Proteomic Contributions to Medicinal Plant Research: From Plant Metabolism to Pharmacological Action
Abstract
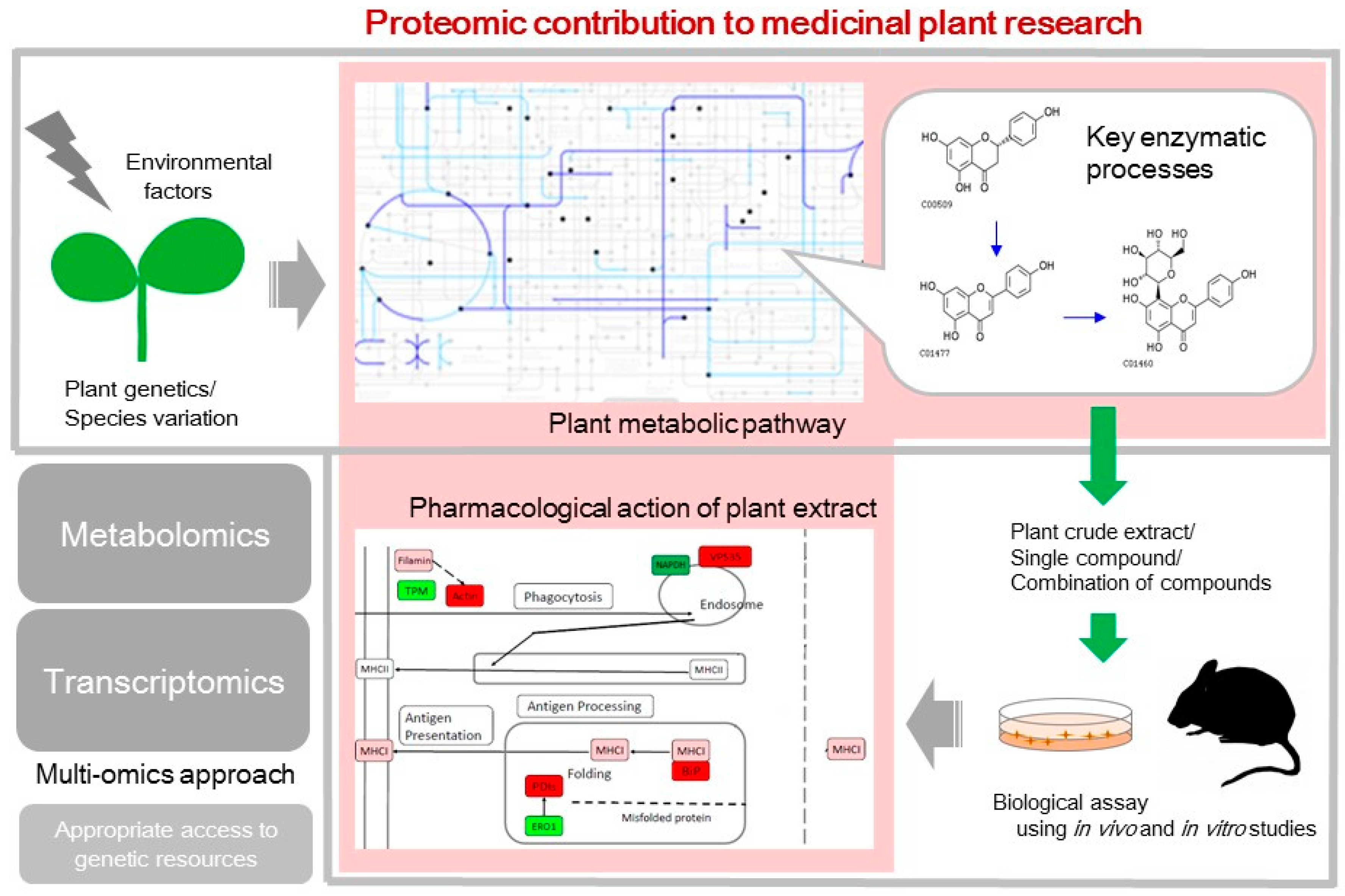
1. Introduction
2. Plant Metabolic Pathways that Synthesize an Array of Bioactive Compounds
3. Pharmacological Action of Plants Tested Using In Vivo and In Vitro Studies
4. Application of Proteomics to Indigenous Plants
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of interest
Abbreviations
| iTRAQ | isobaric tags for relative and absolute protein quantitation |
| UV | ultraviolet |
| CPLL | combinatorial peptide ligand libraries |
| iBAQ | intensity-based absolute quantification |
| NF-κB | nuclear factor-kappa B |
| AKT1 | RAC serine/threonine-protein kinase |
| ABS | access and benefit-sharing |
References
- Cragg, G.M.; Boyd, M.R.; Cardellina, J.H., II; Newman, D.J.; Snader, K.M.; McCloud, T.G. Ethnobotany and drug discovery: The experience of the US National Cancer Institute. Ciba Found. Symp. 1994, 185, 178–190; discussion 190–196. [Google Scholar] [PubMed]
- Weaver, B.A. How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell. 2014, 25, 2677–2681. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, A.; Zhu, W.; Tian, J.; Komatsu, S. Proteomics and metabolomics-driven pathway reconstruction of mung bean for nutraceutical evaluation. Biochim. Biophys. Acta 2017, 1865, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Park, H.S.; Lee, D.K.; Jayakodi, M.; Kim, N.H.; Koo, H.J.; Lee, S.C.; Kim, Y.J.; Kwon, S.W.; Yang, T.J. Integrated transcriptomic and metabolomic analysis of five Panax ginseng cultivars reveals the dynamics of ginsenosidebiosynthesis. Front. Plant Sci. 2017, 8, 1048. [Google Scholar] [CrossRef] [PubMed]
- Plaza, M.; Pozzo, T.; Liu, J.; Gulshan Ara, K.Z.; Turner, C.; Nordberg Karlsson, E. Substituent effects on in vitro antioxidizing properties, stability, and solubility in flavonoids. J. Agric. Food Chem. 2014, 62, 3321–3333. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate: Coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T. From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin Cancer Biol. 2017, 46, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Makarević, J.; Rutz, J.; Juengel, E.; Kaulfuss, S.; Reiter, M.; Tsaur, I.; Bartsch, G.; Haferkamp, A.; Blaheta, R.A. Amygdalin blocks bladder cancer cell growth in vitro by diminishing cyclin A and cdk2. PLoS ONE 2014, 9, e105590. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Y.; Tsai, A.C.; Chen, M.C.; Chang, L.H.; Sun, H.L.; Chang, Y.L.; Chen, C.C.; Teng, C.M.; Pan, S.L. Aciculatin induces p53-dependent apoptosis via MDM2 depletion in human cancer cells in vitro and in vivo. PLoS ONE 2012, 7, e42192. [Google Scholar] [CrossRef] [PubMed]
- Sok, S.P.; Arshad, N.M.; Azmi, M.N.; Awang, K.; Ozpolat, B.; Hasima Nagoor, N. The apoptotic effect of 1′S-1′-acetoxychavicol acetate (ACA) enhanced by inhibition of non-canonical autophagy in human non-small cell lung cancer cells. PLoS ONE 2017, 12, e0171329. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhou, Y.; Jiang, Z.; Zhao, Y.; Zhang, D.; Cong, X.; Cao, R.; Li, H.; Tian, W. Cytotoxic and chemosensitization effects of Scutellarin from traditional Chinese herb Scutellaria altissima L. in human prostate cancer cells. Oncol. Rep. 2017, 38, 1491–1499. [Google Scholar] [PubMed]
