [6,7,8,9]. Gel-based proteomics is one of the most multipurpose means for the analysis of complex proteomes, such as seen in skeletal muscle tissues [10,11,12]. Two-dimensional gel electrophoresis (2DGE) exemplifies an exceptionally robust strategy to separate, identify, and quantify proteins when used in combination with MS analysis or various immunological tests [13,14,15]. While 2DGE is an effective technique for the systematic analysis of complex protein lysates, the ability to directly visualise changes in the abundance of proteins/post-translational modifications (PTM) is a powerful application.
Since 2DGE approaches present a visual platform for the efficient separation of individual proteoforms prior to MS analysis, the availability of internet-based comparisons via international 2D-gel databases is a strength of gel-based proteomics. A considerable number of 2DGE databases were established, especially in the period 1993–2008 [16,17,18,19,20,21,22,23,24,25,26,27,28], and include SWISS-2DPAGE, World-2DPAGE, Human 2-D PAGE biobase, Flicker, UAB Proteomics Database, AGML Central, GELBANK, WEB P.A.G.E, Open2Dprot Project, LECB 2-D PAGE Gel Images Data Sets, ProteomeWeb, PHProteomicsDB, pProRep and DynaProt 2D. These extensive collections of 2DGE images and bioinformatic analysis tools can be highly useful for the comparative analysis of newly established 2DGE images, the systematic cataloguing of proteoforms using 2DGE techniques and the integration of gel-based proteomic findings and mass spectral data [28]. Comparative image analyses can also be helpful for the swift identification of one-dimensional (1D) protein bands and 2D protein spots within archived gels that were produced prior to the establishment of MS-based proteomics [29,30]. Unfortunately, many of these 2DGE image databases are no longer actively maintained, which constitutes a drawback in the further development of the field of gel-based proteomics and hampers the efficient sharing of proteomic data generated by 2DGE analyses. Hopefully, international efforts will be able to re-establish functional and internet-based databanks for future gel electrophoretic studies and the efficient comparison of proteomic maps.
Enormous progress has been made in the establishment of the tissue-based map of the human proteome [31], including the systematic cataloguing of the protein constituents of skeletal muscles [32,33,34,35]. Large-scale biochemical studies with a focus on the protein composition of voluntary contractile tissues were carried out by various proteomic techniques with a peptide-centric approach following the digestion of muscle proteins extracted from various animal species prior to peptide MS analysis [10,11,12]. These proteomic surveys have investigated in detail the complex effects of fibre transitions, neuromuscular disease, exercise, disuse atrophy and aging on the skeletal muscle proteome [36,37,38,39]. Efficient muscle protein separation by liquid chromatography (LC) or diverse gel electrophoretic techniques have been employed in combination with in-solution or in-gel digestion protocols, whereby 2DGE plays a key role in proteome science [13,14,15,40,41]. The bioanalytical advantages versus potential technical limitations of gel electrophoretic approaches, as applied to the highly complex and dynamic proteome of skeletal muscle tissues, have been previously discussed in an extensive review on comparative skeletal muscle proteomics using 2DGE [42]. The recent introduction of the photo-cleavable anionic surfactant 4-hexylphenylazosulfonate as a potential sodium dodecyl sulfate replacement in polyacrylamide GE [43] might be suitable for improved gel-based and top-down proteomic workflows [44].
Using a variety of GE- and LC-based protein separation techniques in combination with MS analysis, over 10,000 muscle-associated proteins have been identified [32,33,34,35,45,46,47,48]. Since peptide-based and bottom-up bioanalytical methods can often not properly distinguish among discrete proteoforms, such as individual isoforms or post-translationally modified protein variants, biochemical approaches using top-down proteomics with a focus on the mass spectrometric characterization of intact protein species can be advantageous [49,50,51]. Importantly, specific forms of top-down proteomics can provide data for accurate protein mass determination and detailed information on the identity of specific protein isoforms and their PTMs [52,53,54], making it an ideal investigative tool of modern protein biochemistry for determining the structure and function of macromolecular protein complexes [55,56,57]. In this review, the usage of top-down proteomics in the field of skeletal muscle biochemistry is outlined with special emphasis on gel-based techniques, especially 2DGE, for studying normal, adapting and aging muscles. Detailed descriptions and critical assessments of the establishment of the skeletal muscle proteome and the usage of comparative MS-based proteomics for studying pathobiochemical changes in neuromuscular disorders have already been extensively reviewed [10,11,12,58,59,60].
The contractile function of the skeletal muscle system is based on the dynamic arrangement of an unusual cell type that is very large, multi-nucleated and extremely heterogeneous in its protein isoform composition [61]. A striking biochemical feature of contractile tissues is the presence of a considerable number of very high-molecular-mass protein species [62], which are occasionally difficult to separate for proteomic studies [63]. The physiological coupling of individual muscle fibres to its innervating motor neuron within distinct motor units influences skeletal muscle mass, fibre size, contractile kinetics, energy metabolism, muscle cell specificity and fibre type-related protein expression pattern [64]. Besides a mixture of fast-twitching, slow-twitching and hybrid fibres [65], individual skeletal muscles also contain several layers of connective tissue, capillaries and myospecific stem cells. Although this heterogeneous cellular composition often complicates the systematic mass spectrometric profiling of the skeletal muscle proteome in health and disease [66], skeletal muscle proteomics has clearly confirmed the multi-functionality and high adaptive capacity of the voluntary contractile system. This brief review on representative top-down proteomic studies of contractile proteins from skeletal muscle fibres illustrates how the systematic application of gel-based and MS approaches has improved our biochemical knowledge of protein structure-function relationships and the dynamic nature of the neuromuscular proteome.
2. Top-Down Proteomics
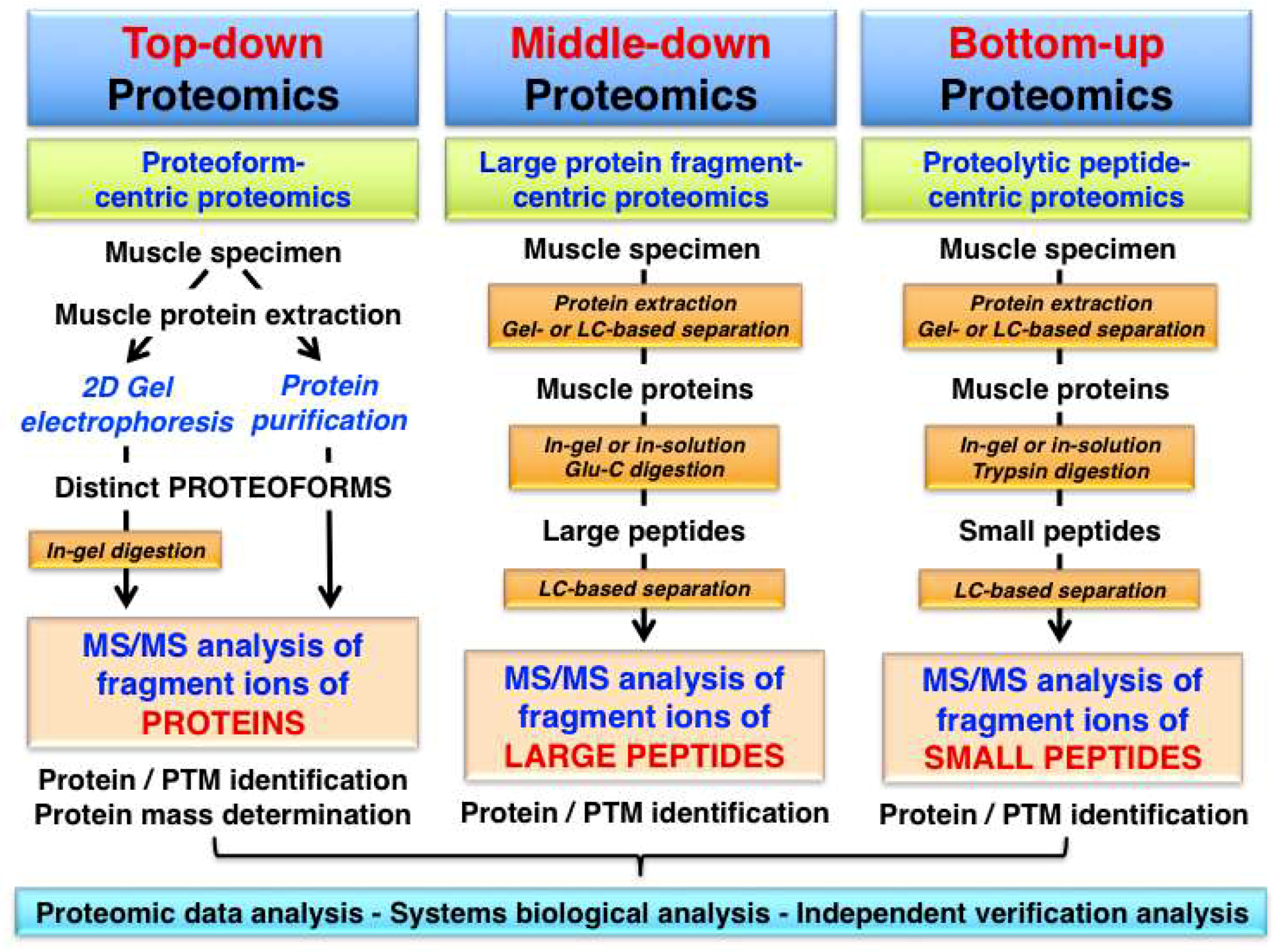
Top-down proteomics is an advanced biochemical method for the quantitative analysis of specific proteoforms leading to the identification and thorough characterization of intact protein species by tandem MS/MS analysis. Fundamental differences in the analytical approach used in top-down proteomics, as compared to middle-down proteomics or bottom-up proteomics are described in Figure 1.
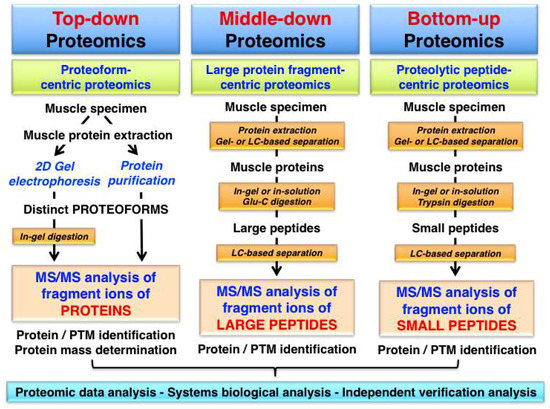
Figure 1.
Overview of proteomic approaches used for the identification and characterization of skeletal muscle proteins and their post-translational modifications (PTMs). The diagram shows differences in the bioanalytical workflow involved in top-down proteomics, middle-down proteomics and bottom-up proteomics, which is characterized by the mass spectrometry (MS) analysis of intact proteins/proteoforms, protein fragments and peptides, respectively.
Comprehensive guides for sample preparation and discussions of technical issues in relation to specific approaches in top-down protein analysis have been published [67,68,69,70,71,72]. The main analytical advantages of this approach are the determination of distinct characteristics of a particular protein isoform and the capability to distinguish degradation products, as well as identify sequence variants and combinations of dynamic PTMs [73].
While top-down proteomics focuses on the MS analysis of intact proteins with a proteoform-centric approach, the other proteomic strategies involve the study of differently sized protein fragments following protein digestion and peptide generation. Middle-down proteomics is used for the analysis of large protein fragments and the most frequently employed bottom-up proteomics approach is a peptide-centric technique [74]. In this article, we define ‘top-down proteomics’ in its broadest sense and use the term not just in a focused way describing exclusively the MS/MS analysis of an intact protein, but also include the characterization of distinct proteoforms using gel-based separation technology in combination with routine proteomic identification methods. This review includes therefore bioanalytical approaches that initiate the proteomic analysis with a separated and thus isolated proteoform of a particular protein and then employ differing MS methods for protein/peptide identification and/or PTM analysis.
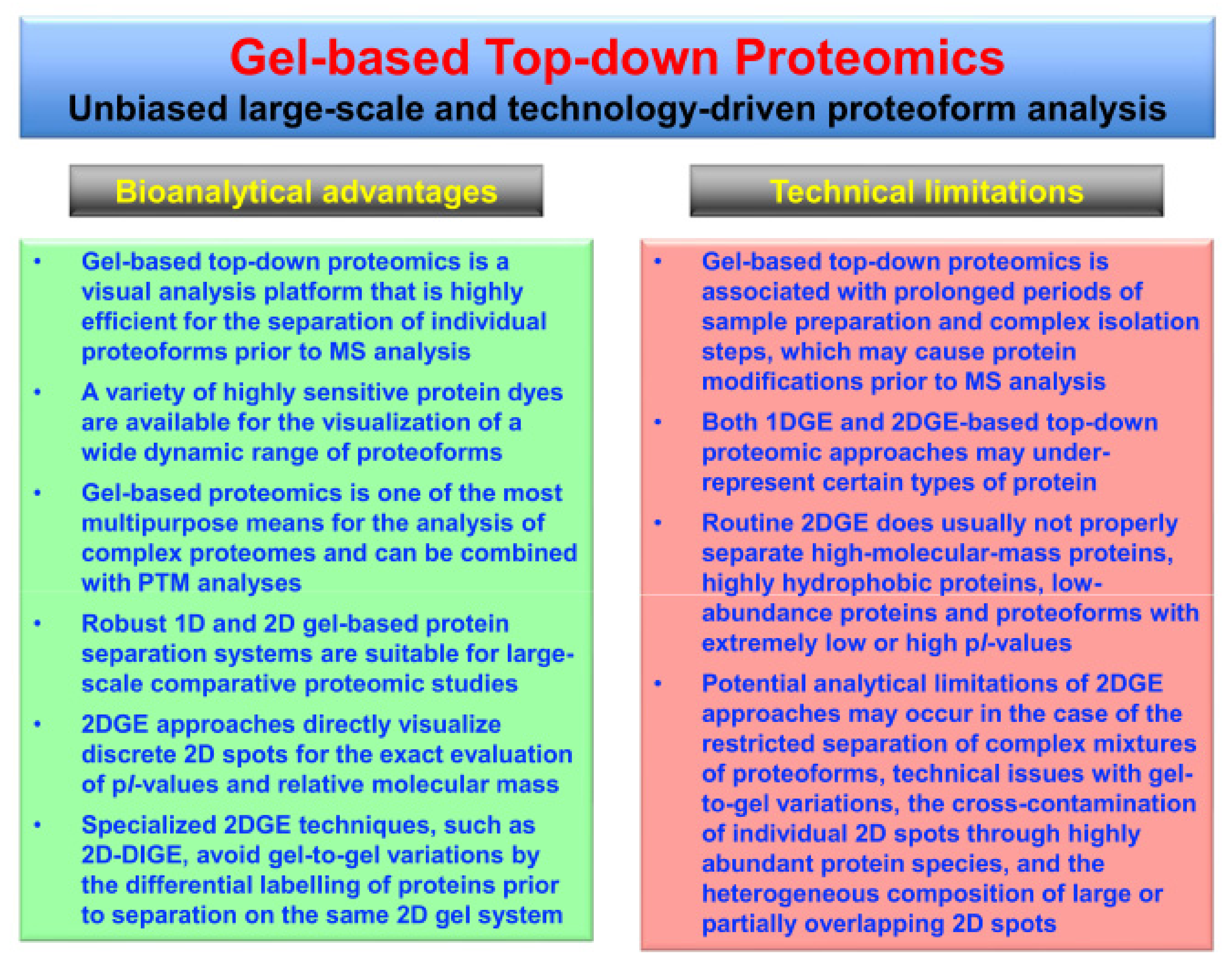
Bioanalytical advantages versus potential technical limitations of gel-based top-down proteomics, especially those issues associated with 2DGE approaches [42], are listed in Figure 2.
