Differential Proteome Analysis of Extracellular Vesicles from Breast Cancer Cell Lines by Chaperone Affinity Enrichment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Sample Collection
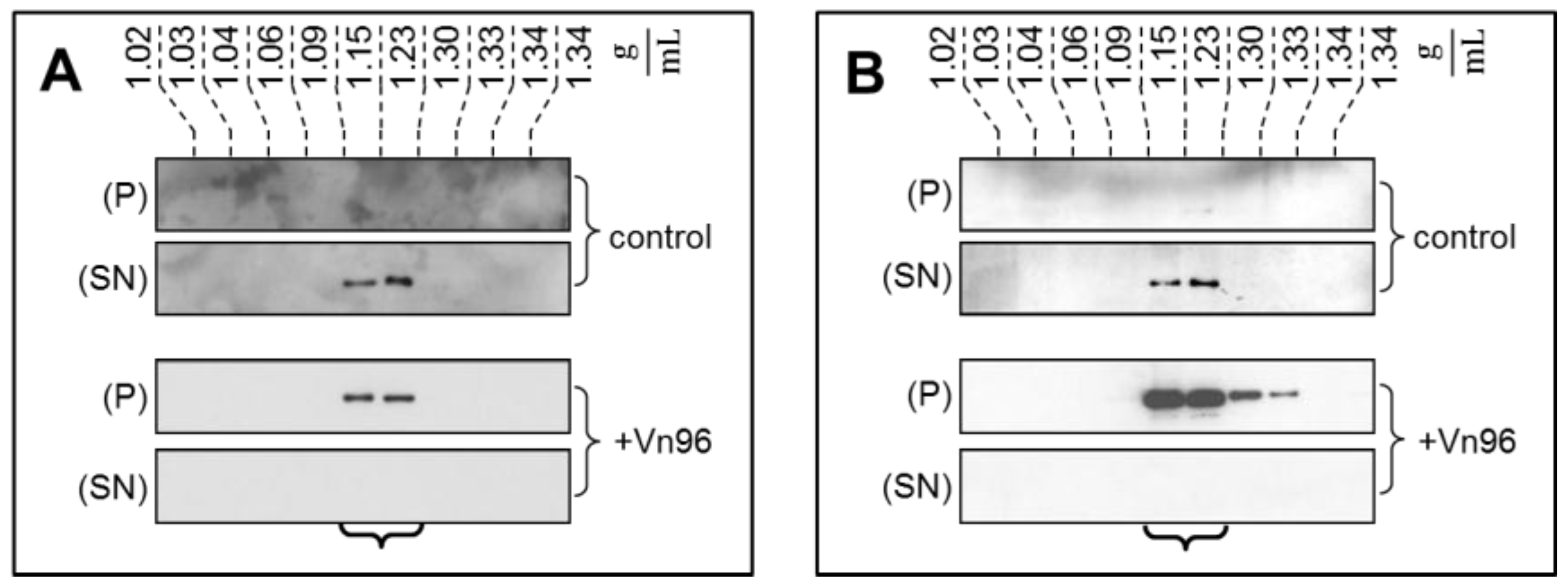
2.2. Ultracentrifugation and Sucrose Density Gradient Fractionation