- Brantley, S.J.; Argikar, A.A.; Lin, Y.S.; Nagar, S.; Paine, M.F. Herb-drug interactions: Challenges and opportunities for improved predictions. Drug Metab. Dispos. 2014, 42, 301–317. [Google Scholar] [CrossRef] [PubMed]
- García, B.F.; Torres, A.; Macías, F.A. Synergy and other interactions between polymethoxyflavones from citrus byproducts. Molecules 2015, 20, 20079–20106. [Google Scholar] [CrossRef] [PubMed]
- Komape, N.P.; Bagla, V.P.; Kabongo-Kayoka, P.; Masoko, P. Anti-mycobacteria potential and synergistic effects of combined crude extracts of selected medicinal plants used by Bapedi traditional healers to treat tuberculosis related symptoms in Limpopo Province, South Africa. BMC Complement. Altern. Med. 2017, 17, 128. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, A.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [PubMed]
- Giovannoni, J.J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Cereal crop proteomics: Systemic analysis of crop drought stress responses towards marker-assisted selection breeding. Front. Plant Sci. 2017, 8, 757. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, A.; Komatsu, S. Proteomics of soybean plant. In Proteomics in Food Science: From Farm to Fork; Colgrave, M., Ed.; Elsevier: Amsterdam, Netherlands, 2017; Chapter 6; pp. 89–105. [Google Scholar]
- De Steur, H.; Gellynck, X.; Storozhenko, S.; Liqun, G.; Lambert, W.; Van Der Straeten, D.; Viaene, J. Health impact in China of folate-biofortified rice. Nat. Biotechnol. 2010, 28, 554–556. [Google Scholar] [CrossRef] [PubMed]
- Blancquaert, D.; Van Daele, J.; Strobbe, S.; Kiekens, F.; Storozhenko, S.; De Steur, H.; Gellynck, X.; Lambert, W.; Stove, C.; Van Der Straeten, D. Improving folate (vitamin B9) stability in biofortified rice through metabolic engineering. Nat. Biotechnol. 2015, 33, 1076–1078. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Gupta, R.; Lee, S.H.; Min, C.W.; Agrawal, G.K.; Rakwal, R.; Kim, J.B.; Jo, I.H.; Park, S.Y.; Kim, J.K.; et al. An integrated biochemical, proteomics, and metabolomics approach for supporting medicinal value of Panax ginseng fruits. Front. Plant Sci. 2016, 7, 994. [Google Scholar] [CrossRef] [PubMed]
- Romero-Sandoval, E.A.; Kolano, A.L.; Alvarado-Vázquez, P.A. Cannabis and Cannabinoids for Chronic Pain. Curr. Rheumatol. Rep. 2017, 19, 67. [Google Scholar] [CrossRef] [PubMed]
- Raharjo, T.J.; Widjaja, I.; Roytrakul, S.; Verpoorte, R. Comparative proteomics of Cannabis sativa plant tissues. J. Biomol. Tech. 2004, 15, 97–106. [Google Scholar] [PubMed]