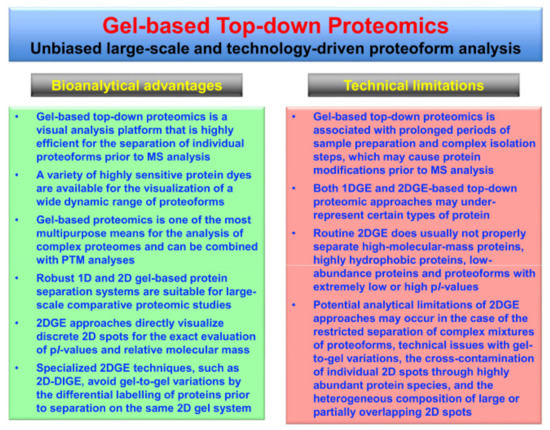
Figure 2.
Overview of the bioanalytical advantages versus potential technical limitations of gel-based top-down proteomics, especially in relation to two-dimensional gel electrophoresis (2DGE) approaches.
Gel-based methods are robust, highly reproducible and reliable protein separation techniques that are ideally suited for large-scale surveys of proteoforms. Staining of gel electrophoretically separated proteins can be achieved with a wide range of highly sensitive dyes, such as colloidal Coomassie Brilliant Blue, silver stains and various fluorescent dyes, as extensively reviewed [75,76,77,78,79,80]. The analytical strength of gel-based methods, as compared to protein separation by LC techniques, is the direct visualization of proteins within 1D gel bands or 2D gel spots. Thus, the position of discrete protein spots in 2D gels provides a swift and reliable way for determining the characteristic combination of the relative molecular mass and the pI (isoelectric point) value of a specific proteoform of interest. Potential bioanalytical complications of gel-based top-down proteomics are associated with the fact that prolonged periods of sample preparation and complex isolation steps may introduce protein modifications prior to MS analysis. Routine 2DGE analyses may also under-estimate the presence of certain types of proteins, such as high-molecular-mass proteins, very hydrophobic proteoforms and low-abundance proteins [42].
3. Top-Down Proteomics of Contractile Proteins from Skeletal Muscle
3.1. Acto-Myosin Apparatus and Auxilary Components
In the longitudinal direction, mature skeletal muscle fibres exhibit a characteristic striated appearance due to the regular patterning of anisotropic A-bands and isotropic I-bands, which form sarcomeric units between two neighbouring Z-disks [81]. A major feature of muscle genetics is the fact that the isoform-specific expression of a limited number of genes that encode contractile components leads to the production of the majority of the protein content of contractile cells [61]. Within the sarcomere structure, the contractile apparatus of skeletal muscle fibres consists of two main contractile filaments forming the acto-myosin system [82,83,84] and two auxiliary filamentous systems acting as myofibrillar gap structures [85,86,87,88]. The complex network of the sarcomere units consists mainly of myosin light and heavy chains, actin and the regulatory tropomyosin/troponin complex, the giant proteins titin and nebulin, as well as the structural and functional support units presented by the M-band zone and the Z-disk complex [89].
The principal motor molecules of the contractile protein assembly, which produce force via cross-bridge/swinging lever-arm mechanisms [90], are filamentous actin and the myosin heavy chains [91]. The entire myosin complex exists as a hexameric structure of two myosin heavy chains (MyHC) and four myosin light chains (MLC) [92]. The MyHC head structure mediates the reversible coupling and cross-bridging process between the thick filaments located in the A-band region and the actin filaments [93]. The regulatory and catalytic light chains provide critical phosphorylation sites, which are essential for the movement of phosphorylated myosin cross-bridges away from the thick filament, as well as the fine tuning of myosin motor function by providing structural stability to the lever arm domain of the myosin head [94]. Below listed are the main components of the thick myosin-containing filament, the thin actin-containing filament, the auxiliary titin filament, the auxiliary nebulin filament, the sarcomeric M-band complex and the sarcomeric Z-disk complex [66,95]:
- Thick myosin-containing filament: hexameric myosin complex consisting of myosin heavy chains (MyHC I, IIa, IId/x, and IIb), myosin light chains (fast and slow MLC isoforms; regulatory and catalytic subunits) and myosin binding proteins
- Thin actin-containing filament: α-actin, α/β-tropomyosin, troponin complex (fast and slow troponin-I, troponin-T and troponin-C isoforms)
- Auxiliary titin filament: half-sarcomere spanning titin and muscle ankyrin repeat protein
- Auxiliary nebulin filament: nebulin, in close contact to actin-containing thin filament
- Sarcomeric M-band complex: myomesin and obscurin, linked to titin filament
- Sarcomeric Z-disk complex: α-actinin, plectin, telethonin, desmin, myozenin, myotilin, synemin and filamin
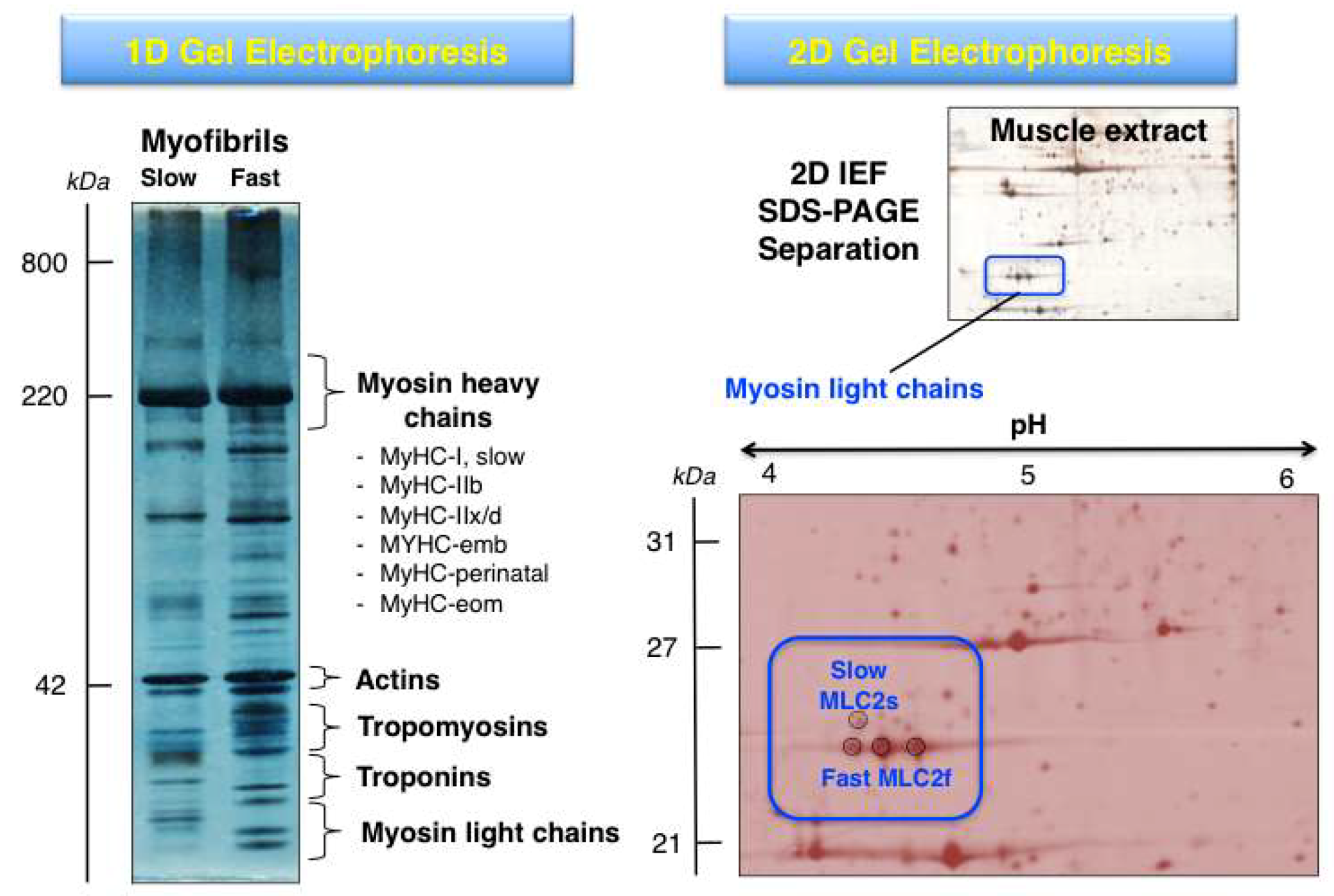
The systematic 2DGE separation of protein isoforms from skeletal muscle subtypes has led, in combination with MS analysis, to the identification of distinct isoforms of myosins, actins, troponins and tropomyosins, as well as many associated proteins of the sarcomeric structure [10,42]. Building on the findings from initial gel-based studies of the skeletal muscle proteome [96,97,98], high-resolution 2DGE was applied to the cataloguing of human vastus lateralis, deltoideus and laryngeal muscles [99,100,101,102] and a variety of animal species including rat, rabbit, cow and fish [103,104,105,106,107,108,109,110]. The differential expression patterns of contractile proteins in predominantly fast- versus slow-twitching skeletal muscles was established for various muscle subtypes, such as gastrocnemius, extensor digitorium longus, longissimus dorsi, semitendinosus and soleus muscles, using 2DGE [111,112,113,114,115]. The gel-based fibre specification maps agree with the distribution of fast versus slow muscle protein isoforms as determined by in vivo stable isotope labelling with amino acids in the SILAC mouse model [116]. Figure 3 shows representative 1DGE and 2DGE images of separated muscle protein populations. These approaches are routinely used for the proteomic identification of distinct proteoforms of contractile components.
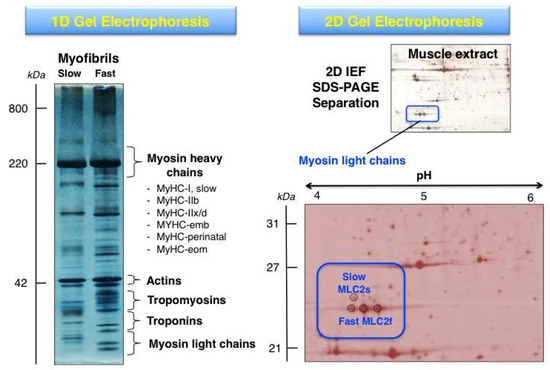
Figure 3.
Comparison of gel electrophoretic images using 1D (one-dimensional) GE versus 2D (two-dimensional) GE analysis of contractile proteins from rodent skeletal muscle. The 1D gel shows the separation of isolated myofibrils from slow versus fast muscles and the relative position of key contractile proteins from the thick and thin filaments. The MS analysis of 1D gel zones has established the presence of isoforms of myosin heavy chain, myosin light chain, actin, troponin and tropomyosin. The 2DGE image of a separated skeletal muscle extract and its enlargement highlights the MS-based identification of proteoforms of myosin light chain (slow MLC2/P51667 and fast MLC2/P97457).