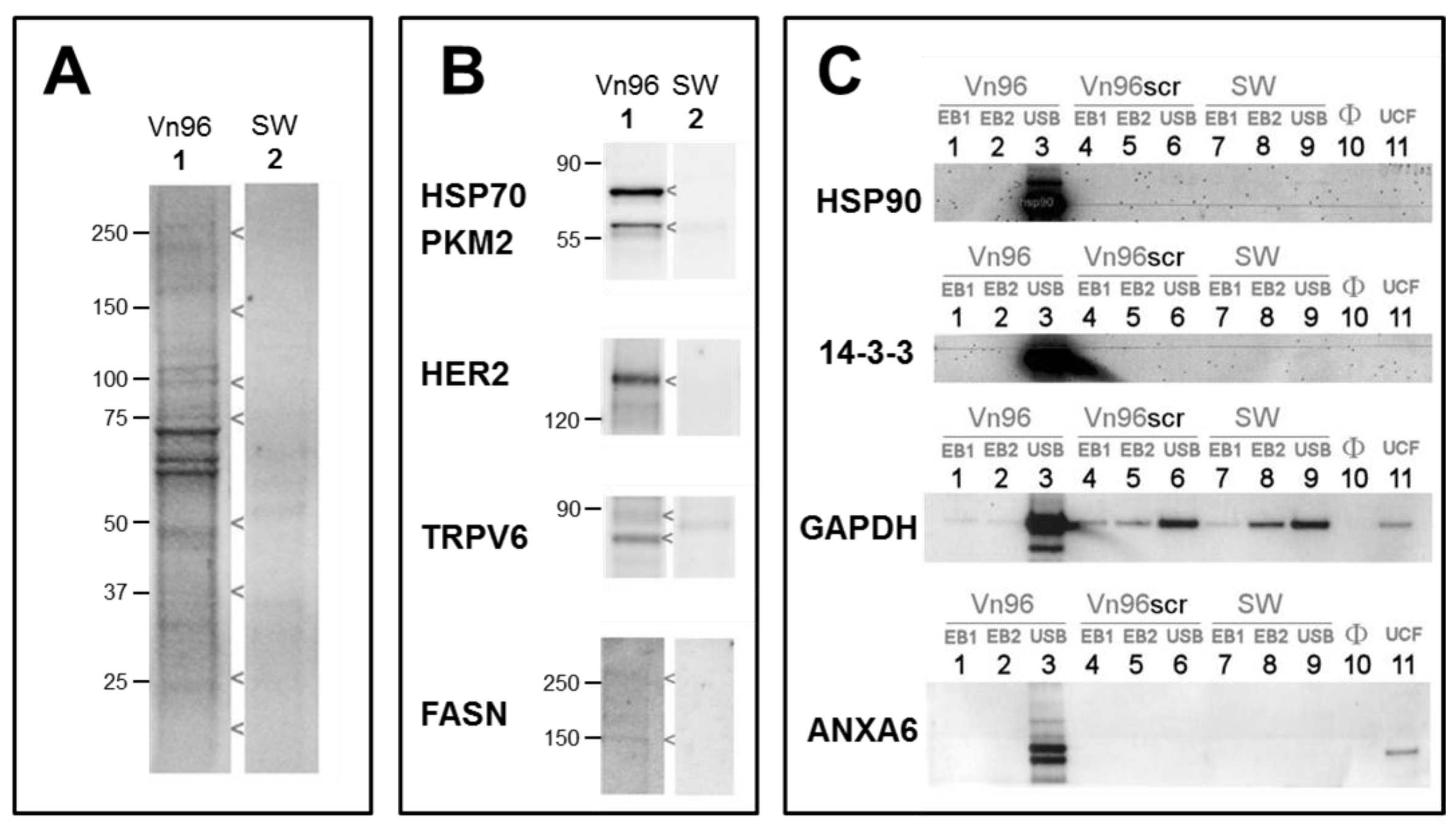
2.3. Vn96 Affinity Capture of EVs
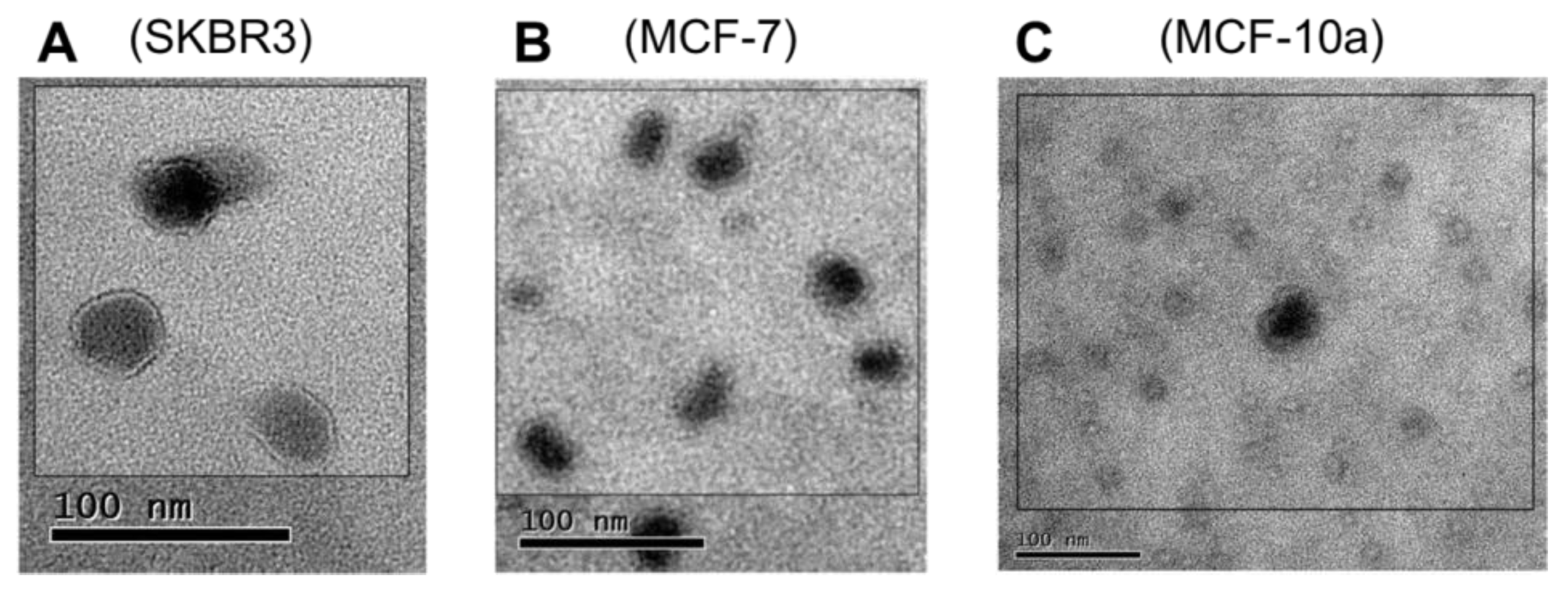
2.4. EV Purity by Immunobloting and Transmission Electron Microscopy
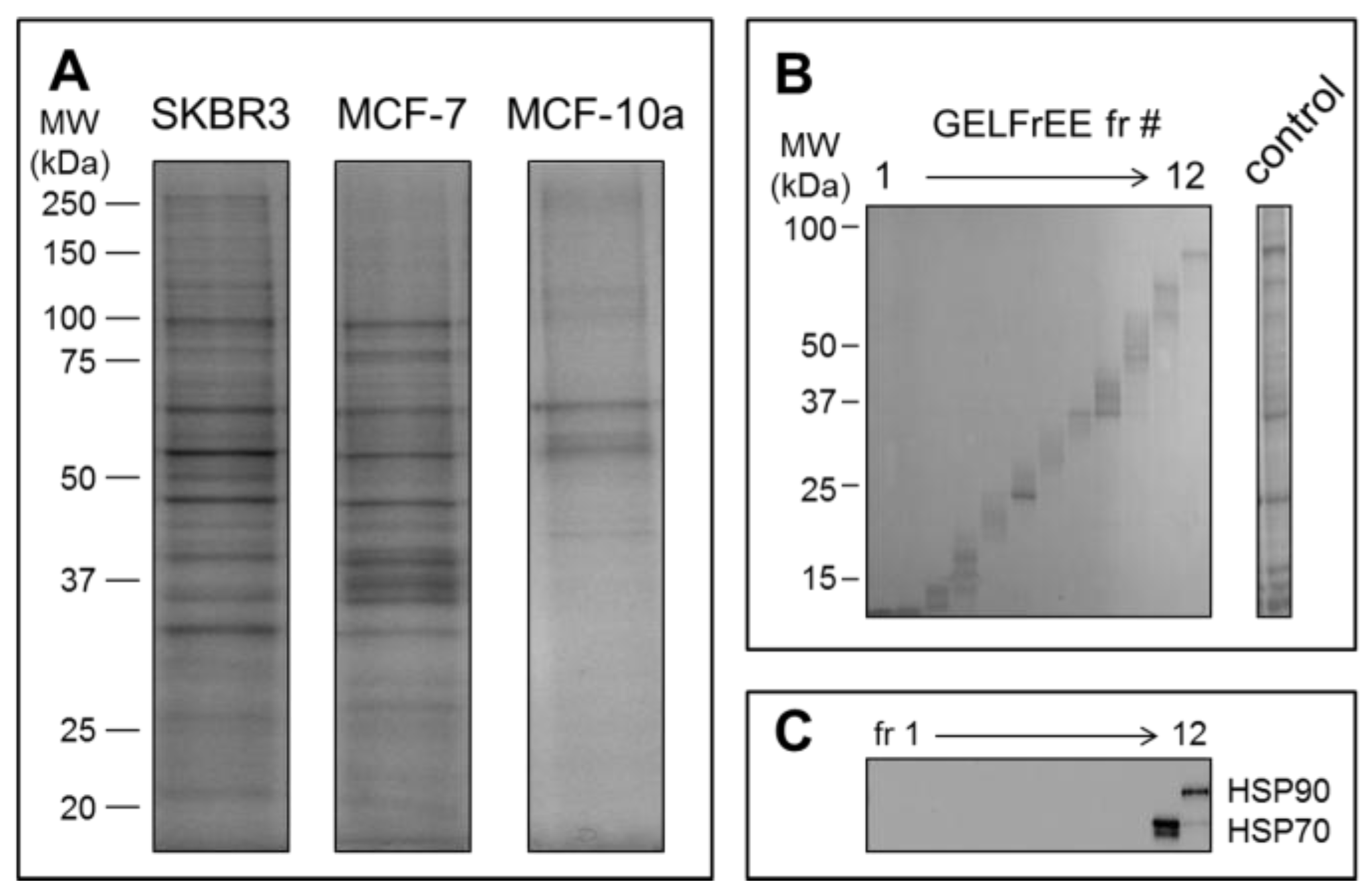
2.5. Proteome Analysis
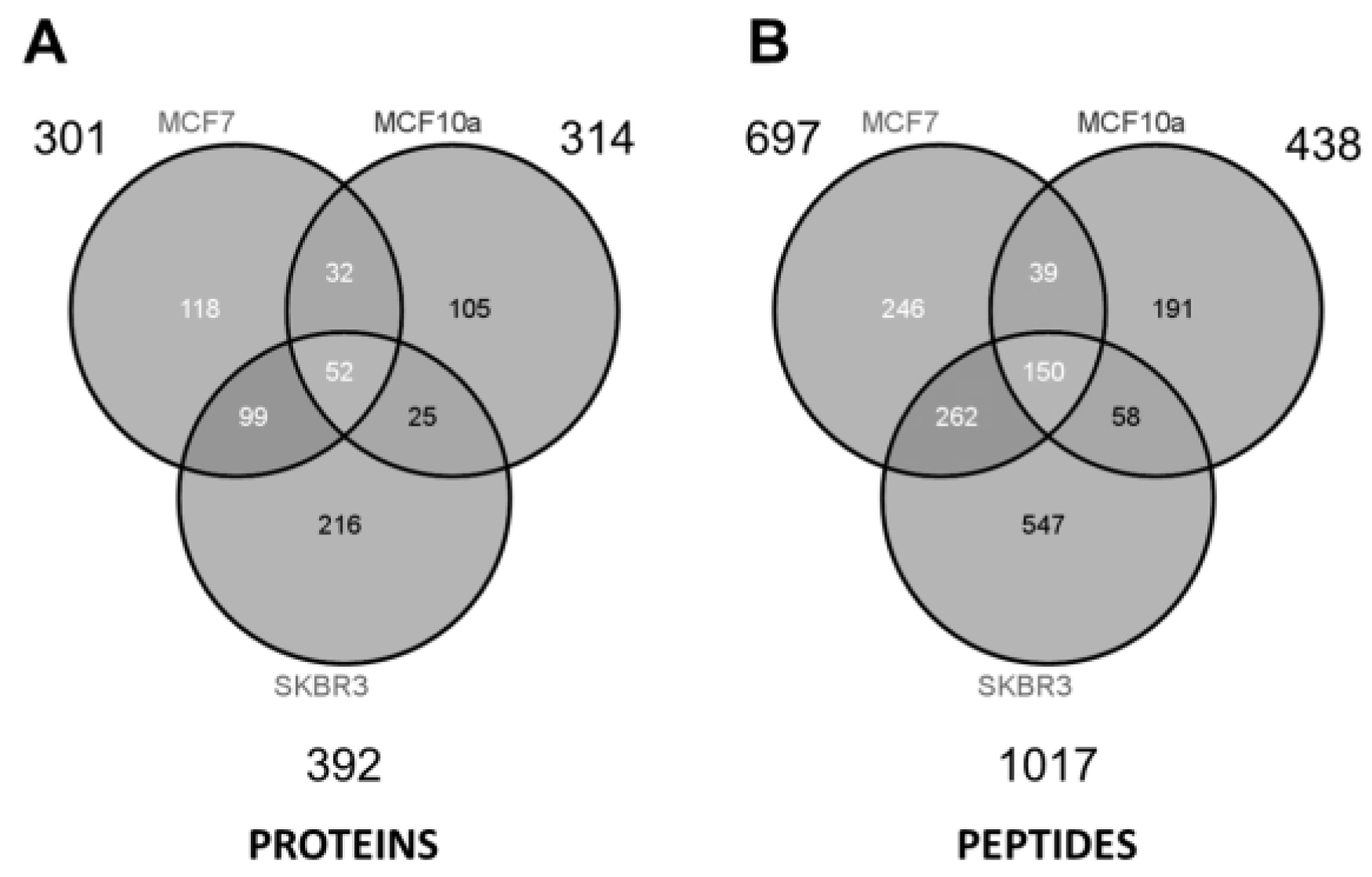
3. Results and Discussion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Americal Cancer Society. Available online: https://www.cancer.org/cancer/breast-cancer/understanding-a-breast-cancer-diagnosis/breast-cancer-survival-rates.html (accessed on 25 July 2017).
