Abstract
The Golgi apparatus (GA) is a crucial organelle in the biosynthesis of non-cellulosic polysaccharides, glycoproteins and proteoglycans that are primarily destined for secretion to the cell surface (plasma membrane, cell wall and apoplast). Only a small proportion of the proteins involved in these processes have been identified in plants, with the majority of their functions still unknown. The availability of a GA proteome would greatly assist plant biochemists, cell and molecular biologists in determining the precise function of the cell wall-related proteins. There has been some progress towards defining the GA proteome in the model plant system Arabidopsis thaliana, yet in commercially important species, such as either the cereals or woody species there has been relatively less progress. In this study, we applied discontinuous sucrose gradient centrifugation to partially enrich GA from suspension cell cultures (SCCs) and combined this with stable isotope labelling (iTRAQ) to determine protein sub-cellular locations. Results from a representative grass species, Italian ryegrass (Lolium multiflorum) and a dicot species, black cottonwood (Populus trichocarpa) are compared. The results confirm that membrane fractionation approaches that provide effective GA-enriched fractions for proteomic analyses in Arabidopsis are much less effective in the species examined here and highlight the complexity of the GA, both within and between species.
1. Introduction
The Golgi apparatus (GA) is the cell’s “engine room” for glycosylation and hence plays an important role in the biosynthesis of non-cellulosic polysaccharides, glycoproteins and proteoglycans, the major macromolecular components of the plant cell surface. The GA is vital for plant growth and development, as well as for responses to abiotic and biotic stress [1]. It is an important component of a very dynamic membranous secretory pathway, which contributes significantly to the challenges of defining the GA proteome [2]. While a number of proteins are resident to the GA, many are transiently associated with the organelle and exert their function elsewhere in the cell. This is the case for all proteins that transit through the GA during secretion to their final destination at either the vacuole or the cell surface. The GA is embedded in a matrix and is also tightly associated with the endoplasmic reticulum (ER), although the way these two organelles are associated is still disputed [3]. Undoubtedly, the similarity of the structural properties of the GA and ER makes them difficult to separate
GA enrichment was successfully used to study enzymes involved in cell wall biosynthesis [4,5] well before attempts were made to define the GA proteome. There has been some progress towards mapping the GA proteome in the model plant system Arabidopsis [6,7,8,9,10]. However, it has been estimated that out of the 2239 ± 465 predicted GA proteins only 20% have been experimentally identified thus far by combining all published Arabidopsis GA proteomes [2]. In contrast, there has been little progress in experimentally defining the GA proteome of commercially important plant species, such as agricultural crops and trees. For example, a total of only 106 and 32 GA proteins have been identified in rice [11,12] and the conifer Pinus radiata [13,14], respectively.
One of the difficulties faced in studying the GA proteome is that there is not one universal purification/fractionation method that works equally for all plant systems. For example, the Free Flow Electrophoresis (FFE) method used in Arabidopsis to produce a GA-enriched fraction of 80% purity [9], performed poorly when applied to Pinus radiata [13], with only 5%–10% of the identified proteins predicted to belong to the GA. Furthermore, the FFE method involves specialized and expensive instrumentation that requires challenging optimization and as a consequence has not been widely adopted. The most straightforward and widely adopted method is sucrose gradient centrifugation even though significant contamination from other organelles is unavoidable. An extension of the sucrose gradient centrifugation technique is the Localization of Organelle Proteins by Isotope Tagging (LOPIT) method which makes the assumption that proteins from the same organelle fractionate similarly on a density gradient; it is typically combined with the use of stable isotope tagging and predictive bioinformatic tools to identify potential GA-resident proteins [6].
Here we have chosen to study suspension cell cultures (SCCs) as they provide a ready source of a homogenous single cell type and overcome the difficult extraction of proteins from resilient woody tissues. Two different plant systems were used, i.e., L. multiflorum and Populus trichocarpa, a commelinid monocot and a woody dicot, respectively, that differ markedly in the polysaccharide composition of their cell walls. The L. multiflorum cell wall is rich in β-1,3;1,4-glucans (mixed linked glucans (MLGs)) and hetero (arabino) xylans [15], while the P. trichocarpa cell wall lacks MLGs but contains other cell wall carbohydrates typical of eudicots, i.e., xyloglucans and pectins rather than heteroxylans. We have used a similar but somewhat simpler approach to LOPIT [6,7], with discontinuous sucrose gradient centrifugation and stable isotope labelling (iTRAQ) to fractionate the sub-cellular membranes and determine protein identity and sub-cellular locations.
2. Materials and Methods
2.1. Plant Material
All chemicals were from Sigma-Aldrich (St. Louis, MO, USA), unless specified. L. multiflorum and P. trichocarpa SCCs were grown as described earlier [16,17].