3.2. Top-Down Proteomics of Myosins
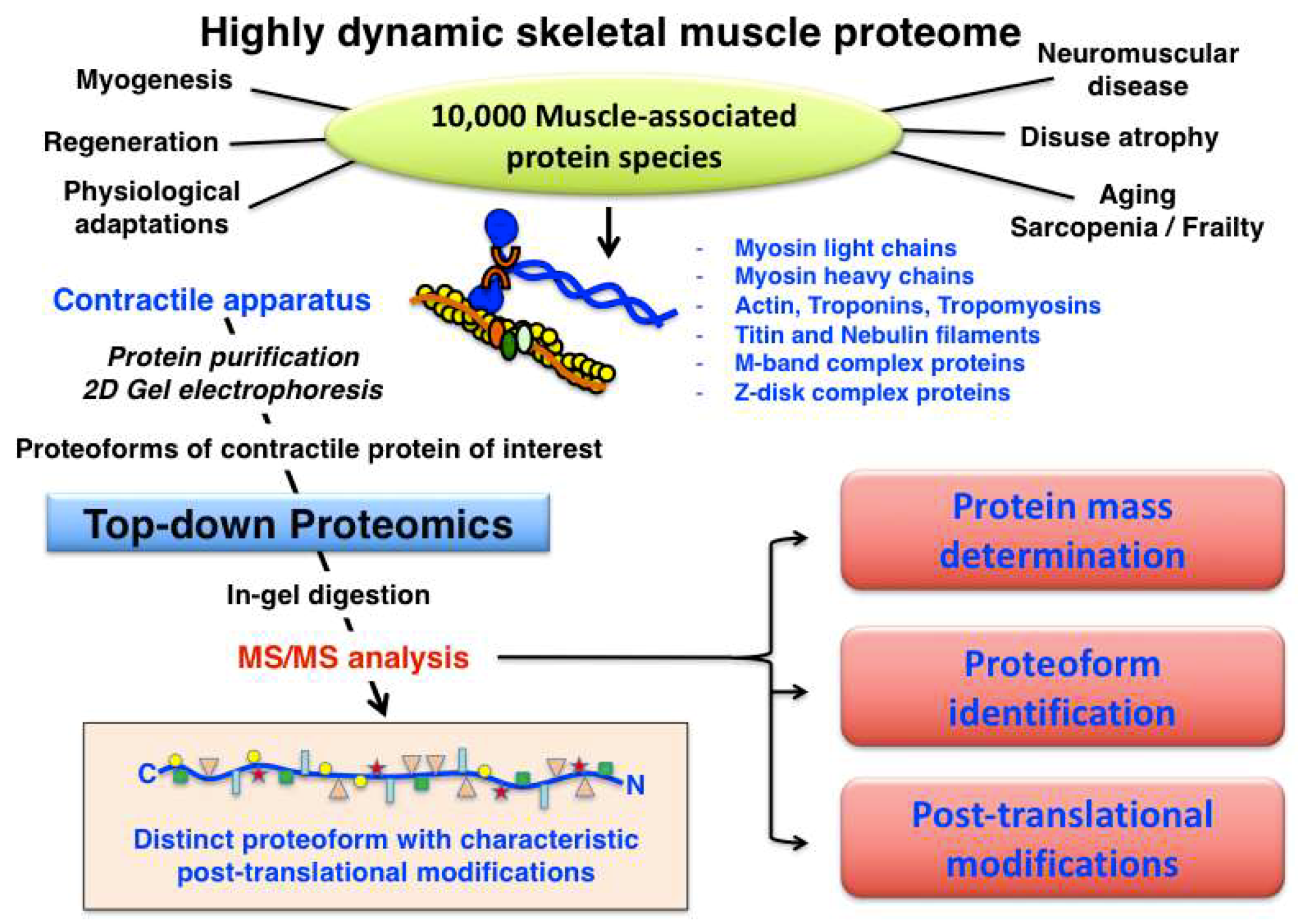
Muscle fibre type classification can be conveniently carried out by determining the distribution of myosin light and heavy chain isoforms, making this large family of contractile proteins an essential class of fibre type markers [92]. The expression pattern of MyHC molecules is often performed by immunofluorescence microscopy of transverse cryosections from individual skeletal muscles [117]. Most adult skeletal muscles contain a mixture of the main fibre types I, IIa, IIx, IIb, as well as the mixed hybrid fibre types I/IIa, IIa/IIx and IIx/IIb. However, considerable species-specific differences exist in relation to the distribution of cellular subtypes within large fibre populations [118]. Although at least 11 MyHC isoforms and nine MLC isoforms appear to exist in developing and matured skeletal muscles [119,120], the spectrum of slow-twitching oxidative fibres towards faster-twitching and more glycolytic fibres can be accurately evaluated by the abundance of the main human slow/cardiac MyHC-I-beta (P12883; MYH7 gene), fast MyHC-2a (Q9UKX2; MYH2 gene), fast MyHC-2x (P12882; MYH1 gene) and fast MyHC-2b (Q9Y623; MYH4 gene) isoforms [117,118]. In addition, developing and regenerating muscles contain MyHC-neo (P13535; MYH8 gene) and MyHC-emb (P11055; MYH3 gene), and the highly specialist extraocular muscle fibres are characterized by the presence of MyHC-EO (Q9UKX3; MYH13 gene), MyHC-I-ton (A7E2Y1; MYH7b gene) and MyHC-15 (Q9Y2K3; MYH15 gene) [92]. Human MLC molecules exist mainly as essential catalytic subunits and regulatory/phosphorylatable subunits, i.e., as fast MLC1/MLC3 (P05976; MYL1 gene) and fast MLC2 (Q96A32; MYLPF gene) or slow MLC1 (MLC3) (P08590; MYL3 gene) and slow MLC2 (P10916; MYL2 gene) isoforms. The combination of various MyHC and MLC subunits form a very large number of hexameric iso-myosins in the sarcomere [92,121,122]. In contrast, human filamentous F-actin polymers in developing and mature fibres consist of only two G-actin isoforms, i.e., cardiac alpha-actin (P68032; ACTC1 gene) and muscle alpha-actin (P68133; ACTA1 gene). An overview of the systematic characterization of contractile proteins from skeletal muscle using top-down proteomics is provided in Figure 4.
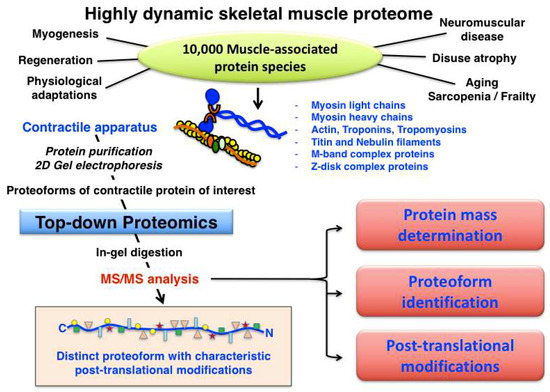
Figure 4.
Overview of top-down proteomic analysis of contractile proteins from skeletal muscle.
The detailed proteomic analysis of fast versus slow skeletal muscles has confirmed the complexity of myosin expression patterns. Due to their differing molecular mass and electrophoretic mobility, heavy chains and light chains of myosin complexes are routinely studied by 1DGE versus 2DGE, respectively. The 2DGE top-down proteomic analysis of rat gastrocnemius versus soleus muscle extracts has clearly shown the differential expression pattern of myosin light chains MLC1s and MLC2s in slow muscle versus MLC1f, MLC2f and MLC3f in fast muscle [114]. Thus, myosin subunits represent reliable proteomic markers of fibre type specification and the expression of the main fast versus slow isoforms correlates well with major histological, physiological and biochemical parameters, such as tissue colour, fibre diameter, contraction time, power output, endurance type, aerobic versus anaerobic activities, degree of resistance to fatigue and capillary density, as well as glycolytic versus oxidative metabolism. During fibre type transitions [123], triggered by graded exercise or chronic neuromuscular stimulation, myosin subunits show characteristic shifts in density and isoform expression patterns [36,124,125]. Detailed adaptations of the acto-myosin apparatus have been examined in a large number of gel-based proteomic studies, which have employed different types of physical activity (interval training, endurance exercise, vibration exercise during long-term bed rest, repeated eccentric exercise, downhill running, bouts of exhaustive exercise) in skeletal muscles from human, pig, rat and mouse [39,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140], as well as chronic low-frequency electro-stimulated rabbit muscle [141,142].
Fast-to-slow transitions are clearly associated with the up-regulation of the slow/cardiac isoform MyHC-I and MLC1s, and a concomitant decrease in MLC1f, MLC2f and MLC3f [141] showing the usefulness of myosin changes for determining molecular adaptations during skeletal muscle transformation [11,36]. The specific effects of myostatin-related hypertrophy [143,144], hypoxia-induced skeletal muscle adaptations [145,146,147,148] and disuse-associated muscular atrophy (due to immobilization, denervation or neuromuscular unloading) [149,150,151,152,153,154,155,156,157,158] could also confirm distinct shifts in sarcomeric proteins due to fibre type transitions. The 2DGE top-down proteomic analysis of skeletal muscle development [159,160,161,162,163] versus muscle aging [164,165,166,167,168,169,170,171,172,173] has revealed interesting alterations in key proteins of the contractile apparatus [174,175]. For example, the gel-based analysis of fast-to-slow fibre transitions during muscle aging demonstrated a drastic increase of slow myosin light chain MLC2 in senescent skeletal muscle tissue [170], establishing this myosin subunit as an excellent marker of age-related fibre type shifting [176]. PTMs, such as altered phosphorylation levels of MLCs, appear to play a crucial role during skeletal muscle adaptations and fibre aging [177,178].
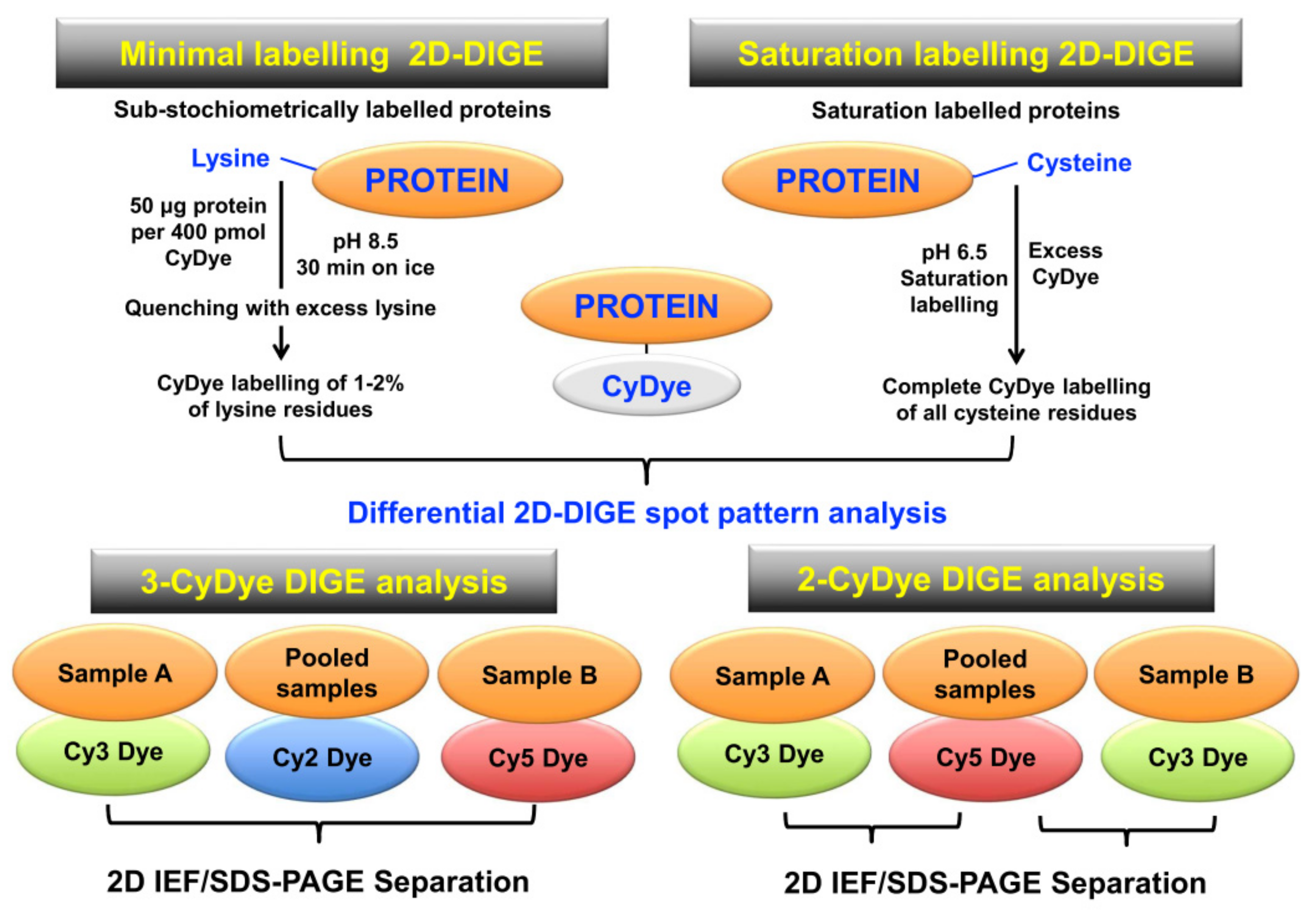
A key 2DGE technique is represented by fluorescence difference in-gel electrophoresis (DIGE) [179,180,181]. Figure 5 compares minimal protein labelling on lysine residues versus saturation protein labelling on cysteine residues prior to gel electrophoretic separation, and also illustrates the different CyDye labeling protocols used in fluorescent two-dye versus three-dye systems [182,183,184].
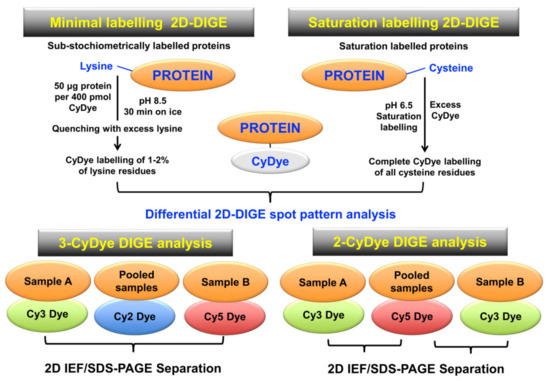
Figure 5.
Overview of fluorescence two-dimensional difference in-gel electrophoresis (2D-DIGE) techniques for the comparative analysis of proteoforms. The upper panel shows the two main DIGE approaches for the systematic pre-electrophoretic labelling of protein fractions using fluorescent CyDyes. These methods utilise minimal sub-stochiometric labelling of assessable lysines in proteins or saturation labelling of assessable cysteines in proteins. The lower panel outlines three-dye (Cy2, Cy3, Cy5) versus two-dye (Cy3, Cy5) DIGE labelling methodology.
The 2D-DIGE method was originally developed by Minden and co-workers [185,186,187] and has been frequently modified as an efficient protein biochemical tool for studying large-scale changes in protein expression and interaction patterns [182,188,189,190]. The application of the 2D-DIGE technique for the comparative analysis of the skeletal muscle proteome from human and animal specimens has been described in detail in various methods papers [191,192,193,194,195]. For the optimum pre-electrophoretic DIGE labelling of protein fractions from tissue extracts, 50 μg protein aliquots can be differentially labelled with Cy2, Cy3 or Cy5 dyes and then separated on the same 2D gel, which eliminates gel-to-gel variations [181]. The development of sophisticated DIGE analysis software programs and improved protein identification approaches has greatly increased the quality of quantitative assessments of multiple proteomes on the same 2D gel [196,197,198].