- Lehman, C.D.; Isaacs, C.; Schnall, M.D.; Pisano, E.D.; Ascher, S.M.; Weatherall, P.T.; Bluemke, D.A.; Owen, D.J.; Marcom, P.K.; Armstrong, D.K.; et al. Cancer yield of mammography, MR, and US in high-risk women: Prospective multi-institution breast cancer screening study. Radiology 2007, 244, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Berg, W.A.; Zhang, Z.; Lehrer, D.; Jong, R.A.; Pisano, E.D.; Barr, R.G.; Böhm-Vélez, M.; Mahoney, M.C.; Evans, W.P., 3rd; Larsen, L.H.; et al. Detection of breast cancer with addition of annual screening ultrasound or a single screening MRI to mammography in women with elevated breast cancer risk. JAMA 2012, 307, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.G.; Barton, M.B.; Moceri, V.M.; Polk, S.; Arena, P.J.; Fletcher, S.W. Ten-year risk of false positive screening mammograms and clinical breast examinations. N. Engl. J. Med. 1998, 338, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Gøtzsche, P.C.; Jørgensen, K.J.; Maehlen, J.; Zahl, P.-H. Estimation of lead time and overdiagnosis in breast cancer screening. Br. J. Cancer 2009, 100, 219. [Google Scholar] [CrossRef] [PubMed]
- Keen, J.D.; Keen, J.E. What is the point: Will screening mammography save my life? BMC Med. Inform. Decis. Mak. 2009, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S.M.; Pitteri, S.J.; Faca, V.M. Mining the plasma proteome for cancer biomarkers. Nature 2008, 452, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Meng, R.; Gormley, M.; Bhat, V.B.; Rosenberg, A.; Quong, A.A. Low abundance protein enrichment for discovery of candidate plasma protein biomarkers for early detection of breast cancer. J. Proteom. 2011, 75, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, Y.S.; Moresco, J.J.; Yates, J.R.; Nardulli, A.M. Integration of breast cancer secretomes with clinical data elucidates potential serum markers for disease detection, diagnosis, and prognosis. PLoS ONE 2016, 11, e0158296. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef] [PubMed]
- Abramowicz, A.; Widlak, P.; Pietrowska, M. Proteomic analysis of exosomal cargo: The challenge of high purity vesicle isolation. Mol. Biosyst. 2016, 12, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Clayton, A.; Amigorena, S.; Raposo, G. Isolation and characterization of exosomes from cell culture supernatants. Curr. Protoc. Cell Biol. 2006. [Google Scholar] [CrossRef]
- Mitchell, P.J.; Welton, J.; Staffurth, J.; Court, J.; Mason, M.D.; Tabi, Z.; Clayton, A. Can urinary exosomes act as treatment response markers in prostate cancer? J. Transl. Med. 2009, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome isolation for proteomic analyses and RNA profiling. In Serum/Plasma Proteomics: Methods and Protocols; Simpson, R.J., Greening, D.W., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 235–246. ISBN 978-1-61779-068-3. [Google Scholar]
- Griffiths, S.G.; Lewis, S.E. Polypeptides with affinity for heat shock proteins (HSPs) and HSP associated complexes (HACS) and their use in diagnosis and therapy. U.S. Patent 8,956,878, 17 February 2015. [Google Scholar]
- Melendez, K.; Wallen, E.S.; Edwards, B.S.; Mobarak, C.D.; Bear, D.G.; Moseley, P.L. Heat shock protein 70 and glycoprotein 96 are differentially expressed on the surface of malignant and nonmalignant breast cells. Cell Stress Chaperones 2006, 11, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.G.; Martins, V.R.; Hajj, G.N.M. Unconventional secretion of heat shock proteins in cancer. Int. J. Mol. Sci. 2017, 18, 946. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef] [PubMed]
- Bijnsdorp, I.V.; Maxouri, O.; Kardar, A.; Schelfhorst, T.; Piersma, S.R.; Pham, T.V.; Vis, A.; van Moorselaar, R.J.; Jimenez, C.R. Feasibility of urinary extracellular vesicle proteome profiling using a robust and simple, clinically applicable isolation method. J. Extracell. Vesicles 2017, 6, 1313091. [Google Scholar] [CrossRef] [PubMed]
- Knol, J.C.; de Reus, I.; Schelfhorst, T.; Beekhof, R.; de Wit, M.; Piersma, S.R.; Pham, T.V.; Smit, E.F.; Verheul, H.M.W.; Jiménez, C.R. Peptide-mediated “miniprep” isolation of extracellular vesicles is suitable for high-throughput proteomics. EuPA Open Proteom. 2016, 11, 11–15. [Google Scholar] [CrossRef]
- Mitchell, J.P.; Court, J.; Mason, M.D.; Tabi, Z.; Clayton, A. Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J. Immunol. Methods 2008, 335, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Botelho, D.; Wall, M.J.; Vieira, D.B.; Fitzsimmons, S.; Liu, F.; Doucette, A. Top-down and bottom-up proteomics of SDS-containing solutions following mass-based separation. J. Proteom. Res. 2010, 9, 2863–2870. [Google Scholar] [CrossRef] [PubMed]
- Orton, D.J.; Doucette, A.A. A universal, high recovery assay for protein quantitation through temperature programmed liquid chromatography (TPLC). J. Chromatogr. B 2013, 921–922, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sadygov, R.G.; Yates, J.R., III. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004, 76, 4193–4201. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Gastpar, R.; Gehrmann, M.; Bausero, M.A.; Asea, A.; Gross, C.; Schroeder, J.A.; Multhoff, G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005, 65, 5238–5247. [Google Scholar] [CrossRef] [PubMed]
- Németh, A.; Orgovan, N.; Sódar, B.W.; Osteikoetxea, X.; Pálóczi, K.; Szabó-Taylor, K.É.; Vukman, K.V.; Kittel, Á.; Turiák, L.; Wiener, Z.; et al. Antibiotic-induced release of small extracellular vesicles (exosomes) with surface-associated DNA. Sci. Rep. 2017, 7, 8202. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.G.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- ExoCarta. Available online: http://www.exocarta.org/ (accessed on 26 July 2017).