2.2. Tissue Extraction
Seven-day-old L. multiflorum and P. trichocarpa SCCs were used for protein extraction. SCCs (100 g) were ground in a mortar and pestle with an extraction buffer consisting of 5% (v/v) 0.1 M MES buffer pH 6.5; 5% (w/v) sucrose; 1 mM EDTA; 2 mM EGTA; Roche EDTA-free protease inhibitor cocktail (1 tablet per 50 mL of buffer; Roche Applied Science, Penzburg, Germany) at 4 °C and filtered through two layers of Miracloth (Calbiochem, San Deigo, CA, USA). The filtrates were centrifuged in a Heraeus Multifuge 3 SR (ThermoFisher Scientific, Waltham, MA, USA) at 1000× g for 10 min at 4 °C to remove cell wall debris.
2.3. Microsomal Membrane Preparation
A portion of the tissue extract was used to prepare the microsomal membrane (MM) fractions by centrifuging the previous filtrates at 100,000× g for 60 min in an Optima L-80 XP ultracentrifuge (Beckman Coulter, Brea, CA, USA) and re-suspending the resulting pellet in 0.1 M MES pH 6.5, 1 mM EDTA and 2 mM EGTA.
2.4. Membrane Fractionation
These membrane extracts were layered onto discontinuous gradients consisting of 5 mL each of 20%, 35% and 50% sucrose (w/v), prepared in 0.1 M MES buffer pH 6.5; 5% (w/v) sucrose; 1 mM EDTA; 2 mM EGTA. The discontinuous gradients were centrifuged at 100,000× g for 60 min at 4 °C. Membrane samples were collected from the supernatant/20% (w/v) sucrose interface (S1 fraction), 20% (w/v)/35% (w/v) sucrose interface (S2 fraction) and 35% (w/v)/50% (w/v) sucrose interface (S3 fraction). Each of the fractions was diluted at least 5 times with 0.1 M MES buffer pH 6.5; 5% (w/v) sucrose; 1 mM EDTA; 2 mM EGTA and pelleted by centrifugation at 50,000× g for 45 min at 4 °C. The pellets were washed twice with 0.1 M MES, pH 6.5; 5% (w/v) sucrose; 1 mM EDTA; 2 mM EGTA and re-suspended in the same mixture devoid of sucrose. Protein concentrations were determined using the BCA protein assay kit (ThermoFisher Scientific).
2.5. Enzyme Marker Assays
The latent inosine diphosphatase (IDPase) activity was used as a marker for the GA, the vanadate-sensitive Mg2+-dependent ATPase activity was used as a plasma membrane (PM) marker and the nitrate-sensitive ATPase activity was used as a vacuolar marker as described previously [18,19]. The mitochondrial cytochrome-c oxidase (Sigma-Aldrich, St. Louis, MO, USA), and the endoplasmic reticulum (ER) cytochrome-c reductase (Sigma) assays, were performed following the manufacturer’s instructions.
2.6. SDS-PAGE and Western Blot Analysis
Protein fractions (10 μg of each) were separated using NuPAGE Novex 4%–12% Bis-Tris Protein SDS-PAGE Gels (ThermoFisher Scientific) and proteins transferred onto nitrocellulose membranes using the iBLOT 2 dry blot system (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. Western blotting was performed using the SNAP2 blot system (Merck Millipore, Billerica, MA, USA). Membranes were probed with 1:1000 dilutions of antibodies directed against the following proteins: reversibly glycosylated protein 1 (α-RGP 1) from pea (kindly provided by Kanwarpal Dhugga, Dupont Pioneer, Aurelia, IA, USA) as a marker for the GA, the PM H+-ATPase (Agrisera, Vännäs, Sweden) as a PM marker, cytochrome-c oxidase subunit II (anti-COX II) (Agrisera) as a marker for mitochondria (MT), luminal-binding protein (anti-BiP) (Agrisera) as a marker for ER and the vanadate-sensitive ATPase epsilon subunit (V-ATPase) (Agrisera) as a vacuole marker. Primary antibodies were detected with goat anti-rabbit IgG secondary antibody (1:2000 dilution) conjugated to Pierce horseradish peroxidase (ThermoFisher Scientific) and SuperSignal West Femto Maximum Sensitivity chemiluminescent substrate (ThermoFisher Scientific), then digitally captured using a Chemi-Doc MP imager (Bio-Rad, Hercules, CA, USA) and analyzed using the Image Lab software version 4.1 (Bio-Rad).