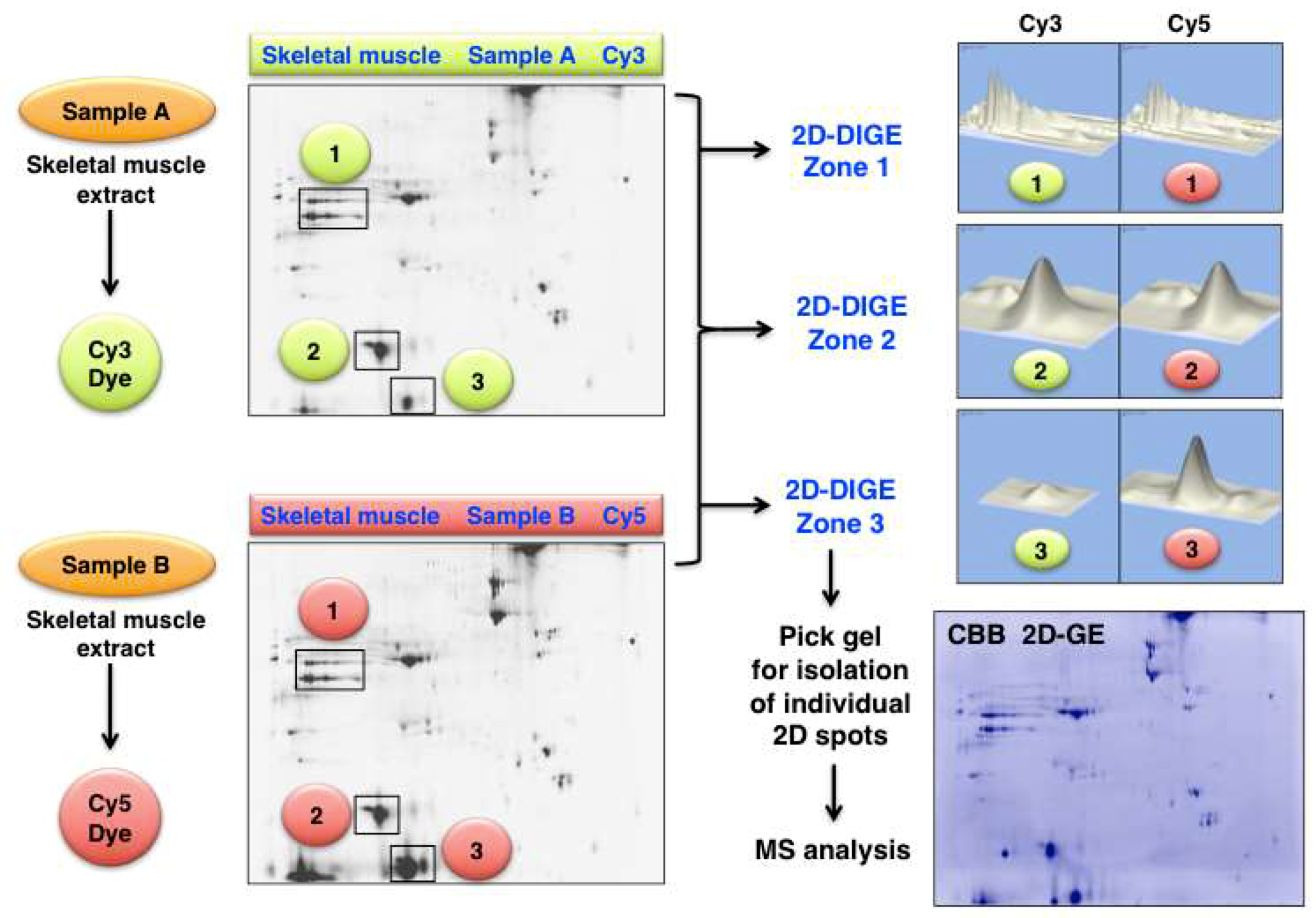
An illustrative example of analysing changes in the skeletal muscle proteome by 2D-DIGE methodology is presented in Figure 6. Shown are 2D gel images from an optimization study of human gastrocnemius muscle. The figure clearly demonstrates the differing expression levels of distinct 2D protein spots in the gel electrophoretically separated Cy3-labelled controls (Sample A) versus physiologically challenged and Cy5-labelled specimens (Sample B). The protein clusters in the marked 2D-DIGE Zones 1 to 3 were densitometrically analysed and show unaltered proteoforms (Zone 1 and 2) versus drastically changed expression levels of distinct proteoforms (Zone 3).
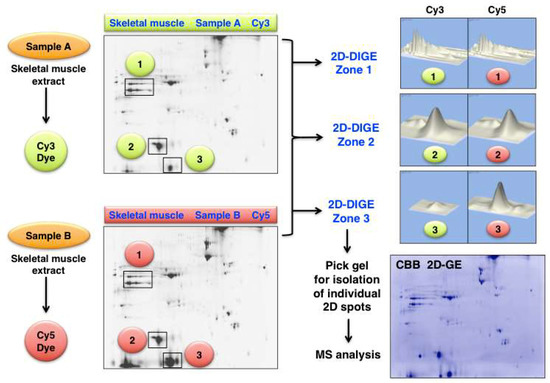
Figure 6.
Fluorescence two-dimensional difference in-gel electrophoresis (2D-DIGE) of human gastrocnemius muscle. The 2DGE images on the left show Cy3 (Sample A) versus Cy5 (Sample B) labelled and separated muscle proteins. The densitometric analysis of the marked 2D-DIGE Zones 1 to 3 is shown on the right and depicts clusters of mostly unaltered proteins (Zone 1 and 2) versus significantly altered expression levels (Zone 3). The routine identification of distinct proteoforms is performed by MS analysis of individual 2D spots from Coomassie Brilliant Blue (CBB) stained pick gels.
3.3. Top-Down Proteomics of Troponins and Tropomyosins
As already described above for isoforms of myosins, the application of 2DGE top-down proteomics has also established regulatory proteins of the actin-containing thin filament as robust markers of fibre type shifting. Fast versus slow fibre specification in gastrocnemius and soleus muscle is clearly reflected by altered expression levels of the fast versus the slow isoform of tropomyosin-1-alpha [114]. During chronic electro-stimulation induced fast-to-slow muscle transitions, an increase in slow troponin TnT and TnI and concomitant decrease in fast troponin TnI were established by fluorescence 2D-DIGE analysis [142]. Troponins and tropomyosins are critical factors of the calcium-associated regulation of the excitation-contraction-relaxation cycle [199,200]. Tropomyosin interacts directly with actin filaments in an inhibitory role and is linked to troponin TnT, while inhibitory troponin TnI is regulated by the calcium-sensing TnC subunit [201].
In mature skeletal muscles, tropomyosin (TM) exists as slow and fast isoform combinations and troponin (TN) can be categorised as fast TnCf, slow TnCs, fast TnT1f to TnT4f, slow TnT1s, slow TnT2s, fast TnIf and slow TnIs [200]. These individual subunits relate in human muscle to slow TnT (P13805; TNNT1 gene), fast TnT (P45378; TNNT3 gene), slow TnI (P19237; TNNI1 gene), fast TnI (P48788; TNNI2 gene), slow TnC (P63316; TNNC1 gene), fast TnC (P02585; TNNC2 gene), TM-alpha-1 (P09493; TPM1 gene), TM-alpha-3 (P06753; TPM3 gene) and TM-alpha-4 (P67936; TPM4 gene). The recent top-down proteomic analysis of tropomyosins in rat, pig and human muscle has established that tropomyosin isoforms Tpm1.1 and Tpm2.2 are the two major tropomyosin isoforms in swine and rat skeletal fibres [202]. In contrast, isoforms Tpm1.1, Tpm2.2, and Tpm3.12 were identified in human muscles. Since the age-related loss of skeletal muscle mass and contractile strength [203] plays an essential role in the frailty syndrome [204,205,206], top-down proteomics is instrumental for studying skeletal muscle aging. Gel-based proteomic surveys of senescent human skeletal muscles and established animal models of sarcopenia strongly suggest that a gradual shift to more oxidative metabolism and a slower mode of fibre contraction occurs during aging [176]. Besides myosin heavy and light chains, the regulatory elements tropomyosin and troponin also showed distinct fast-to-slow changes in proteoform expression levels [164,165,166,168,207]. Decreases in fast TnT and tropomyosin TM-alpha agree with the idea that age-associated abnormalities in the peripheral nervous system trigger denervation, faulty patterns of re-innervation and excitation-contraction-uncoupling, which indirectly affects the skeletal muscle phenotype and causes a gradual transformation to a slower-contracting muscle [176]. The phosphorylation status of the sarcomeric Z-disk component cypher, as well as PTMs in other sarcomeric proteins, was determined to be altered in aged muscle by quantitative top-down proteomics [208].
3.4. Analysis of Post-Translational Modifications Using Two-Dimensional Gel Electrophoresis
There are many PTMs including phosphorylation, glycosylation, acetylation and ubiquitination that are involved in a large variety of processes such as signal transduction pathways, protein trafficking, enzymatic activity or cell motility to name but a few [209]. Determining the precise role of PTMs in physiology and pathophysiology can decisively increase our biochemical understanding and help to take the correct approach when trying to modulate these modifications with respect to therapeutic actions. 2DGE is a highly effective top-down approach to resolve proteoforms, including post-translationally modified proteins. Thousands of proteins can be identified using optimized 2DGE when combined with advanced MS, and potentially many PTMs can be detected. Specific stains have been developed for phosphoproteins (Pro-Q Diamond) and glycoproteins (Pro-Q Emerald) to study specific PTMs using fluorescence 2DGE [210,211,212,213,214]. By using a combination of background-adjusted Pro-Q Diamond staining/total protein ratio values, differential phosphorylation for specific proteins can be detected. Phosphorylated proteins of interest can subsequently be digested, phospho-peptides enriched using titanium dioxide (TiO2) chromatography and specific phosphorylation sites identified by MS analysis. The advantages of separating protein by 2DGE and then identifying augmented phosphorylation signals means that researchers can focus on individual proteins and characterise their PTM status specifically, as opposed to high-throughput LC-MS/MS analysis of enriched peptides that represents a more global approach.
Reversible protein phosphorylation, predominantly involving serine, threonine or tyrosine residues, is one of the most important and intensely investigated PTMs [215]. As an established model system of sarcopenia of old age, 30-month old rat gastrocnemius muscle fibres were evaluated using phospho-specific Pro-Q Diamond staining of 2D gels [216]. Increased phosphorylation levels were shown for various contractile proteins including slow myosin light chain MLC2, tropomyosin alpha and actin, providing evidence that age-related muscle wasting has a complex pathology that is associated with significant changes in the abundance of phosphorylated proteins [177,178,208]. The importance of contractile protein phosphorylation was confirmed by a study on myosin binding protein MyBP-C. Verduyn and co-workers [217] induced differences in the degree of phosphorylation of the myofilaments to explore the role of phosphorylation of the cMyBP-C isoform in isolated rat ventricular myocytes under different physiological conditions, namely phosphorylation of myosin binding protein in quiescent hearts perfused with lidocaine at a relatively low calcium concentration, higher levels of phosphorylation obtained by β-adrenergic stimulation, enhancement of calmodulin-dependent protein kinase CaMK II activity by an increase in the time averaged intracellular calcium concentration, and/or by blockade of phosphatase activity. Using 2DGE and ProQ Diamond staining procedures, the endogenous phosphorylation levels of contractile proteins was determined for troponin cTnT, myosin light chain MLC1 and myosin light chain MLC2. Phosphorylation levels of the contractile proteins varied considerably with the experimental conditions used, with an approximate 2-fold increase in MLC2 phosphorylation in perfused contractile tissues stimulated at 5 Hz. Addition of the beta-agonist isoprenaline to the stimulated hearts resulted in 3.7-fold increases in both cMyBP-C and troponin cTnI phosphorylation [217].
Protein acetylation is involved in the regulation of protein stability and function during various cellular and physiological processes [218,219], including skeletal muscle atrophy [220] and age-related fibre wasting [221]. Acetylation of lysine residues on both histones and non-histone proteins is a reversible, dynamic protein modification regulated by lysine acetyltransferases and deacetylases, controlling a range of different diverse biological functions including protein-protein interactions, protein-DNA interactions, enzymatic activity and subcellular localization. High levels of dihydrolipoyllysine-residue acetyltransferase were detected in the nucleus in aged rat triceps muscle by fluorescence 2D-DIGE analysis and shown to be associated with increased protein acetylation [221]. Acetylation of lysine residues removes the positive charge of the side chain and therefore directly impacts the electrostatic status of the modified protein, allowing for separation associated with isoelectric focusing. Protein pI-shifts due to PTMs revealed that the theoretical pI-value of the non-acetylated form of tropomyosin-beta is close to the pI-value of its experimentally acetylated counterpart, while additional acetylation of tropomyosin (LEKTIDDLEETLASAK + acetyl (K); acetyl (N-term)) does not significantly affect the pI-value [222]. Interestingly, a variety of contractile proteins are changed in chronic Chagas disease, an often fatal outcome of Trypanosoma cruzi infection, which is characterized by severe cardiomyopathy and chronic skeletal muscle myositis and vasculitis [223,224]. Protein changes were evaluated using 2DGE of specimens from end-stage chronic Chagas disease patients to gain insight into its pathophysiology [225]. Several gel spots with differing pI-values and comparable molecular mass were identified by peptide mass fingerprinting as proteoforms of key structural and contractile proteins, including several forms of actin (ACTA1, ACTA2, ACTC, ACTG2, ACTN2), desmin, myosin (MYL3 and MYL7) and vimentin. The variations in their position within 2D slab gels could be a result of PTMs, such as acetylation or other processes.
Glycosylation is one of the most complex PTMs and is involved in many biological mechanisms including cell differentiation, intracellular signalling and protein trafficking [226]. Cieniewski-Bernard and co-workers [227] demonstrated the utility of 2DGE for the analysis of O-linked N-acetylglucosaminylation (O-GlcNAc) in skeletal muscle proteins, focusing on rat gastrocnemius muscle that is composed of both fast- and slow-contracting fibres. O-GlcNAc is an abundant and reversible type of glycosylation and especially found within the cytosolic and the nuclear compartments. The total level of O-GlcNAc proteins in rat after hind-limb unloading, a model of skeletal muscle atrophy, were also evaluated. The O-GlcNAc modification of myosin was found to be significant and verified using immunoblot analysis with an anti-O-GlcNAc antibody. The authors suggested that O-GlcNAc could be involved in the regulation of the polymerization of myosin in the thick filament assembly. The variation in the O-GlcNAc level measured after skeletal muscle unloading suggests a role for dynamic glycosylation patterns in muscle plasticity [227].
Ubiquitination is a key type of reversible PTM in proteins and plays a significant role in the regulation of many biological processes, including protein degradation and signal transduction [228]. Ubiquitin, a highly conserved 76-amino acid protein, can be added to substrate protein as a protein tag by the sequential actions of ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2) and ubiquitin protein ligase (E3). Using fractions of gastrocnemius muscles isolated from Trim32 knockout mice (T32KO) compared to wild type, 36 proteins with altered abundance in T32KO muscle were identified using a combination of fluorescence 2D-DIGE and LC-MS/MS analysis [229]. Mutations in the TRIM32 gene, which encodes an E3 ubiquitin ligase containing the tripartite motif, have been implicated in the pathogenesis of limb girdle muscular dystrophies. Of the significant proteins identified using this model system, the fast muscle isoform of troponin TnT was shown to decrease in abundance in T32KO muscle, a result that supports selective type II (fast) fibre atrophy in T32KO mice.