- Olofsson, M.H.; Ueno, T.; Pan, Y.; Xu, R.; Cai, F.; van der Kuip, H.; Muerdter, T.E.; Sonnenberg, M.; Aulitzky, W.E.; Schwarz, S.; et al. Cytokeratin-18 is a useful serum biomarker for early determination of response of breast carcinomas to chemotherapy. Clin. Cancer Res. 2007, 13, 3198–3206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-H.; Tai, L.K.; Wong, L.L.; Sethi, S.K.; Koay, E.S.C. Proteomics of breast cancer: Enhanced expression of cytokeratin19 in human epidermal growth factor receptor type 2 positive breast tumors. Proteomics 2005, 5, 1797–1805. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Jurisic, V.; Radenkovic, S.; Konjevic, G. The Actual Role of LDH as Tumor Marker, Biochemical and Clinical Aspects. In Advances in Cancer Biomarkers: From Biochemistry to Clinic for a Critical Revision; Scatena, R., Ed.; Springer: Dordrecht, The Netherlands, 2015; pp. 115–124. ISBN 978-94-017-7215-0. [Google Scholar]
- Xu, R.; Pelicano, H.; Zhou, Y.; Carew, J.S.; Feng, L.; Bhalla, K.N.; Keating, M.J.; Huang, P. Inhibition of glycolysis in cancer cells: A novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005, 65, 613–621. [Google Scholar] [PubMed]
- Muñoz-Pinedo, C.; Mjiyad, N.; Ricci, J.-E. Cancer metabolism: Current perspectives and future directions. Cell Death Dis. 2012, 3, e248. [Google Scholar] [CrossRef] [PubMed]
- Bayley, J.-P.; Devilee, P. The Warburg effect in 2012. Curr. Opin. Oncol. 2012, 24, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Young, C.D.; Anderson, S.M. Sugar and fat—That’s where it’s at: Metabolic changes in tumors. Breast Cancer Res. 2008, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Deng, C.; Yang, T.; Dong, Q.; Chen, Y.; Nice, E.C.; Huang, C.; Wei, Y. Proteomics revisits the cancer metabolome. Expert Rev. Proteom. 2011, 8, 505–533. [Google Scholar] [CrossRef] [PubMed]
- Kamili, A.; Roslan, N.; Frost, S.; Cantrill, L.C.; Wang, D.; Della-Franca, A.; Bright, R.K.; Groblewski, G.E.; Straub, B.K.; Hoy, A.J.; et al. TPD52 expression increases neutral lipid storage within cultured cells. J. Cell Sci. 2015, 128, 3223–3238. [Google Scholar] [CrossRef] [PubMed]
- Flavin, R.; Peluso, S.; Nguyen, P.; Loda, M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010, 6, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Adam, R.M.; Kim, J.; Kim, R.; Sotgia, F.; Williams, T.; Demichelis, F.; Solomon, K.R.; Loda, M.; Rubin, M.A.; et al. Caveolin-1 interacts with a lipid raft-associated population of fatty acid synthase. Cell Cycle 2008, 7, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kuhajda, F.P.; Li, J.N.; Pizer, E.S.; Han, W.F.; Sokoll, L.J.; Chan, D.W. Fatty acid synthase (FAS) expression in human breast cancer cell culture supernatants and in breast cancer patients. Cancer Lett. 2001, 167, 99–104. [Google Scholar] [CrossRef]
- Notarnicola, M.; Tutino, V.; Calvani, M.; Lorusso, D.; Guerra, V.; Caruso, M.G. Serum levels of fatty acid synthase in colorectal cancer patients are associated with tumor stage. J. Gastrointest. Cancer 2012, 43, 508–511. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Martin, A.; Colomer, R.; Brunet, J.; Lupu, R.; Menendez, J.A. Overexpression of fatty acid synthase gene activates HER1/HER2 tyrosine kinase receptors in human breast epithelial cells. Cell Prolif. 2008, 41, 59–85. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Cappello, F.; Macario, A.J.L.; Conway de Macario, E.; Logozzi, M.; Fais, S.; Campanella, C. Exosomal HSP60: A potentially useful biomarker for diagnosis, assessing prognosis, and monitoring response to treatment. Expert Rev. Mol. Diagn. 2017, 17, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Bhatia, A.; Dong, H.; Jayaprakash, P.; Guo, J.; Sahu, D.; Hou, Y.; Tsen, F.; Tong, C.; O’Brien, K.; et al. Evolutionarily conserved dual lysine motif determines the non-chaperone function of secreted Hsp90alpha in tumour progression. Oncogene 2017, 36, 2160–2171. [Google Scholar] [CrossRef] [PubMed]
- Graner, M.W.; Cumming, R.I.; Bigner, D.D. The heat shock response and chaperones/heat shock proteins in brain tumors: Surface expression, release, and possible immune consequences. J. Neurosci. 2007, 27, 11214–11227. [Google Scholar] [CrossRef] [PubMed]