- Xu, W.; Choi, H.K.; Huang, L. State of Panax ginseng research: A global analysis. Molecules 2017, 22, 1518. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.W.; Wong, A.S. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Mathur, A.; Mathur, A.K.; Pal, M.; Uniyal, G.C. Comparison of qualitative and quantitative in vitro ginsenoside production in callus cultures of three Panax species. Planta Med. 1999, 65, 484–486. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhao, S. Progress in understanding of ginsenoside biosynthesis. Plant Biol. 2008, 10, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Sun, L.; Chen, X.; Jiang, R.; Sun, H.; Zhao, D. Proteomic changes in different growth periods of ginseng roots. Plant Physiol. Biochem. 2013, 67, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Jayakodi, M.; Lee, S.C.; Lee, Y.S.; Park, H.S.; Kim, N.H.; Jang, W.; Lee, H.O.; Joh, H.J.; Yang, T.J. Comprehensive analysis of Panax ginseng root transcriptomes. BMC Plant Biol. 2015, 15, 138. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Sun, L.; Chen, X.; Mei, B.; Chang, G.; Wang, M.; Zhao, D. Proteomic analyses provide novel insights into plant growth and ginsenoside biosynthesis in forest cultivated Panax ginseng (F. Ginseng). Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Wang, Y.; Abozeid, A.; Zu, Y.G.; Tang, Z.H. The integration of GC-MS and LC-MS to assay the metabolomics profiling in Panax ginseng and Panax quinquefolius reveals a tissue- and species-specific connectivity of primary metabolites and ginsenosides accumulation. J. Pharm. Biomed. Anal. 2017, 135, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Xu, S.; Liu, G.; Liu, X.; Xu, X.; Wen, B.; Liu, S. Improvement of peptide identification with considering the abundance of mRNA and peptide. BMC Bioinform. 2017, 18, 109. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Li, X.; Zhao, Z.; Yang, T.; Wang, X.; Luo, B.; Zhang, Q.; Hu, Y.; Hu, X. Comprehensive analysis of the triterpenoid saponins biosynthetic pathway in Anemone flaccida by transcriptome and proteome profiling. Front. Plant Sci. 2016, 7, 1094. [Google Scholar] [CrossRef] [PubMed]
- Bryant, L.; Flatley, B.; Patole, C.; Brown, G.D.; Cramer, R. Proteomic analysis of Artemisia annua—Towards elucidating the biosynthetic pathways of the antimalarial pro-drug artemisinin. BMC Plant Biol. 2015, 15, 175. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Matsuda, F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Xiao, C.; Liu, H.; Hao, F.; Tang, H. Combined NMR and LC-DAD-MS analysis reveals comprehensive metabonomic variations for three phenotypic cultivars of Salvia miltiorrhiza Bunge. J. Proteome Res. 2010, 9, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Song, Z.; Fang, X.; Pan, Y.; Guo, L.; Liu, T.; Wang, J. Effect of genotype and environment on Salvia miltiorrhiza roots using LC/MS-based metabolomics. Molecules 2016, 21, 414. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Komatsu, S.; Zhu, W.; Zhang, L.; Li, X.; Cui, L.; Tian, J. Response and defense mechanisms of Taxus chinensis leaves under UV-A radiation are revealed using comparative proteomics and metabolomics analyses. Plant Cell Physiol. 2016, 57, 1839–1853. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Yang, B.; Zhang, D.; Chen, M.; Tian, J. Enhanced metabolic process to indole alkaloids in Clematis terniflora DC. after exposure to high level of UV-B irradiation followed by the dark. BMC Plant Biol. 2016, 16, 231. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Guan, Q.; Tian, J.; Komatsu, S. Transcriptomic and proteomic analyses of leaves from Clematis terniflora DC. under high level of ultraviolet-B irradiation followed by dark treatment. J. Proteom. 2017, 150, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, X.; Gao, C.; Chen, M.; Guan, Q.; Tian, J.; Komatsu, S. Proteomic and metabolomic analyses of leaf from Clematis terniflora DC. exposed to high-level ultraviolet-B irradiation with dark treatment. J. Proteome Res. 2016, 15, 2643–2657. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Yang, B.; Komatsu, S.; Lu, X.; Li, X.; Tian, J. Binary stress induces an increase in indole alkaloid biosynthesis in Catharanthus roseus. Front. Plant Sci. 2015, 6, 582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, X.; Zheng, W.; Fu, Z.; Li, W.; Ma, L.; Li, K.; Sun, L.; Tian, J. Proteomics analysis of UV-irradiated Lonicera japonica Thunb. with bioactive metabolites enhancement. Proteomics 2013, 13, 3508–3522. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Li, X.; Zhang, L.; Zhang, Y.; Lu, X.; Tian, J. Improved metabolites of pharmaceutical ingredient grade Ginkgo biloba and the correlated proteomics analysis. Proteomics 2015, 15, 1868–1883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhu, W.; Zhang, Y.; Yang, B.; Fu, Z.; Li, X.; Tian, J. Proteomics analysis of Mahonia bealei leaves with induction of alkaloids via combinatorial peptide ligand libraries. J. Proteom. 2014, 110, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Sarkar, A.; Righetti, P.G.; Pedreschi, R.; Carpentier, S.; Wang, T.; Barkla, B.J.; Kohli, A.; Ndimba, B.K.; Bykova, N.V.; et al. A decade of plant proteomics and mass spectrometry: Translation of technical advancements to food security and safety issues. Mass Spectrom. Rev. 2013, 32, 335–365. [Google Scholar] [CrossRef] [PubMed]
- Firenzuoli, F.; Gori, L. Herbal medicine today: Clinical and research issues. Evid.-Based Complement. Altern. Med. 2007, 4 (Suppl. 1), 37–40. [Google Scholar] [CrossRef] [PubMed]
- Anquez-Traxler, C. The legal and regulatory framework of herbal medicinal products in the European Union: A focus on the traditional herbal medicines category. Drug Inf. J. 2011, 45, 15–23. [Google Scholar] [CrossRef]
- Wang, H.; Ye, Y.; Pan, S.Y.; Zhu, G.Y.; Li, Y.W.; Fong, D.W.; Yu, Z.L. Proteomic identification of proteins involved in the anticancer activities of oridonin in HepG2 cells. Phytomedicine 2011, 18, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Qiao, W.; Zhao, Y.; Fang, H.; Xu, D.; Xia, Q. Curdione attenuates thrombin-induced human platelet activation: β1-tubulin as a potential therapeutic target. Fitoterapia 2017, 116, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhang, F.; Zhang, Y.; Cao, C.; Li, X.; Li, D.; Liu, X.; Yang, H.; Huang, L. Proteomic investigation of signatures for geniposide-induced hepatotoxicity. J. Proteome Res. 2014, 13, 5724–5733. [Google Scholar] [CrossRef] [PubMed]