4. Conclusions
MS-based proteomics has increasingly become the most used method for analysing complex protein populations in biological samples, such as skeletal muscle tissues, facilitated by rapidly advancing hardware and software products. Bioanalytical MS analysis can be employed for experiments focusing on muscle protein profiling, protein quantification, analysis of muscle protein interactions and studying PTMs. Within the field of skeletal muscle biochemistry, there is still a position in proteomics for 2D-gels, both in terms of a standalone technology and as a complementary approach to MS analysis. When used in combination with narrow range pH-gels, 2DGE can resolve more than 5000 distinct protein spots, supplying the end-user with a significant amount of data to interrogate specific biological features of interest, such as muscle fibre type specification or skeletal muscle transitions. The ease of the quantitative analysis of intact muscle proteins using approaches like fluorescence 2D-DIGE makes the comparison of normal versus adapting tissues or healthy versus diseased skeletal muscle samples very achievable. As 2D-gel patterns provide detailed information regarding the position of protein spots on large slab gels (isoelectric point and molecular mass), identifying PTMs that affect their charge and molecular mass is a particular niche application for 2DGE. Compatibility with a variety of proteomic workflows for the systematic identification of skeletal muscle proteins and to characterize their amino acid sequences, and using antibody-based analysis before electrophoresis (such as immuno-affinity purification) or after electrophoretic separation (such as immunoblotting), will ensure that 2DGE remains a relevant laboratory techniques for the next generation of muscle researchers.
Author Contributions
P.D., M.Z., D.S. and K.O. discussed this review article in detail and wrote it collectively.
Funding
Research was supported by grants from Muscular Dystrophy Ireland and the Irish Health Research Board (HRB/MRCG-2016-20) and the Human Health Research Institute of Maynooth University.
Acknowledgments
The authors wish to thank their many national and international colleagues for their continued support of our skeletal muscle proteomics project.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| 1D | one-dimensional |
| 2D | two-dimensional |
| DIGE | difference in-gel electrophoresis |
| GE | gel electrophoresis |
| IEF | isoelectric focusing |
| LC | liquid chromatography |
| MLC | myosin light chain |
| MS | mass spectrometry |
| MyHC | myosin heavy chain |
| PAGE | polyacrylamide gel electrophoresis |
| pI | isoelectric point |
| PTM | post-translational modification |
| TM | tropomyosin |
| TN | troponin |
References
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Mann, M. Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 2010, 190, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Eliuk, S.; Makarov, A. Evolution of Orbitrap Mass Spectrometry Instrumentation. Annu. Rev. Anal. Chem. (Palo Alto Calif) 2015, 8, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.M. Mass Analyzers and Mass Spectrometers. Adv. Exp. Med. Biol. 2016, 919, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Vidova, V.; Spacil, Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal. Chim. Acta 2017, 964, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Lössl, P.; van de Waterbeemd, M.; Heck, A.J. The diverse and expanding role of mass spectrometry in structural and molecular biology. EMBO J. 2016, 35, 2634–2657. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Levy, E.; Slavov, N. Single cell protein analysis for systems biology. Essays Biochem. 2018, 62, 595–605. [Google Scholar] [CrossRef]
- Chen, Z.A.; Rappsilber, J. Protein Dynamics in Solution by Quantitative Crosslinking/Mass Spectrometry. Trends Biochem. Sci. 2018, 43, 908–920. [Google Scholar] [CrossRef]
- Gelfi, C.; Vasso, M.; Cerretelli, P. Diversity of human skeletal muscle in health and disease: Contribution of proteomics. J. Proteomics 2011, 74, 774–795. [Google Scholar] [CrossRef]
- Ohlendieck, K. Skeletal muscle proteomics: Current approaches, technical challenges and emerging techniques. Skelet. Muscle 2011, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Freire, M.; Semba, R.D.; Ubaida-Mohien, C.; Fabbri, E.; Scalzo, P.; Højlund, K.; Dufresne, C.; Lyashkov, A.; Ferrucci, L. The Human Skeletal Muscle Proteome Project: A reappraisal of the current literature. J. Cachexia Sarcopenia Muscle 2017, 8, 5–18. [Google Scholar] [CrossRef]
- Görg, A.; Weiss, W.; Dunn, M.J. Current two-dimensional electrophoresis technology for proteomics. Proteomics 2004, 4, 3665–3685. [Google Scholar] [CrossRef]
- Rabilloud, T.; Chevallet, M.; Luche, S.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteomics 2010, 73, 2064–2077. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.M.; Coorssen, J.R.; Martins-de-Souza, D. 2DE: The phoenix of proteomics. J. Proteomics 2014, 104, 140–150. [Google Scholar] [CrossRef]
- Appel, R.D.; Sanchez, J.C.; Bairoch, A.; Golaz, O.; Miu, M.; Vargas, J.R.; Hochstrasser, D.F. SWISS-2DPAGE: A database of two-dimensional gel electrophoresis images. Electrophoresis 1993, 14, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Celis, J.E.; Gromov, P.; Ostergaard, M.; Madsen, P.; Honoré, B.; Dejgaard, K.; Olsen, E.; Vorum, H.; Kristensen, D.B.; Gromova, I.; et al. Human 2-D PAGE databases for proteome analysis in health and disease: http://biobase.dk/cgi-bin/celis. FEBS Lett. 1996, 398, 129–134. [Google Scholar] [CrossRef]
- Hoogland, C.; Mostaguir, K.; Sanchez, J.C.; Hochstrasser, D.F.; Appel, R.D. SWISS-2DPAGE, ten years later. Proteomics 2004, 4, 2352–2356. [Google Scholar] [CrossRef]
- Lemkin, P.F. The 2DWG meta-database of two-dimensional electrophoretic gel images on the Internet. Electrophoresis 1997, 18, 2759–2773. [Google Scholar] [CrossRef]
- Lemkin, P.F.; Thornwall, G. Flicker image comparison of 2-D gel images for putative protein identification using the 2DWG meta-database. Mol. Biotechnol. 1999, 12, 159–172. [Google Scholar] [CrossRef]
- Babnigg, G.; Giometti, C.S. ProteomeWeb: A web-based interface for the display and interrogation of proteomes. Proteomics 2003, 3, 584–600. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.; Kim, H. The UAB Proteomics Database. Bioinformatics 2003, 19, 2149–2151. [Google Scholar] [CrossRef] [PubMed]
- Babnigg, G.; Giometti, C.S. GELBANK: A database of annotated two-dimensional gel electrophoresis patterns of biological systems with completed genomes. Nucleic Acids Res. 2004, 32, D582–D585. [Google Scholar] [CrossRef][Green Version]
- Drews, O.; Görg, A. DynaProt 2D: An advanced proteomic database for dynamic online access to proteomes and two-dimensional electrophoresis gels. Nucleic Acids Res. 2005, 33, D583–D587. [Google Scholar] [CrossRef] [PubMed]
- Stanislaus, R.; Chen, C.; Franklin, J.; Arthur, J.; Almeida, J.S. AGML Central: Web based gel proteomic infrastructure. Bioinformatics 2005, 21, 1754–1757. [Google Scholar] [CrossRef] [PubMed]
- Pernet, P.; Bruneel, A.; Baudin, B.; Vaubourdolle, M. PHProteomicDB: A module for two-dimensional gel electrophoresis database creation on personal web sites. Genomics Proteomics Bioinform. 2006, 4, 134–136. [Google Scholar] [CrossRef]
- Laukens, K.; Matthiesen, R.; Lemière, F.; Esmans, E.; Onckelen, H.V.; Jensen, O.N.; Witters, E. Integration of gel-based proteome data with pProRep. Bioinformatics 2006, 22, 2838–2840. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hoogland, C.; Mostaguir, K.; Appel, R.D.; Lisacek, F. The World-2DPAGE Constellation to promote and publish gel-based proteomics data through the ExPASy server. J. Proteomics 2008, 71, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.; Ohlendieck, K. Proteomic profiling of large myofibrillar proteins from dried and long-term stored polyacrylamide gels. Anal. Biochem. 2018, 543, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Takemori, N.; Komori, N. Identification of Proteins on Archived 2D Gels. Methods Mol. Biol. 2019, 1855, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Højlund, K.; Yi, Z.; Hwang, H.; Bowen, B.; Lefort, N.; Flynn, C.R.; Langlais, P.; Weintraub, S.T.; Mandarino, L.J. Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol. Cell. Proteomics 2008, 7, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.C.; Walsh, R.J.; Salajegheh, M.; Amato, A.A.; Krastins, B.; Sarracino, D.A.; Greenberg, S.A. Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J. Proteome Res. 2009, 8, 3265–3277. [Google Scholar] [CrossRef] [PubMed]
- Lefort, N.; Yi, Z.; Bowen, B.; Glancy, B.; De Filippis, E.A.; Mapes, R.; Hwang, H.; Flynn, C.R.; Willis, W.T.; Civitarese, A.; et al. Proteome profile of functional mitochondria from human skeletal muscle using one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J. Proteomics 2009, 72, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
- Højlund, K.; Bowen, B.P.; Hwang, H.; Flynn, C.R.; Madireddy, L.; Geetha, T.; Langlais, P.; Meyer, C.; Mandarino, L.J.; Yi, Z. In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC-ESI-MS/MS. J. Proteome Res. 2009, 8, 4954–4965. [Google Scholar] [CrossRef] [PubMed]
- Ohlendieck, K. Proteomic profiling of skeletal muscle plasticity. Muscles Ligaments Tendons J. 2012, 1, 119–126. [Google Scholar] [PubMed]
- Dowling, D.; Murphy, S.; Ohlendieck, K. Proteomic profiling of muscle fibre type shifting in neuromuscular diseases. Expert Rev. Proteomics 2016, 13, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Murgia, M.; Toniolo, L.; Nagaraj, N.; Ciciliot, S.; Vindigni, V.; Schiaffino, S.; Reggiani, C.; Mann, M. Single Muscle Fiber Proteomics Reveals Fiber-Type-Specific Features of Human Muscle Aging. Cell Rep. 2017, 19, 2396–2409. [Google Scholar] [CrossRef]
- Cho, Y.; Ross, R.S. A mini review: Proteomics approaches to understand disused vs. exercised human skeletal muscle. Physiol. Genomics 2018, 50, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Westermeier, R. 2D gel-based Proteomics: there’s life in the old dog yet. Arch. Physiol. Biochem. 2016, 122, 236–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.I.; Cho, J.Y. Gel-based proteomics in disease research: Is it still valuable? Biochim. Biophys. Acta Proteins Proteomics 2019, 1867, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.; Dowling, P.; Ohlendieck, K. Comparative Skeletal Muscle Proteomics Using Two-Dimensional Gel Electrophoresis. Proteomes 2016, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.A.; Chen, B.; Guardado-Alvarez, T.M.; Lin, Z.; Hwang, L.; Ayaz-Guner, S.; Jin, S.; Ge, Y. A photocleavable surfactant for top-down proteomics. Nat. Methods 2019, 16, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Brown, K.A.; Lin, Z.; Ge, Y. Top-Down Proteomics: Ready for Prime Time? Anal. Chem. 2018, 90, 110–127. [Google Scholar] [CrossRef]
- Deshmukhm, A.S.; Murgia, M.; Nagaraj, N.; Treebak, J.T.; Cox, J.; Mann, M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell. Proteomics 2015, 14, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Murgia, M.; Nagaraj, N.; Deshmukh, A.S.; Zeiler, M.; Cancellara, P.; Moretti, I.; Reggiani, C.; Schiaffino, S.; Mann, M. Single muscle fiber proteomics reveals unexpected mitochondrial specialization. EMBO Rep. 2015, 16, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Burniston, J.G.; Connolly, J.; Kainulainen, H.; Britton, S.L.; Koch, L.G. Label-free profiling of skeletal muscle using high-definition mass spectrometry. Proteomics 2014, 14, 2339–2344. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.; Zweyer, M.; Raucamp, M.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Proteomic profiling of the mouse diaphragm and refined mass spectrometric analysis of the dystrophic phenotype. J. Muscle Res. Cell Motil. 2019, 40, 9–28. [Google Scholar] [CrossRef]
- Patrie, S.M. Top-Down Mass Spectrometry: Proteomics to Proteoforms. Adv. Exp. Med. Biol. 2016, 919, 171–200. [Google Scholar] [CrossRef]
- Toby, T.K.; Fornelli, L.; Kelleher, N.L. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. (Palo Alto Calif) 2016, 9, 499–519. [Google Scholar] [CrossRef]
- Tholey, A.; Becker, A. Top-down proteomics for the analysis of proteolytic events - Methods, applications and perspectives. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Gregorich, Z.R.; Ge, Y. Top-down proteomics in health and disease: Challenges and opportunities. Proteomics 2014, 14, 1195–1210. [Google Scholar] [CrossRef]
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top Down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Tucholski, T.; Chen, B.; Alpert, A.J.; McIlwain, S.; Kohmoto, T.; Jin, S.; Ge, Y. Top-Down Proteomics of Large Proteins up to 223 kDa Enabled by Serial Size Exclusion Chromatography Strategy. Anal. Chem. 2017, 89, 5467–5475. [Google Scholar] [CrossRef] [PubMed]