- Rodina, A.; Wang, T.; Yan, P.; Gomes, E.D.; Dunphy, M.P.S.; Pillarsetty, N.; Koren, J.; Gerecitano, J.F.; Taldone, T.; Zong, H.; et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature 2016, 538, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Grzegrzolka, J.; Kurnol, K.; Piotrow, P.; Pula, B.; Kobierzycki, C.; Piotrowska, A.; Jablonska, K.; Wojnar, A.; Rys, J.; Dziegiel, P.; et al. Hsp27 expression in invasive ductal breast carcinoma. Folia Histochem. Cytobiol. 2012, 50, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Wallenberg, M.; Misra, S.; Björnstedt, M. Selenium cytotoxicity in cancer. Basic Clin. Pharmacol. Toxicol. 2014, 114, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Wise, R.; Duhachek-Muggy, S.; Qi, Y.; Zolkiewski, M.; Zolkiewska, A. Protein disulfide isomerases in the endoplasmic reticulum promote anchorage-independent growth of breast cancer cells. Breast Cancer Res. Treat. 2016, 157, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Ramos, F.S.; Serino, L.T.R.; Carvalho, C.M.S.; Lima, R.S.; Urban, C.A.; Cavalli, I.J.; Ribeiro, E.M.S.F. PDIA3 and PDIA6 gene expression as an aggressiveness marker in primary ductal breast cancer. Genet. Mol. Res. 2015, 14, 6960–6967. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Jin, Q.; Wang, X.-D.; Zhu, H.-J.; Ni, Q.-C. Aldehyde dehydrogenase 1 expression is correlated with poor prognosis in breast cancer. 2017, 96, e7171. [Google Scholar] [CrossRef] [PubMed]
- Matta, A.; Siu, K.W.M.; Ralhan, R. 14-3-3 zeta as novel molecular target for cancer therapy. Expert Opin. Ther. Targets 2012, 16, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.P.; Evans, R.L.; Egland, K.A. Multiple functions of suchi domain containing 2 (SUSD2) in breast tumorigenisis. Mol. Cancer Res. 2013, 11, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, R.; Gogliettino, M.; Cocca, E.; Iannitti, R.; Sandomenico, A.; Ruvo, M.; Balestrieri, M.; Rossi, M.; Palmieri, G. APEH inhibition affects osteosarcoma cell viability via downregulation of the proteasome. Int. J. Mol. Sci. 2016, 17, 1614. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhu, L.; Jiang, Y.; Xu, J.; Wang, F.; He, Z. miR-219-5p suppresses the proliferation and invasion of colorectal cancer cells by targeting calcyphosin. Oncol. Lett. 2017, 13, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; He, R.; Liu, Y.; Wu, Y.; Kang, L. UPregulated single-stranded DNA-binding protein 1 induces cell chemoresistance to cisplatin in lung cancer cell lines. Mol. Cell. Biochem. 2017, 431, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, E.; Tinari, N.; Semeraro, D.; Traini, S.; Fichera, I.; Cumashi, A.; La Sorda, R.; Spinella, F.; Bagnato, A.; Lattanzio, R.; et al. LGALS3BP, lectin galactoside-binding soluble 3 binding protein, induces vascular endothelial growth factor in human breast cancer cells and promotes angiogenesis. J. Mol. Med. 2013, 91, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, M.; Qader Hamadneh, L.A.; Veerakumarasivam, A.; Abdul Rahman, S.; Shohaimi, S.; Rosli, R. Calreticulin mediates an invasive breast cancer phenotype through the transcriptional dysregulation of p53 and MAPK pathways. Cancer Cell Int. 2016, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Rahimipour, S.; Ben-Aroya, N.; Koch, Y.; Kaganovsky, E.; Okon, E. Two forms of gonadotropin-releasing hormone (GnRH) are expressed in human breast tissue and overexpressed in breast cancer: A putative mechanism for the antiproliferative effect of GnRH by down-regulation of acidic ribosomal phosphoproteins P1 and P2. Cancer Res. 2002, 62, 1036–1044. [Google Scholar] [PubMed]