- Ocak, S.; Friedman, D.B.; Chen, H.; Ausborn, J.A.; Hassanein, M.; Detry, B.; Weynand, B.; Aboubakar, F.; Pilette, C.; Sibille, Y.; et al. Discovery of new membrane-associated proteins overexpressed in small-cell lung cancer. J. Thorac. Oncol. 2014, 9, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.J.; Yang, Z.; Lu, D.Z.; Wo, X.D.; Shi, J.J.; Lin, T.Q.; Wang, M.M.; Li, Y.; Tang, L.H. Dihydroartemisinin-induced inhibition of proliferation in BEL-7402 cells: An analysis of the mitochondrial proteome. Mol. Med. Rep. 2012, 6, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.M. Synergy and other interactions in phytomedicines. Phytomedicine 2001, 8, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Lu, X.; Fu, Z.; Zhang, L.; Li, X.; Xu, X.; Ren, Y.; Lu, Y.; Fu, H.; Tian, J. Identification of candidate synovial membrane biomarkers after Achyranthes aspera treatment for rheumatoid arthritis. Biochim. Biophys. Acta 2016, 1864, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.E.; Kim, J.A.; Nagappan, A.; Yumnam, S.; Lee, H.J.; Kim, E.H.; Lee, W.S.; Shin, S.C.; Park, H.S.; Kim, G.S. Flavonoids identified from Korean Scutellaria baicalensis Georgi inhibit inflammatory signaling by suppressing activation of NF-κB and MAPK in RAW 264.7 cells. Evid.-Based Complement. Altern. Med. 2013, 2013, 912031. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Nagappan, A.; Park, H.S.; Saralamma, V.V.; Hong, G.E.; Yumnam, S.; Lee, H.J.; Raha, S.; Kim, E.H.; Young, P.S.; et al. Proteome profiling of lipopolysaccharide induced L6 rat skeletal muscle cells response to flavonoids from Scutellaria baicalensis Georgi. BMC Complement. Altern. Med. 2014, 14, 379. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, A.; Hitachi, K.; Zhu, W.; Tian, J.; Tsuchida, K.; Komatsu, S. Mung bean (Vigna radiata (L.)) coat extract modulates macrophage functions to enhance antigen presentation: A proteomic study. J. Proteom. 2017, 161, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, W.; Li, J.; Jundoria, A.; Sama, A.E.; Wang, H. It is not just folklore: The aqueous extract of mung bean coat is protective against sepsis. Evid.-Based Complement. Altern. Med. 2012, 2012, 498467. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.L.; Teng, H.J.; Yin, B.; Xu, Y.; Du, Y.; He, F.P.; Chu, K.T.; Luo, B.Y.; Zheng, G.Q. A systematic review and meta-analysis of buyang huanwu decoction in animal model of focal cerebral ischemia. Evid.-Based Complement. Altern. Med. 2013, 2013, 138484. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.D.; Wang, J.H.; Jin, G.R.; Zhao, Y.; Zhang, H.J. Neuroprotective effect of Buyang Huanwu decoction against focal cerebral ischemia/reperfusion injury in rats—Time window and mechanism. J. Ethnopharmacol. 2012, 140, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Cai, J.; Zhan, L.; Guo, Y.; Huang, R.Y.; Li, X.; Zhou, M.; Xu, D.; Zhan, J.; Chen, H. Buyang Huanwu decoction facilitates neurorehabilitation through an improvement of synaptic plasticity in cerebral ischemic rats. BMC Complement. Altern. Med. 2017, 17, 173. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Shen, Y.C.; Shiao, Y.J.; Liou, K.T.; Hsu, W.H.; Hsieh, P.H.; Lee, C.Y.; Chen, Y.R.; Lin, Y.L. Multiplex brain proteomic analysis revealed the molecular therapeutic effects of Buyang Huanwu decoction on cerebral ischemic stroke mice. PLoS ONE 2015, 10, e0140823. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shergis, J.L.; Wu, L.; Yu, X.; Zeng, Q.; Xu, Y.; Guo, X.; Zhang, A.L.; Xue, C.C.; Lin, L. A systematic review and meta-analysis of the herbal formula Buzhong Yiqi Tang for stable chronic obstructive pulmonary disease. Complement. Ther. Med. 2016, 29, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Liu, Z.; Gao, M.; Zhang, Y.; Li, Y.; Ling, S.; Zhang, P.; Zhao, C.; Jiang, L.; Liu, Y.; et al. The effect of Bu Zhong Yi Qi decoction on simulated weightlessness-induced muscle atrophy and its mechanisms. Mol. Med. Rep. 2017, 16, 5165–5174. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Meng, X.; Han, Y.Q.; Xue, M.M.; Wu, S.; Wu, P.; Yuan, Y.; Zhu, Q.; Zhang, T.J.; Wong, C.C.L. Therapeutic mechanistic studies of ShuFengJieDu capsule in an acute lung injury animal model using quantitative proteomics technology. J. Proteome Res. 2017, 16, 4009–4019. [Google Scholar] [CrossRef] [PubMed]
- Andreu, G.L.; Delgado, R.; Velho, J.A.; Curti, C.; Vercesi, A.E. Mangiferin, a natural occurring glucosyl xanthone, increases susceptibility of rat liver mitochondria to calcium-induced permeability transition. Arch. Biochem. Biophys. 2005, 439, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Renuse, S.; Harsha, H.C.; Kumar, P.; Acharya, P.K.; Sharma, J.; Goel, R.; Kumar, G.S.; Raju, R.; Prasad, T.S.; Slotta, T.; et al. Proteomic analysis of an unsequenced plant—Mangifera indica. J. Proteom. 2012, 75, 5793–5796. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.J.; Lu, X.P.; Chen, H.S.; Lu, Y.; Mo, Z.Y. Evaluation of four protein extraction methods for proteomic analysis of mango peel. Genet. Mol. Res. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Esteve, C.; D’Amato, A.; Marina, M.L.; García, M.C.; Righetti, P.G. In-depth proteomic analysis of banana (Musa spp.) fruit with combinatorial peptide ligand libraries. Electrophoresis 2013, 34, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Kong, K.W.; Ismail, A. Phytochemicals and medicinal properties of indigenous tropical fruits with potential for commercial development. Evid.-Based Complement. Altern. Med. 2016, 2016, 7591951. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, I.O.; Benabdelkamel, H.; Alfadda, A.A.; AlYahya, S.A.; Alghamdi, W.M.; Aljohi, H.A.; Almalik, A.; Masood, A. Proteomic analysis of the protein expression profile in the mature Nigella sativa (black seed). Appl. Biochem. Biotechnol. 2016, 179, 1184–1201. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zou, Q.; Wang, J.; Zhang, J.; Liu, Z.; Chen, X. Proteomic profiles reveal the function of different vegetative tissues of Moringa oleifera. Protein J. 2016, 35, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Wang, S.; Liu, Z.; Liu, X.; Zou, L.; Gu, W.; Hou, Y.; Ma, Y.; Luo, Y.; Liu, J. iTRAQ-based quantitative proteomic analysis of cultivated Pseudostellaria heterophylla and its wild-type. J. Proteom. 2016, 139, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Kumar, M.; Padula, M.P.; Pernice, M.; Kahlke, T.; Kim, M.; Ralph, P.J. Development of an efficient protein extraction method compatible with LC-MS/MS for proteome mapping in two Australian seagrasses Zostera muelleri and Posidonia australis. Front. Plant Sci. 2017, 8, 1416. [Google Scholar] [CrossRef] [PubMed]
- Zidorn, C. Secondary metabolites of seagrasses (Alismatales and Potamogetonales; Alismatidae): Chemical diversity, bioactivity, and ecological function. Phytochemistry 2016, 124, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, C.; Jaleel, A.; Deb, L.; Thomas, G.; Sakuntala, M. Development of an efficient virus induced gene silencing strategy in the non-model wild ginger-Zingiber zerumbet and investigation of associated proteome changes. PLoS ONE 2015, 10, e0124518. [Google Scholar] [CrossRef] [PubMed]
- Gray, N.E.; Zweig, J.A.; Matthews, D.G.; Caruso, M.; Quinn, J.F.; Soumyanath, A. Centella asiatica attenuates mitochondrial dysfunction and oxidative stress in Aβ-exposed hippocampal neurons. Oxid. Med. Cell. Longev. 2017, 2017, 7023091. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Sparavigna, A. The 24-hour skin hydration and barrier function effects of a hyaluronic 1%, glycerin 5%, and Centella asiatica stem cells extract moisturizing fluid: An intra-subject, randomized, assessor-blinded study. Clin. Cosmet. Investig. Dermatol. 2017, 10, 311–315. [Google Scholar] [CrossRef] [PubMed]
- De Costa, F.; Barber, C.J.S.; Kim, Y.B.; Reed, D.W.; Zhang, H.; Fett-Neto, A.G.; Covello, P.S. Molecular cloning of an ester-forming triterpenoid: UDP-glucose 28-O-glucosyltransferase involved in saponin biosynthesis from the medicinal plant Centella asiatica. Plant Sci. 2017, 262, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Hodzic, J.; Gurbeta, L.; Omanovic-Miklicanin, E.; Badnjevic, A. Overview of next-generation sequencing platforms used in published draft plant genomes in light of genotypization of immortelle plant (Helichrysium arenarium). Med. Arch. 2017, 71, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Um, Y.; Kim, S.; Kim, O.T.; Koo, S.C.; Reddy, C.S.; Kim, S.C.; Hong, C.P.; Park, S.G.; Kim, H.B.; et al. Development of genome-wide SSR markers from Angelica gigas Nakai using next generation sequencing. Genes (Basel) 2017, 8, 238. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Rai, A.; Nakamura, M.; Suzuki, H.; Takahashi, H.; Yamazaki, M.; Saito, K. De novo deep transcriptome analysis of medicinal plants for gene discovery in biosynthesis of plant natural products. Methods Enzymol. 2016, 576, 19–45. [Google Scholar] [PubMed]