- Boeri Erba, E. Investigating macromolecular complexes using top-down mass spectrometry. Proteomics 2014, 14, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Kar, U.K.; Simonian, M.; Whitelegge, J.P. Integral membrane proteins: Bottom-up, top-down and structural proteomics. Expert Rev. Proteomics 2017, 14, 715–723. [Google Scholar] [CrossRef]
- Li, H.; Nguyen, H.H.; Ogorzalek Loo, R.R.; Campuzano, I.D.G.; Loo, J.A. An Integrated Native Mass Spectrometry and Top-Down Proteomics Method that Connects Sequence to Structure and Function of Macromolecular Complexes. Nat. Chem. 2018, 10, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.; Murphy, S.; Dowling, P.; Ohlendieck, K. Pathoproteomic profiling of the skeletal muscle matrisome in dystrophinopathy associated myofibrosis. Proteomics 2016, 16, 345–366. [Google Scholar] [CrossRef]
- Carr, S.J.; Zahedi, R.P.; Lochmüller, H.; Roos, A. Mass spectrometry-based protein analysis to unravel the tissue pathophysiology in Duchenne muscular dystrophy. Proteomics Clin. Appl. 2018, 12. [Google Scholar] [CrossRef]
- Kruse, R.; Højlund, K. Proteomic study of skeletal muscle in obesity and type 2 diabetes: Progress and potential. Expert Rev. Proteomics 2018, 15, 817–828. [Google Scholar] [CrossRef]
- Nishikawa, K.C.; Monroy, J.A.; Tahir, U. Muscle Function from Organisms to Molecules. Integr. Comp. Biol. 2018, 58, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.C.; Wright, N.T. Structure of giant muscle proteins. Front. Physiol. 2013, 4, 368. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.; Dowling, P.; Zweyer, M.; Swandulla, D.; Ohlendieck, K. Proteomic profiling of giant skeletal muscle proteins. Expert Rev. Proteomics 2019, 16, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Blaauw, B.; Schiaffino, S.; Reggiani, C. Mechanisms modulating skeletal muscle phenotype. Compr. Physiol. 2013, 3, 1645–1687. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Holland, A.; Ohlendieck, K. Proteomic profiling of the contractile apparatus from skeletal muscle. Expert Rev. Proteomics 2013, 10, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Ntai, I.; Toby, T.K.; LeDuc, R.D.; Kelleher, N.L. A Method for Label-Free, Differential Top-Down Proteomics. Methods Mol. Biol. 2016, 1410, 121–133. [Google Scholar] [CrossRef]
- Wright, E.P.; Prasad, K.A.; Padula, M.P.; Coorssen, J.R. Deep imaging: How much of the proteome does current top-down technology already resolve? PLoS ONE 2014, 9, e86058. [Google Scholar] [CrossRef]
- Wright, E.P.; Partridge, M.A.; Padula, M.P.; Gauci, V.J.; Malladi, C.S.; Coorssen, J.R. Top-down proteomics: Enhancing 2D gel electrophoresis from tissue processing to high-sensitivity protein detection. Proteomics 2014, 14, 872–889. [Google Scholar] [CrossRef]
- Padula, M.P.; Berry, I.J.; O Rourke, M.B.; Raymond, B.B.; Santos, J.; Djordjevic, S.P. A Comprehensive Guide for Performing Sample Preparation and Top-Down Protein Analysis. Proteomes 2017, 5, 11. [Google Scholar] [CrossRef]
- Noaman, N.; Abbineni, P.S.; Withers, M.; Coorssen, J.R. Coomassie staining provides routine (sub) femtomole in-gel detection of intact proteoforms: Expanding opportunities for genuine Top-down Proteomics. Electrophoresis. 2017, 38, 3086–3099. [Google Scholar] [CrossRef] [PubMed]
- Kachuk, C.; Doucette, A.A. The benefits (and misfortunes) of SDS in top-down proteomics. J. Proteomics 2018, 175, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.C.; Zamdborg, L.; Ahlf, D.R.; Lee, J.E.; Catherman, A.D.; Durbin, K.R.; Tipton, J.D.; Vellaichamy, A.; Kellie, J.F.; Li, M.; et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 2011, 480, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Moradian, A.; Kalli, A.; Sweredoski, M.J.; Hess, S. The top-down, middle-down, and bottom-up mass spectrometry approaches for characterization of histone variants and their post-translational modifications. Proteomics 2014, 14, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.; Crawford, J.; Gianazza, E. Protein stains for proteomic applications: Which, when, why? Proteomics 2006, 6, 5385–5408. [Google Scholar] [CrossRef] [PubMed]
- Riederer, B.M. Non-covalent and covalent protein labeling in two-dimensional gel electrophoresis. J. Proteomics 2008, 71, 231–244. [Google Scholar] [CrossRef]
- Weiss, W.; Weiland, F.; Görg, A. Protein detection and quantitation technologies for gel-based proteome analysis. Methods Mol. Biol. 2009, 564, 59–82. [Google Scholar] [CrossRef]
- Gauci, V.J.; Wright, E.P.; Coorssen, J.R. Quantitative proteomics: Assessing the spectrum of in-gel protein detection methods. J. Chem. Biol. 2011, 4, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Panfoli, I.; Calzia, D.; Santucci, L.; Ravera, S.; Bruschi, M.; Candiano, G. A blue dive: From ‘blue fingers’ to ‘blue silver’. A comparative overview of staining methods for in-gel proteomics. Expert Rev. Proteomics 2012, 9, 627–634. [Google Scholar] [CrossRef]
- Butt, R.H.; Coorssen, J.R. Coomassie blue as a near-infrared fluorescent stain: A systematic comparison with Sypro Ruby for in-gel protein detection. Mol. Cell. Proteomics 2013, 12, 3834–3850. [Google Scholar] [CrossRef]
- Millman, B.M. The filament lattice of striated muscle. Physiol. Rev. 1998, 78, 359–391. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.M.; Homsher, E.; Regnier, M. Regulation of contraction in striated muscle. Physiol. Rev. 2000, 80, 853–924. [Google Scholar] [CrossRef] [PubMed]
- Lehman, W. Thin Filament Structure and the Steric Blocking Model. Compr. Physiol. 2016, 6, 1043–1069. [Google Scholar] [CrossRef] [PubMed]
- Irving, M. Regulation of Contraction by the Thick Filaments in Skeletal Muscle. Biophys. J. 2017, 113, 2579–2594. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Geist, J.; Grogan, A.; Hu, L.R.; Kontrogianni-Konstantopoulos, A. Thick Filament Protein Network, Functions, and Disease Association. Compr. Physiol. 2018, 8, 631–709. [Google Scholar] [CrossRef] [PubMed]
- Krüger, M.; Kötter, S. Titin, a Central Mediator for Hypertrophic Signaling, Exercise-Induced Mechanosignaling and Skeletal Muscle Remodeling. Front. Physiol. 2016, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Gregorio, C.C.; Pappas, C.T. Nebulin, a multi-functional giant. J. Exp. Biol. 2016, 219, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Tskhovrebova, L.; Trinick, J. Titin and Nebulin in Thick and Thin Filament Length Regulation. Subcell. Biochem. 2017, 82, 285–318. [Google Scholar] [CrossRef]
- Gautel, M.; Djinović-Carugo, K. The sarcomeric cytoskeleton: From molecules to motion. J. Exp. Biol. 2016, 219, 135–145. [Google Scholar] [CrossRef]
- Holmes, K.C. The swinging lever-arm hypothesis of muscle contraction. Curr. Biol. 1997, 7, R112–R118. [Google Scholar] [CrossRef]
- Guhathakurta, P.; Prochniewicz, E.; Thomas, D.D. Actin-Myosin Interaction: Structure, Function and Drug Discovery. Int. J. Mol. Sci. 2018, 19, 628. [Google Scholar] [CrossRef] [PubMed]
- Pette, D.; Staron, R.S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 2000, 50, 500–509. [Google Scholar] [CrossRef]
- Walklate, J.; Ujfalusi, Z.; Geeves, M.A. Myosin isoforms and the mechanochemical cross-bridge cycle. J. Exp. Biol. 2016, 219, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Katayama, E.; Kodera, N. Unconventional Imaging Methods to Capture Transient Structures during Actomyosin Interaction. Int. J. Mol. Sci. 2018, 19, 1402. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.L.; Song, T.; Sadayappan, S. Myofilaments: Movers and Rulers of the Sarcomere. Compr. Physiol. 2017, 7, 675–692. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.C.; Chiappe, D.; Converset, V.; Hoogland, C.; Binz, P.A.; Paesano, S.; Appel, R.D.; Wang, S.; Sennitt, M.; Nolan, A.; et al. The mouse SWISS-2D PAGE database: A tool for proteomics study of diabetes and obesity. Proteomics 2001, 1, 136–163. [Google Scholar] [CrossRef]
- Yan, J.X.; Harry, R.A.; Wait, R.; Welson, S.Y.; Emery, P.W.; Preedy, V.R.; Dunn, M.J. Separation and identification of rat skeletal muscle proteins using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 2001, 1, 424–434. [Google Scholar] [CrossRef]
- Isfort, R.J. Proteomic analysis of striated muscle. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 771, 155–165. [Google Scholar] [CrossRef]
- Li, Z.B.; Lehar, M.; Braga, N.; Westra, W.; Liu, L.H.; Flint, P.W. Study of human laryngeal muscle protein using two-dimensional electrophoresis and mass spectrometry. Proteomics 2003, 3, 1325–1334. [Google Scholar] [CrossRef]
- Gelfi, C.; De Palma, S.; Cerretelli, P.; Begum, S.; Wait, R. Two-dimensional protein map of human vastus lateralis muscle. Electrophoresis 2003, 24, 286–295. [Google Scholar] [CrossRef]
- Capitanio, D.; Viganò, A.; Ricci, E.; Cerretelli, P.; Wait, R.; Gelfi, C. Comparison of protein expression in human deltoideus and vastus lateralis muscles using two-dimensional gel electrophoresis. Proteomics 2005, 5, 2577–2586. [Google Scholar] [CrossRef]
- Kovalyova, M.A.; Kovalyov, L.I.; Toropygin, I.Y.; Shigeev, S.V.; Ivanov, A.V.; Shishkin, S.S. Proteomic analysis of human skeletal muscle (m. vastus lateralis) proteins: Identification of 89 gene expression products. Biochemistry (Moscow) 2009, 74, 1239–1252. [Google Scholar] [CrossRef] [PubMed]
- Bouley, J.; Chambon, C.; Picard, B. Mapping of bovine skeletal muscle proteins using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 2004, 4, 1811–1824. [Google Scholar] [CrossRef] [PubMed]
- Chaze, T.; Bouley, J.; Chambon, C.; Barboiron, C.; Picard, B. Mapping of alkaline proteins in bovine skeletal muscle. Proteomics 2006, 6, 2571–2575. [Google Scholar] [CrossRef] [PubMed]
- Chiu, K.H.; Huang, H.W.; Mok, H.K. Differential proteome analysis of hagfish dental and somatic skeletal muscles. Mar. Biotechnol. (N.Y.) 2007, 9, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Raddatz, K.; Albrecht, D.; Hochgräfe, F.; Hecker, M.; Gotthardt, M. A proteome map of murine heart and skeletal muscle. Proteomics 2008, 8, 1885–1897. [Google Scholar] [CrossRef]
- Almeida, A.M.; Campos, A.; van Harten, S.; Cardoso, L.A.; Coelho, A.V. Establishment of a proteomic reference map for the gastrocnemius muscle in the rabbit (Oryctolagus cuniculus). Res. Vet. Sci. 2009, 87, 196–199. [Google Scholar] [CrossRef]
- Addis, M.F.; Tanca, A.; Pagnozzi, D.; Rocca, S.; Uzzau, S. 2-D PAGE and MS analysis of proteins from formalin-fixed, paraffin-embedded tissues. Proteomics 2009, 9, 4329–4339. [Google Scholar] [CrossRef]
- Lu, J.; Zheng, J.; Liu, H.; Li, J.; Chen, H.; Chen, K. Protein profiling analysis of skeletal muscle of a pufferfish, Takifugu rubripes. Mol. Biol. Rep. 2010, 37, 2141–2147. [Google Scholar] [CrossRef]
- Abbaraju, N.V.; Boutaghou, M.N.; Townley, I.K.; Zhang, Q.; Wang, G.; Cole, R.B.; Rees, B.B. Analysis of tissue proteomes of the Gulf killifish, Fundulus grandis, by 2D electrophoresis and MALDI-TOF/TOF mass spectrometry. Integr. Comp. Biol. 2012, 52, 626–635. [Google Scholar] [CrossRef][Green Version]
- Kim, N.K.; Joh, J.H.; Park, H.R.; Kim, O.H.; Park, B.Y.; Lee, C.S. Differential expression profiling of the proteomes and their mRNAs in porcine white and red skeletal muscles. Proteomics 2004, 4, 3422–3428. [Google Scholar] [CrossRef] [PubMed]
- Okumura, N.; Hashida-Okumura, A.; Kita, K.; Matsubae, M.; Matsubara, T.; Takao, T.; Nagai, K. Proteomic analysis of slow- and fast-twitch skeletal muscles. Proteomics 2005, 5, 2896–2906. [Google Scholar] [CrossRef] [PubMed]
- Le Bihan, M.C.; Hou, Y.; Harris, N.; Tarelli, E.; Coulton, G.R. Proteomic analysis of fast and slow muscles from normal and kyphoscoliotic mice using protein arrays, 2-DE and MS. Proteomics 2006, 6, 4646–4661. [Google Scholar] [CrossRef] [PubMed]
- Gelfi, C.; Viganò, A.; De Palma, S.; Ripamonti, M.; Begum, S.; Cerretelli, P.; Wait, R. 2-D protein maps of rat gastrocnemius and soleus muscles: A tool for muscle plasticity assessment. Proteomics 2006, 6, 321–340. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R.; Ferreira, R.; Neuparth, M.; Guedes, S.; Williams, J.; Tomer, K.B.; Domingues, P.M.; Appell, H.J.; Duarte, J.A.; Amado, F.M. Subcellular proteomics of mice gastrocnemius and soleus muscles. Anal. Biochem. 2007, 366, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Drexler, H.C.; Ruhs, A.; Konzer, A.; Mendler, L.; Bruckskotten, M.; Looso, M.; Günther, S.; Boettger, T.; Krüger, M.; Braun, T. On marathons and sprints: An integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers. Mol. Cell. Proteomics 2012, 11, M111.010801. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S. Fibre types in skeletal muscle: A personal account. Acta Physiol. (Oxf.) 2010, 199, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S. Muscle fiber type diversity revealed by anti-myosin heavy chain antibodies. FEBS J. 2018, 285, 3688–3694. [Google Scholar] [CrossRef]