- Timpe, L.C.; Yen, R.; Haste, N.V.; Litsakos-Cheung, C.; Yen, T.Y.; Macher, B.A. Systemic alteration of cell-surface and secreted glycoprotein expression in malignant breast cancer cell lines. Glycobiology 2013, 23, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Bateman, N.W.; Sun, M.; Hood, B.L.; Flint, M.S.; Conrads, T.P. Defining central themes in breast cancer biology by differential proteomics: Conserved regulation of cell spreading and focal adhesion kinase. J. Proteome Res. 2010, 9, 5311–5324. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Kim, G.; Lee, J.W.; Lee, J.E.; Kim, Y.S.; Yu, J.H.; Lee, S.T.; Ahn, S.H.; Kim, H.; Lee, C. Identification of ganglioside GM2 activator playing a role in cancer cell migration through proteomic analysis of breast cancer secretomes. Cancer Sci. 2016, 107, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tai, L.K.; Wong, L.L.; Chiu, L.-L.; Sethi, S.K.; Koay, E.S.C. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol. Cell. Proteom. 2005, 4, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
| SKBR3 | ||||
|---|---|---|---|---|
| Protein Name | ID | Unique Peptides | PSMs | Function |
| Enolase | P06733 | 13 | 197 | metabolism |
| Fatty acid synthase | P49327 | 20 | 139 | metabolism |
| Phosphoglycerate kinase | P00558 | 11 | 82 | metabolism |
| Fructose bisphosphatase 1 | P09467 | 11 | 70 | metabolism |
| GAPDH | E7EUT5 | 7 | 64 | metabolism |
| Malate dehydrogenase | E9PDB2 | 10 | 59 | metabolism |
| L-lactate dehydrogenase | P00338 | 8 | 50 | metabolism |
| Aldehyde dehydrogenase | F8W0A9 | 8 | 48 | metabolism |
| Aldolase | H3BPS8 | 6 | 33 | metabolism |
| Triosephosphate isomerase | P60174 | 6 | 31 | metabolism |
| Glucosidase 2 subunit beta | P14314 | 6 | 28 | metabolism |
| Selenium-binding protein 1 | Q13228 | 6 | 106 | binding |
| 60 kDa heat shock protein | P10809 | 8 | 101 | binding |
| Protein disulfide-isomerase | I3L2P8 | 9 | 63 | binding |
| 14-3-3 protein zeta/delta | P63104 | 6 | 49 | binding |
| Lamin A/C | Q6UYC3 | 6 | 49 | binding |
| Tumor protein D52 | D3YTI0 | 7 | 44 | binding |
| 14-3-3 protein epsilon | P62258 | 8 | 38 | binding |
| TER ATPase | P55072 | 6 | 25 | binding |
| Cytokeratin 19 | P08727 | 18 | 635 | assembly |
| Cytokeratin 8 | P05787 | 20 | 350 | assembly |
| Cytokeratin 18 | F8VZY9 | 8 | 112 | assembly |
| Cytokeratin 16 | P08779 | 7 | 109 | assembly |
| alpha-Actinin-4 | O43707 | 11 | 72 | assembly |
| Myosin-9 | P35579 | 11 | 26 | assembly |
| MCF-7 | ||||
|---|---|---|---|---|
| Protein Name | ID | Unique Peptides | PSMs | Function |
| Enolase | P06733 | 9 | 103 | metabolism |
| Aldolase | H3BQN4 | 7 | 62 | metabolism |
| Fructose bisphosphatase 1 | P09467 | 11 | 59 | metabolism |
| Triosephosphate isomerase | P60174 | 8 | 49 | metabolism |
| Pyruvate kinase | P14618 | 7 | 45 | metabolism |
| Phosphoglycerate kinase | B7Z7A9 | 8 | 38 | metabolism |
| GAPDH | E7EUT5 | 6 | 27 | metabolism |
| Tryptophan-tRNA ligase | P23381 | 8 | 23 | metabolism |
| Cathepsin D | P07339 | 5 | 20 | metabolism |
| Kynureninase | Q16719 | 6 | 15 | metabolism |
| TER ATPase | P55072 | 5 | 15 | metabolism |
| Lactoferroxin-C | B7Z4X2 | 9 | 14 | metabolism |
| Hexokinase-1 | B4DG62 | 7 | 13 | metabolism |
| 14-3-3 protein zeta/delta | P63104 | 7 | 83 | binding |
| 14-3-3 protein epsilon | P62258 | 9 | 63 | binding |
| Selenium-binding protein 1 | Q13228 | 6 | 38 | binding |
| HSP 90-a | P07900 | 5 | 29 | binding |
| Agrin | O00468 | 5 | 25 | binding |
| Protein SET | Q01105 | 5 | 21 | binding |
| Cytokeratin 8 | P05787 | 23 | 557 | assembly |
| Cytokeratin 19 | P08727 | 11 | 241 | assembly |
| Cytokeratin 18 | P05783 | 13 | 197 | assembly |
| Actin, cytoplasmic 1 | P60709 | 9 | 103 | assembly |
| Filamin A | A6NDY9 | 7 | 38 | assembly |
| Lamin A/C | Q6UYC3 | 6 | 38 | assembly |
| MCF-10a | ||||
|---|---|---|---|---|
| Protein Name | ID | Unique Peptides | PSMs | Function |
| Enolase | P06733 | 5 | 29 | metabolism |
| GAPDH | E7EUT5 | 4 | 15 | metabolism |
| Matrix metalloproteinase-2 | B4DWH3 | 4 | 7 | metabolism |
| Tripeptidyl-peptidase 1 | O14773 | 3 | 7 | metabolism |
| Cathepsin D | P07339 | 3 | 5 | metabolism |
| Complement component 1r | H0YFH3 | 3 | 4 | metabolism |
| Protein disulfide-isomerase | P13667 | 3 | 4 | metabolism |
| Anastellin | F8W7G7 | 12 | 63 | binding |
| Periostin | F5H628 | 4 | 29 | binding |
| Serpin B5 | P36952 | 4 | 15 | binding |
| TGF-β-induced protein ig-h3 | G8JLA8 | 6 | 14 | binding |
| Actin, cytoplasmic 1 | F5GYT4 | 4 | 70 | assembly |
| Laminin subunit gamma-2 | F5H430 | 4 | 53 | assembly |
| Filamin A | A6NDY9 | 6 | 39 | assembly |