- Shen, C.; Guo, H.; Chen, H.; Shi, Y.; Meng, Y.; Lu, J.; Feng, S.; Wang, H. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci. Rep. 2017, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wang, B.; Yang, J.; Wang, W.; Liu, A.; Leng, L.; Xiang, L.; Song, C.; Chen, S. Weighted gene co-expression network analysis of the dioscin rich medicinal plant Dioscorea nipponica. Front. Plant Sci. 2017, 8, 789. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Takahashi, H.; Suzuki, Y.; Sugano, S.; Noji, M.; Kenmoku, H.; Toyota, M.; Kanaya, S.; Kawahara, N.; Asakawa, Y.; et al. Comparative analysis of transcriptomes in aerial stems and roots of Ephedra sinica based on high-throughput mRNA sequencing. Genom. Data 2016, 10, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Kamochi, H.; Suzuki, H.; Nakamura, M.; Takahashi, H.; Hatada, T.; Saito, K.; Yamazaki, M. De novo transcriptome assembly and characterization of nine tissues of Lonicera japonica to identify potential candidate genes involved in chlorogenic acid, luteolosides, and secoiridoid biosynthesis pathways. J. Nat. Med. 2017, 71, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Loke, K.K.; Rahnamaie-Tajadod, R.; Yeoh, C.C.; Goh, H.H.; Mohamed-Hussein, Z.A.; Mohd Noor, N.; Zainal, Z.; Ismail, I. RNA-seq analysis for secondary metabolite pathway gene discovery in Polygonum minus. Genom. Data 2015, 7, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Zulkapli, M.M.; Rosli, M.A.F.; Salleh, F.I.M.; Mohd Noor, N.; Aizat, W.M.; Goh, H.H. Iso-Seq analysis of Nepenthes ampullaria, Nepenthes rafflesiana and Nepenthes × hookeriana for hybridisation study in pitcher plants. Genom. Data 2017, 12, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Amini, S.; Alias, H.; Aizat-Juhari, M.A.; Mat-Isa, M.N.; Adam, J.H.; Goh, H.H.; Wan, K.L. RNA-seq data from different developmental stages of Rafflesia cantleyi floral buds. Genom. Data 2017, 14, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Sansone, S.A.; Rocca-Serra, P.; Field, D.; Maguire, E.; Taylor, C.; Hofmann, O.; Fang, H.; Neumann, S.; Tong, W.; Amaral-Zettler, L.; et al. Toward interoperable bioscience data. Nat. Genet. 2012, 44, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashiguchi, A.; Tian, J.; Komatsu, S. Proteomic Contributions to Medicinal Plant Research: From Plant Metabolism to Pharmacological Action. Proteomes 2017, 5, 35. https://doi.org/10.3390/proteomes5040035
Hashiguchi A, Tian J, Komatsu S. Proteomic Contributions to Medicinal Plant Research: From Plant Metabolism to Pharmacological Action. Proteomes. 2017; 5(4):35. https://doi.org/10.3390/proteomes5040035
Chicago/Turabian StyleHashiguchi, Akiko, Jingkui Tian, and Setsuko Komatsu. 2017. "Proteomic Contributions to Medicinal Plant Research: From Plant Metabolism to Pharmacological Action" Proteomes 5, no. 4: 35. https://doi.org/10.3390/proteomes5040035
APA StyleHashiguchi, A., Tian, J., & Komatsu, S. (2017). Proteomic Contributions to Medicinal Plant Research: From Plant Metabolism to Pharmacological Action. Proteomes, 5(4), 35. https://doi.org/10.3390/proteomes5040035