- Reiser, P.J. Current understanding of conventional and novel co-expression patterns of mammalian sarcomeric myosin heavy chains and light chains. Arch. Biochem. Biophys. 2019, 662, 129–133. [Google Scholar] [CrossRef]
- Lee, L.A.; Karabina, A.; Broadwell, L.J.; Leinwand, L.A. The ancient sarcomeric myosins found in specialized muscles. Skelet. Muscle 2019, 9, 7. [Google Scholar] [CrossRef]
- Vandenboom, R. Modulation of Skeletal Muscle Contraction by Myosin Phosphorylation. Compr. Physiol. 2016, 7, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Logvinova, D.S.; Levitsky, D.I. Essential Light Chains of Myosin and Their Role in Functioning of the Myosin Motor. Biochemistry (Moscow). 2018, 83, 944–960. [Google Scholar] [CrossRef] [PubMed]
- Olsen, L.A.; Nicoll, J.X.; Fry, A.C. The skeletal muscle fiber: A mechanically sensitive cell. Eur. J. Appl. Physiol. 2019, 119, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Padrão, A.I.; Ferreira, R.; Amado, F.; Vitorino, R.; Duarte, J.A. Uncovering the exercise-related proteome signature in skeletal muscle. Proteomics 2016, 16, 816–830. [Google Scholar] [CrossRef] [PubMed]
- Petriz, B.A.; Gomes, C.P.; Almeida, J.A.; de Oliveira, G.P., Jr.; Ribeiro, F.M.; Pereira, R.W.; Franco, O.L. The Effects of Acute and Chronic Exercise on Skeletal Muscle Proteome. J. Cell. Physiol. 2017, 232, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Holloway, K.V.; O’Gorman, M.; Woods, P.; Morton, J.P.; Evans, L.; Cable, N.T.; Goldspink, D.F.; Burniston, J.G. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics 2009, 9, 5155–5174. [Google Scholar] [CrossRef] [PubMed]
- Egan, B.; Dowling, P.; O’Connor, P.L.; Henry, M.; Meleady, P.; Zierath, J.R.; O’Gorman, D.J. 2-D DIGE analysis of the mitochondrial proteome from human skeletal muscle reveals time course-dependent remodelling in response to 14 consecutive days of endurance exercise training. Proteomics 2011, 11, 1413–1428. [Google Scholar] [CrossRef]
- Moriggi, M.; Vasso, M.; Fania, C.; Capitanio, D.; Bonifacio, G.; Salanova, M.; Blottner, D.; Rittweger, J.; Felsenberg, D.; Cerretelli, P.; et al. Long term bed rest with and without vibration exercise countermeasures: Effects on human muscle protein dysregulation. Proteomics 2010, 10, 3756–3774. [Google Scholar] [CrossRef]
- Salanova, M.; Gelfi, C.; Moriggi, M.; Vasso, M.; Viganò, A.; Minafra, L.; Bonifacio, G.; Schiffl, G.; Gutsmann, M.; Felsenberg, D.; et al. Disuse deterioration of human skeletal muscle challenged by resistive exercise superimposed with vibration: Evidence from structural and proteomic analysis. FASEB J. 2014, 28, 4748–4763. [Google Scholar] [CrossRef]
- Hody, S.; Leprince, P.; Sergeant, K.; Renaut, J.; Croisier, J.L.; Wang, F.; Rogister, B. Human muscle proteome modifications after acute or repeated eccentric exercises. Med. Sci. Sports Exerc. 2011, 43, 2281–2296. [Google Scholar] [CrossRef]
- Malm, C.; Yu, J.G. Exercise-induced muscle damage and inflammation: Re-evaluation by proteomics. Histochem. Cell Biol. 2012, 138, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Burniston, J.G. Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochim. Biophys. Acta 2008, 1784, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Guelfi, K.J.; Casey, T.M.; Giles, J.J.; Fournier, P.A.; Arthur, P.G. A proteomic analysis of the acute effects of high-intensity exercise on skeletal muscle proteins in fasted rats. Clin. Exp. Pharmacol. Physiol. 2006, 33, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, W.; Fujimoto, E.; Higuchi, M.; Tabata, I. A DIGE proteomic analysis for high-intensity exercise-trained rat skeletal muscle. J. Biochem. 2010, 148, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Gandra, P.G.; Valente, R.H.; Perales, J.; Pacheco, A.G.; Macedo, D.V. Proteomic profiling of skeletal muscle in an animal model of overtraining. Proteomics 2012, 12, 2663–2667. [Google Scholar] [CrossRef]
- Gandra, P.G.; Valente, R.H.; Perales, J.; Pacheco, A.G.; Macedo, D.V. Proteomic analysis of rat skeletal muscle submitted to one bout of incremental exercise. Scand. J. Med. Sci. Sports 2012, 22, 207–216. [Google Scholar] [CrossRef]
- Magherini, F.; Abruzzo, P.M.; Puglia, M.; Bini, L.; Gamberi, T.; Esposito, F.; Veicsteinas, A.; Marini, M.; Fiorillo, C.; Gulisano, M.; et al. Proteomic analysis and protein carbonylation profile in trained and untrained rat muscles. J. Proteomics 2012, 75, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, F.G.; van Ginneken, M.M.; Noben, J.P.; Royackers, E.; de Graaf-Roelfsema, E.; Wijnberg, I.D.; van der Kolk, J.H.; Mariman, E.C.; van Breda, E. Differential expression of equine muscle biopsy proteins during normal training and intensified training in young standardbred horses using proteomics technology. Comp. Biochem. Physiol. Part D Genome Proteomics 2010, 5, 55–64. [Google Scholar] [CrossRef]
- Macedo, A.; Moriggi, M.; Vasso, M.; De Palma, S.; Sturnega, M.; Friso, G.; Gelfi, C.; Giacca, M.; Zacchigna, S. Enhanced athletic performance on multisite AAV-IGF1 gene transfer coincides with massive modification of the muscle proteome. Hum. Gene Ther. 2012, 23, 146–157. [Google Scholar] [CrossRef]
- Burniston, J.G.; Kenyani, J.; Gray, D.; Guadagnin, E.; Jarman, I.H.; Cobley, J.N.; Cuthbertson, D.J.; Chen, Y.W.; Wastling, J.M.; Lisboa, P.J.; et al. Conditional independence mapping of DIGE data reveals PDIA3 protein species as key nodes associated with muscle aerobic capacity. J. Proteomics 2014, 106, 230–245. [Google Scholar] [CrossRef]
- Donoghue, P.; Doran, P.; Dowling, P.; Ohlendieck, K. Differential expression of the fast skeletal muscle proteome following chronic low-frequency stimulation. Biochim. Biophys. Acta 2005, 1752, 166–176. [Google Scholar] [CrossRef]
- Donoghue, P.; Doran, P.; Wynne, K.; Pedersen, K.; Dunn, M.J.; Ohlendieck, K. Proteomic profiling of chronic low-frequency stimulated fast muscle. Proteomics 2007, 7, 3417–3430. [Google Scholar] [CrossRef] [PubMed]
- Bouley, J.; Meunier, B.; Chambon, C.; De Smet, S.; Hocquette, J.F.; Picard, B. Proteomic analysis of bovine skeletal muscle hypertrophy. Proteomics 2005, 5, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Keady, S.M.; Kenny, D.A.; Ohlendieck, K.; Doyle, S.; Keane, M.G.; Waters, S.M. Proteomic profiling of bovine M. longissimus lumborum from Crossbred Aberdeen Angus and Belgian Blue sired steers varying in genetic merit for carcass weight. J. Anim. Sci. 2013, 91, 654–665. [Google Scholar] [CrossRef]
- Bosworth, C.A.; Chou, C.W.; Cole, R.B.; Rees, B.B. Protein expression patterns in zebrafish skeletal muscle: Initial characterization and the effects of hypoxic exposure. Proteomics 2005, 5, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- De Palma, S.; Ripamonti, M.; Vigano, A.; Moriggi, M.; Capitanio, D.; Samaja, M.; Milano, G.; Cerretelli, P.; Wait, R.; Gelfi, C. Metabolic modulation induced by chronic hypoxia in rats using a comparative proteomic analysis of skeletal muscle tissue. J. Proteome Res. 2007, 6, 1974–1984. [Google Scholar] [CrossRef]
- Vigano, A.; Ripamonti, M.; De Palma, S.; Capitanio, D.; Vasso, M.; Wait, R.; Lundby, C.; Cerretelli, P.; Gelfi, C. Proteins modulation in human skeletal muscle in the early phase of adaptation to hypobaric hypoxia. Proteomics 2008, 8, 4668–4679. [Google Scholar] [CrossRef] [PubMed]
- Levett, D.Z.; Viganò, A.; Capitanio, D.; Vasso, M.; De Palma, S.; Moriggi, M.; Martin, D.S.; Murray, A.J.; Cerretelli, P.; Grocott, M.P.; et al. Changes in muscle proteomics in the course of the Caudwell Research Expedition to Mt. Everest. Proteomics 2015, 15, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Isfort, R.J.; Hinkle, R.T.; Jones, M.B.; Wang, F.; Greis, K.D.; Sun, Y.; Keough, T.W.; Anderson, N.L.; Sheldon, R.J. Proteomic analysis of the atrophying rat soleus muscle following denervation. Electrophoresis 2000, 21, 2228–2234. [Google Scholar] [CrossRef]
- Isfort, R.J.; Wang, F.; Greis, K.D.; Sun, Y.; Keough, T.W.; Farrar, R.P.; Bodine, S.C.; Anderson, N.L. Proteomic analysis of rat soleus muscle undergoing hindlimb suspension-induced atrophy and reweighting hypertrophy. Proteomics 2002, 2, 543–550. [Google Scholar] [CrossRef]
- Isfort, R.J.; Wang, F.; Greis, K.D.; Sun, Y.; Keough, T.W.; Bodine, S.C.; Anderson, N.L. Proteomic analysis of rat soleus and tibialis anterior muscle following immobilization. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2002, 769, 323–332. [Google Scholar] [CrossRef]
- Seo, Y.; Lee, K.; Park, K.; Bae, K.; Choi, I. A proteomic assessment of muscle contractile alterations during unloading and reloading. J. Biochem. 2006, 139, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Moriggi, M.; Cassano, P.; Vasso, M.; Capitanio, D.; Fania, C.; Musicco, C.; Pesce, V.; Gadaleta, M.N.; Gelfi, C. A DIGE approach for the assessment of rat soleus muscle changes during unloading: Effect of acetyl-L-carnitine supplementation. Proteomics 2008, 8, 3588–3604. [Google Scholar] [CrossRef]
- Ferreira, R.; Vitorino, R.; Neuparth, M.J.; Appell, H.J.; Duarte, J.A.; Amado, F. Proteolysis activation and proteome alterations in murine skeletal muscle submitted to 1 week of hindlimb suspension. Eur. J. Appl. Physiol. 2009, 107, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Basco, D.; Nicchia, G.P.; Desaphy, J.F.; Camerino, D.C.; Frigeri, A.; Svelto, M. Analysis by two-dimensional Blue Native/SDS-PAGE of membrane protein alterations in rat soleus muscle after hindlimb unloading. Eur. J. Appl. Physiol. 2010, 110, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, P.; Liu, H.; Fan, M.; Chen, X. Proteomic analysis of mouse soleus muscles affected by hindlimb unloading and reloading. Muscle Nerve 2015, 52, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.B.; Lehar, M.; Samlan, R.; Flint, P.W. Proteomic analysis of rat laryngeal muscle following denervation. Proteomics 2005, 5, 4764–4776. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Shimizu, M.; Mizunoya, W.; Wariishi, H.; Tatsumi, R.; Buchman, V.L.; Ikeuchi, Y. Differential expression of sarcoplasmic and myofibrillar proteins of rat soleus muscle during denervation atrophy. Biosci. Biotechnol. Biochem. 2009, 73, 1748–1756. [Google Scholar] [CrossRef] [PubMed]
- Tannu, N.S.; Rao, V.K.; Chaudhary, R.M.; Giorgianni, F.; Saeed, A.E.; Gao, Y.; Raghow, R. Comparative proteomes of the proliferating C(2)C(12) myoblasts and fully differentiated myotubes reveal the complexity of the skeletal muscle differentiation program. Mol. Cell. Proteomics 2004, 3, 1065–1082. [Google Scholar] [CrossRef] [PubMed]
- Casadei, L.; Vallorani, L.; Gioacchini, A.M.; Guescini, M.; Burattini, S.; D’Emilio, A.; Biagiotti, L.; Falcieri, E.; Stocchi, V. Proteomics-based investigation in C2C12 myoblast differentiation. Eur. J. Histochem. 2009, 53, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, T.; Ding, F.; Hu, N.; Gu, X. Proteomic studies of rat tibialis anterior muscle during postnatal growth and development. Mol. Cell. Biochem. 2009, 332, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qian, H.; Feng, X.; Xiong, Y.; Lei, M.; Ren, Z.; Zuo, B.; Xu, D.; Ma, Y.; Yuan, H. Differential proteome and transcriptome analysis of porcine skeletal muscle during development. J. Proteomics 2012, 75, 2093–2108. [Google Scholar] [CrossRef] [PubMed]
- Kanski, J.; Alterman, M.A.; Schöneich, C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radic. Biol. Med. 2003, 35, 1229–1239. [Google Scholar] [CrossRef]
- Piec, I.; Listrat, A.; Alliot, J.; Chambon, C.; Taylor, R.G.; Bechet, D. Differential proteome analysis of aging in rat skeletal muscle. FASEB J. 2005, 19, 1143–1145. [Google Scholar] [CrossRef] [PubMed]
- Gelfi, C.; Vigano, A.; Ripamonti, M.; Pontoglio, A.; Begum, S.; Pellegrino, M.A.; Grassi, B.; Bottinelli, R.; Wait, R.; Cerretelli, P. The human muscle proteome in aging. J. Proteome Res. 2006, 5, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, K.; Gannon, J.; Doran, P.; Ohlendieck, K. Proteomic profiling reveals a severely perturbed protein expression pattern in aged skeletal muscle. Int. J. Mol. Med. 2007, 20, 145–153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doran, P.; Gannon, J.; O’Connell, K.; Ohlendieck, K. Aging skeletal muscle shows a drastic increase in the small heat shock proteins alphaB-crystallin/HspB5 and cvHsp/HspB7. Eur. J. Cell Biol. 2007, 86, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Doran, P.; O’Connell, K.; Gannon, J.; Kavanagh, M.; Ohlendieck, K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics 2008, 8, 364–377. [Google Scholar] [CrossRef]
- Gannon, J.; Ohlendieck, K. Subproteomic analysis of basic proteins in aged skeletal muscle following offgel pre-fractionation. Mol. Med. Rep. 2012, 5, 993–1000. [Google Scholar] [CrossRef]