| Cytokeratin 8 | P05787 | 4 | 31 | assembly |
| Alpha-actinin-4 | O43707 | 6 | 23 | assembly |
| Thrombospondin-1 | P07996 | 5 | 20 | assembly |
| Laminin subunit gamma-1 | P11047 | 3 | 16 | assembly |
| alpha-Actinin-1 | H9KV75 | 5 | 13 | assembly |
| Myosin-9 | P35579 | 4 | 13 | assembly |
| Tropomyosin a-1 chain | H0YKP3 | 3 | 10 | assembly |
| Agrin | O00468 | 5 | 9 | assembly |
| Filamin-B | O75369 | 4 | 7 | assembly |
| Desmoplakin | P15924 | 3 | 4 | assembly |
| Laminin subunit alpha-5 | O15230 | 3 | 4 | assembly |
| Protein Description | Process | Protein Spectral Hits (PSMs) | ||
|---|---|---|---|---|
| SKBR3 | MCF7 | MCF10 | ||
| Hexokinase 1 | Glycolysis/Gneo | 1 | 13 | 0 |
| Glucose-6-phosphate isomerase | Glycolysis/Gneo | 1 | 1 | 0 |
| Phosphofructokinase | Glycolysis/Gneo | 0 | 0 | 0 |
| Aldolase (fructose 1,6-bisphosphatase) | Glycolysis/Gneo | 138 | 176 | 12 |
| Triosephosphate isomerase 1 | Glycolysis/Gneo | 31 | 49 | 2 |
| GAPDH | Glycolysis/Gneo | 64 | 27 | 15 |
| Phosphoglycerate kinase | Glycolysis/Gneo | 82 | 38 | 5 |
| Phosphoglycerate mutase | Glycolysis/Gneo | 7 | 3 | 0 |
| Enolase | Glycolysis/Gneo | 197 | 123 | 29 |
| Pyruvate kinase | Glycolysis/Gneo | 17 | 45 | 0 |
| Fatty Acid Synthase | Lipogenesis | 139 | 1 | 0 |
| Tumor Protein D52 | Lipid storage | 44 | 0 | 0 |
| Lactate dehydrogenase A | Glycolysis/Gneo | 50 | 40 | 0 |
| Lactate dehydrogenase B | Glycolysis/Gneo | 11 | 0 | 0 |
| 6-phosphogluconate dehydrogenase | Pentose Phosphate | 27 | 22 | 7 |
| Isocitrate dehydrogenase | TCA | 29 | 14 | 0 |
| Protein Name | SwissProt Accession Number | Peptide Spectral Matches | ||
|---|---|---|---|---|
| SKBR3 | MCF7 | MCF10a | ||
| HSP10 | [B8ZZL8] | 7 | 7 | 0 |
| HSP60 | [P10809] | 101 | 9 | 0 |
| HSP70-1A/1B | [P08107] | 3 | 7 | 1 |
| HSP71 | [E9PKE3], [E9PQQ4] | 0 | 5 | 2 |
| HSP27 | [P04792], [B4DL87] | 32 | 53 | 4 |
| GRP78 (Hsp70-5) | P11021 | 57 | 16 | 0 |
| HSP90-alpha | [P07900] | 77 | 29 | 39 |
| HSP90-beta | [P08238] | 70 | 0 | 41 |
| GRP94 (Hsp90-B1) | [P14625], [E9PEX3] | 12 | 4 | 6 |
| SUM: | 347 | 126 | 87 | |
| Protein Description | Accession | SKBR3 | MCF7 | MCF10 | Function | Ref. |
|---|---|---|---|---|---|---|
| Selenium-binding protein 1 | [Q13228] | 106 | 38 | 0 | Detox | [53] |
| PDIA1 (P4HB) | [I3L2P8] | 63 | 0 | 0 | Invasion | [54] |
| PDIA3 (ERP57, GRP58) | [H7BZJ3] | 52 | 6 | 1 | Invasion | [55] |
| Aldehyde dehydrogenase 1 | [F8W0A9] | 48 | 0 | 0 | Detox | [56] |
| 14-3-3 zeta/delta ζ | [P63104] | 49 | 82 | 0 | Enabler | [57] |
| Sushi Domain Containing D2 | [Q9UGT4] | 48 | 0 | 0 | Invasion | [58] |
| Acylamino-acid-releasing | [P13798] | 41 | 14 | 0 | Detox | [59] |
| Calcyphosin | [Q13938] | 41 | 0 | 0 | Proliferation | [60] |
| SS DNA-binding | [Q04837] | 38 | 0 | 0 | Resistance | [61] |
| Galectin-3-binding | [Q08380] | 36 | 0 | 0 | Invasion | [62] |
| Calreticulin | [P27797] | 33 | 0 | 0 | Protection | [63] |
| 60S ribosomal P2 | [P05387] | 31 | 0 | 0 | Enabler | [64] |
| Glucosidase 2 beta (80-KH) | [P14314] | 28 | 2 | 7 | Invasion | [65] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griffiths, S.G.; Cormier, M.T.; Clayton, A.; Doucette, A.A. Differential Proteome Analysis of Extracellular Vesicles from Breast Cancer Cell Lines by Chaperone Affinity Enrichment. Proteomes 2017, 5, 25. https://doi.org/10.3390/proteomes5040025
Griffiths SG, Cormier MT, Clayton A, Doucette AA. Differential Proteome Analysis of Extracellular Vesicles from Breast Cancer Cell Lines by Chaperone Affinity Enrichment. Proteomes. 2017; 5(4):25. https://doi.org/10.3390/proteomes5040025
Chicago/Turabian StyleGriffiths, Steven G., Michelle T. Cormier, Aled Clayton, and Alan A. Doucette. 2017. "Differential Proteome Analysis of Extracellular Vesicles from Breast Cancer Cell Lines by Chaperone Affinity Enrichment" Proteomes 5, no. 4: 25. https://doi.org/10.3390/proteomes5040025
APA StyleGriffiths, S. G., Cormier, M. T., Clayton, A., & Doucette, A. A. (2017). Differential Proteome Analysis of Extracellular Vesicles from Breast Cancer Cell Lines by Chaperone Affinity Enrichment. Proteomes, 5(4), 25. https://doi.org/10.3390/proteomes5040025