- Gannon, J.; Doran, P.; Kirwan, A.; Ohlendieck, K. Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur. J. Cell Biol. 2009, 88, 685–700. [Google Scholar] [CrossRef]
- Staunton, L.; Zweyer, M.; Swandulla, D.; Ohlendieck, K. Mass spectrometry-based proteomic analysis of middle-aged vs. aged vastus lateralis reveals increased levels of carbonic anhydrase isoform 3 in senescent human skeletal muscle. Int. J. Mol. Med. 2012, 30, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Gueugneau, M.; Coudy-Gandilhon, C.; Gourbeyre, O.; Chambon, C.; Combaret, L.; Polge, C.; Taillandier, D.; Attaix, D.; Friguet, B.; Maier, A.B.; et al. Proteomics of muscle chronological ageing in post-menopausal women. BMC Genomics 2014, 15, 1165. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, D.; Vasso, M.; Fania, C.; Moriggi, M.; Viganò, A.; Procacci, P.; Magnaghi, V.; Gelfi, C. Comparative proteomic profile of rat sciatic nerve and gastrocnemius muscle tissues in ageing by 2-D DIGE. Proteomics 2009, 9, 2004–2020. [Google Scholar] [CrossRef] [PubMed]
- Doran, P.; Donoghue, P.; O’Connell, K.; Gannon, J.; Ohlendieck, K. Proteomics of skeletal muscle aging. Proteomics 2009, 9, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Baraibar, M.A.; Gueugneau, M.; Duguez, S.; Butler-Browne, G.; Bechet, D.; Friguet, B. Expression and modification proteomics during skeletal muscle ageing. Biogerontology 2013, 14, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Ohlendieck, K. Proteomic Profiling of Fast-To-Slow Muscle Transitions during Aging. Front. Physiol. 2011, 2, 105. [Google Scholar] [CrossRef] [PubMed]
- Gregorich, Z.R.; Peng, Y.; Cai, W.; Jin, Y.; Wei, L.; Chen, A.J.; McKiernan, S.H.; Aiken, J.M.; Moss, R.L.; Diffee, G.M.; et al. Top-Down Targeted Proteomics Reveals Decrease in Myosin Regulatory Light-Chain Phosphorylation That Contributes to Sarcopenic Muscle Dysfunction. J. Proteome Res. 2016, 15, 2706–2716. [Google Scholar] [CrossRef]
- Jin, Y.; Diffee, G.M.; Colman, R.J.; Anderson, R.M.; Ge, Y. Top-down Mass Spectrometry of Sarcomeric Protein Post-translational Modifications from Non-human Primate Skeletal Muscle. J. Am. Soc. Mass Spectrom. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Timms, J.F.; Cramer, R. Difference gel electrophoresis. Proteomics 2008, 8, 4886–4897. [Google Scholar] [CrossRef]
- Arentz, G.; Weiland, F.; Oehler, M.K.; Hoffmann, P. State of the art of 2D DIGE. Proteomics Clin. Appl. 2015, 9, 277–288. [Google Scholar] [CrossRef]
- Ohlendieck, K. Comparative DIGE Proteomics. Methods Mol. Biol. 2018, 1664, 17–24. [Google Scholar] [CrossRef]
- Alban, A.; David, S.O.; Bjorkesten, L.; Andersson, C.; Sloge, E.; Lewis, S.; Currie, I. A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 2003, 3, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Robotti, E.; Marengo, E. 2D-DIGE and Fluorescence Image Analysis. Methods Mol. Biol. 2018, 1664, 25–39. [Google Scholar] [CrossRef]
- Ohlendieck, K. Comparative 3-Sample DIGE Analysis of Skeletal Muscles. Methods Mol. Biol. 2018, 1664, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.; Unlü, M.; Minden, J.S. Two-dimensional difference gel electrophoresis. Nat. Protoc. 2006, 1, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Minden, J.S.; Dowd, S.R.; Meyer, H.E.; Stühler, K. Difference gel electrophoresis. Electrophoresis 2009, 30 (Suppl. 1), S156–S161. [Google Scholar] [CrossRef]
- Blundon, M.; Ganesan, V.; Redler, B.; Van, P.T.; Minden, J.S. Two-Dimensional Difference Gel Electrophoresis. Methods Mol. Biol. 2019, 1855, 229–247. [Google Scholar] [CrossRef]
- Karp, N.A.; Kreil, D.P.; Lilley, K.S. Determining a significant change in protein expression with DeCyder during a pair-wise comparison using two-dimensional difference gel electrophoresis. Proteomics 2004, 4, 1421–1432. [Google Scholar] [CrossRef]
- Karp, N.A.; Lilley, K.S. Maximising sensitivity for detecting changes in protein expression: Experimental design using minimal CyDyes. Proteomics 2005, 5, 3105–3115. [Google Scholar] [CrossRef]
- Dani, D.; Dencher, N.A. Native DIGE: Efficient Tool to Elucidate Protein Interactomes. Methods Mol. Biol. 2018, 1664, 53–68. [Google Scholar] [CrossRef]
- Malm, C.; Hadrevi, J.; Bergström, S.A.; Pedrosa-Domellöf, F.; Antti, H.; Svensson, M.; Frängsmyr, L. Evaluation of 2-D DIGE for skeletal muscle: Protocol and repeatability. Scand. J. Clin. Lab. Investig. 2008, 68, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.; Doran, P.; Ohlendieck, K. Proteomic analysis of dystrophic muscle. Methods Mol. Biol. 2012, 798, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Carberry, S.; Zweyer, M.; Swandulla, D.; Ohlendieck, K. Application of fluorescence two-dimensional difference in-gel electrophoresis as a proteomic biomarker discovery tool in muscular dystrophy research. Biology (Basel) 2013, 2, 1438–1464. [Google Scholar] [CrossRef]
- Di Luca, A.; Hamill, R.; Mullen, A.M.; Elia, G. DIGE Analysis of Animal Tissues. Methods Mol. Biol. 2018, 1664, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Gelfi, C.; Capitanio, D. DIGE Analysis of Human Tissues. Methods Mol. Biol. 2018, 1664, 117–136. [Google Scholar] [CrossRef]
- Tonge, R.; Shaw, J.; Middleton, B.; Rowlinson, R.; Rayner, S.; Young, J.; Pognan, F.; Hawkins, E.; Currie, I.; Davison, M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 2001, 1, 377–396. [Google Scholar] [CrossRef]
- Marouga, R.; David, S.; Hawkins, E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal. Bioanal. Chem. 2005, 382, 669–678. [Google Scholar] [CrossRef]
- Hmmier, A.; Dowling, P. DIGE Analysis Software and Protein Identification Approaches. Methods Mol. Biol. 2018, 1664, 41–50. [Google Scholar] [CrossRef]
- Wei, B.; Jin, J.P. Troponin T isoforms and posttranscriptional modifications: Evolution, regulation and function. Arch. Biochem. Biophys. 2011, 505, 144–154. [Google Scholar] [CrossRef]
- El-Mezgueldi, M. Tropomyosin dynamics. J. Muscle Res. Cell Motil. 2014, 35, 203–210. [Google Scholar] [CrossRef]
- Wei, B.; Jin, J.P. TNNT1, TNNT2, and TNNT3: Isoform genes, regulation, and structure-function relationships. Gene 2016, 582, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Peng, Y.; Lin, Z.; Chen, Y.C.; Wei, L.; Hacker, T.A.; Larsson, L.; Ge, Y. Comprehensive analysis of tropomyosin isoforms in skeletal muscles by top-down proteomics. J. Muscle Res. Cell Motil. 2016, 37, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Degens, H.; Li, M.; Salviati, L.; Lee, Y.I.; Thompson, W.; Kirkland, J.L.; Sandri, M. Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 2019, 99, 427–511. [Google Scholar] [CrossRef] [PubMed]
- Keevil, V.L.; Romero-Ortuno, R. Ageing well: A review of sarcopenia and frailty. Proc. Nutr. Soc. 2015, 74, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Kiesswetter, E.; Drey, M.; Sieber, C.C. Nutrition, frailty, and sarcopenia. Aging Clin. Exp. Res. 2017, 29, 43–48. [Google Scholar] [CrossRef]
- Yeung, S.S.Y.; Reijnierse, E.M.; Pham, V.K.; Trappenburg, M.C.; Lim, W.K.; Meskers, C.G.M.; Maier, A.B. Sarcopenia and its association with falls and fractures in older adults: A systematic review and meta-analysis. J. Cachexia Sarcopenia Muscle 2019, in press. [Google Scholar] [CrossRef]
- Donoghue, P.; Staunton, L.; Mullen, E.; Manning, G.; Ohlendieck, K. DIGE analysis of rat skeletal muscle proteins using nonionic detergent phase extraction of young adult versus aged gastrocnemius tissue. J. Proteomics 2010, 73, 1441–1453. [Google Scholar] [CrossRef][Green Version]
- Wei, L.; Gregorich, Z.R.; Lin, Z.; Cai, W.; Jin, Y.; McKiernan, S.H.; McIlwain, S.; Aiken, J.M.; Moss, R.L.; Diffee, G.M.; et al. Novel Sarcopenia-related Alterations in Sarcomeric Protein Post-translational Modifications (PTMs) in Skeletal Muscles Identified by Top-down Proteomics. Mol. Cell. Proteomics 2018, 17, 134–145. [Google Scholar] [CrossRef]
- Pagel, O.; Loroch, S.; Sickmann, A.; Zahedi, R.P. Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Rev. Proteomics 2015, 12, 235–253. [Google Scholar] [CrossRef]
- Hart, C.; Schulenberg, B.; Steinberg, T.H.; Leung, W.Y.; Patton, W.F. Detection of glycoproteins in polyacrylamide gels and on electroblots using Pro-Q Emerald 488 dye, a fluorescent periodate Schiff-base stain. Electrophoresis 2003, 24, 588–598. [Google Scholar] [CrossRef]
- Wu, J.; Lenchik, N.J.; Pabst, M.J.; Solomon, S.S.; Shull, J.; Gerling, I.C. Functional characterization of two-dimensional gel-separated proteins using sequential staining. Electrophoresis 2005, 26, 225–237. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Thelen, J.J. A high-resolution two dimensional Gel- and Pro-Q DPS-based proteomics workflow for phosphoprotein identification and quantitative profiling. Methods Mol. Biol. 2009, 527, 3–19. [Google Scholar] [CrossRef]
- Steinberger, B.; Mayrhofer, C. Principles and examples of gel-based approaches for phosphoprotein analysis. Methods Mol. Biol. 2015, 1295, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Mehta-D’souza, P. Detection of glycoproteins in polyacrylamide gels using Pro-Q Emerald 300 dye, a fluorescent periodate Schiff-base stain. Methods Mol. Biol. 2018, 1853, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.; Shen, H.; Wang, L.; Luo, S.; Lin, L.; Yang, J.; Tian, R. Identification, Quantification, and Site Localization of Protein Posttranslational Modifications via Mass Spectrometry-Based Proteomics. Adv. Exp. Med. Biol. 2016, 919, 345–382. [Google Scholar] [CrossRef]
- Gannon, J.; Staunton, L.; O’Connell, K.; Doran, P.; Ohlendieck, K. Phosphoproteomic analysis of aged skeletal muscle. Int. J. Mol. Med. 2008, 22, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Verduyn, S.C.; Zaremba, R.; van der Velden, J.; Stienen, G.J. Effects of contractile protein phosphorylation on force development in permeabilized rat cardiac myocytes. Basic Res. Cardiol. 2007, 102, 476–487. [Google Scholar] [CrossRef]
- Philp, A.; Rowland, T.; Perez-Schindler, J.; Schenk, S. Understanding the acetylome: Translating targeted proteomics into meaningful physiology. Am. J. Physiol. Cell Physiol. 2014, 307, C763–C773. [Google Scholar] [CrossRef]
- Diallo, I.; Seve, M.; Cunin, V.; Minassian, F.; Poisson, J.F.; Michelland, S.; Bourgoin-Voillard, S. Current trends in protein acetylation analysis. Expert Rev. Proteomics 2019, 16, 139–159. [Google Scholar] [CrossRef]
- Ryder, D.J.; Judge, S.M.; Beharry, A.W.; Farnsworth, C.L.; Silva, J.C.; Judge, A.R. Identification of the Acetylation and Ubiquitin-Modified Proteome during the Progression of Skeletal Muscle Atrophy. PLoS ONE 2015, 10, e0136247. [Google Scholar] [CrossRef]
- Capitanio, D.; Vasso, M.; De Palma, S.; Fania, C.; Torretta, E.; Cammarata, F.P.; Magnaghi, V.; Procacci, P.; Gelfi, C. Specific protein changes contribute to the differential muscle mass loss during ageing. Proteomics 2016, 16, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Zhao, J.; Lubman, D.M.; Miller, F.R.; Barder, T.J. Protein pI shifts due to posttranslational modifications in the separation and characterization of proteins. Anal. Chem. 2005, 77, 2745–2755. [Google Scholar] [CrossRef] [PubMed]
- Báez, A.L.; Reynoso, M.N.; Lo Presti, M.S.; Bazán, P.C.; Strauss, M.; Miler, N.; Pons, P.; Rivarola, H.W.; Paglini-Oliva, P. Mitochondrial dysfunction in skeletal muscle during experimental Chagas disease. Exp. Mol. Pathol. 2015, 98, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.D.; Hoffman, V.J.; Roffe, E.; Murphy, P.M. Low-Level Parasite Persistence Drives Vasculitis and Myositis in Skeletal Muscle of Mice Chronically Infected with Trypanosoma cruzi. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.C.; Iwai, L.K.; Kuramoto, A.C.; Honorato, R.; Fiorelli, A.; Stolf, N.; Kalil, J.; Cunha-Neto, E. Proteomic inventory of myocardial proteins from patients with chronic Chagas’ cardiomyopathy. Braz. J. Med. Biol. Res. 2006, 39, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Qu, Y.; Zhang, Z.; Wang, Z.; Prytkova, I.; Wu, S. Intact glycopeptide characterization using mass spectrometry. Expert Rev. Proteomics 2016, 13, 513–522. [Google Scholar] [CrossRef]
- Cieniewski-Bernard, C.; Bastide, B.; Lefebvre, T.; Lemoine, J.; Mounier, Y.; Michalski, J.C. Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 2004, 3, 577–585. [Google Scholar] [CrossRef]
- Rose, A.; Mayor, T. Exploring the Rampant Expansion of Ubiquitin Proteomics. Methods Mol. Biol. 2018, 1844, 345–362. [Google Scholar] [CrossRef]
- Mokhonova, E.I.; Avliyakulov, N.K.; Kramerova, I.; Kudryashova, E.; Haykinson, M.J.; Spencer, M.J. The E3 ubiquitin ligase TRIM32 regulates myoblast proliferation by controlling turnover of NDRG2. Hum. Mol. Genet. 2015, 24, 2873–2883. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).