2.7. Protein Digestion and iTRAQ Labelling
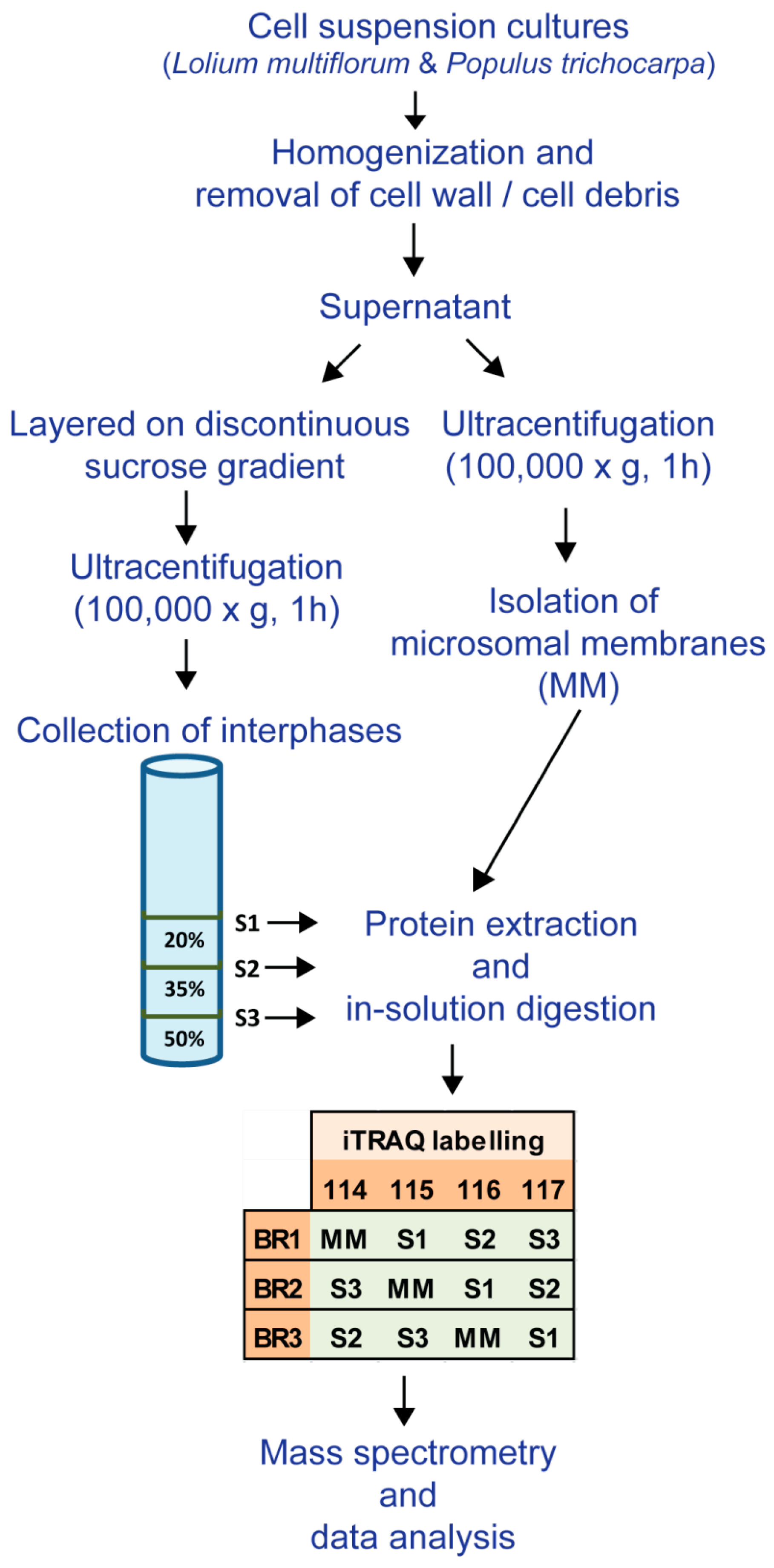
Proteins were solubilized in 6 M urea; 250 mM triethylammonium bicarbonate (pH 8.5), reduced with 5 mM tris-(2-carboxyethyl) phosphine (TCEP) at 60 °C for 1 h, and alkylated with 10 mM methylmethane thiosulphate (MMTS) for 10 min at room temperature (RT). Samples were diluted to 1 M urea with water and proteins were digested with trypsin (4 μg; 1:25 w/w, sequencing grade, Promega, Madison, WI, USA) for 16 h at 37 °C. Proteolysis was stopped with the addition of 10 μL neat formic acid and peptide solutions were desalted using a Sep-Pak C18 Plus Short cartridge (Waters, Milford, MA, USA) as described previously [20]. Samples were concentrated under vacuum to approximately 10 μL and TEAB was added to each tube to reach a final volume of 30 μL. Peptides were labeled with iTRAQ 4plex reagent (Sciex, Framingham, MA, USA) following the manufacturer’s instructions. A separate set of iTRAQ tags were used for each biological replicate and labelled as shown in Figure 1.
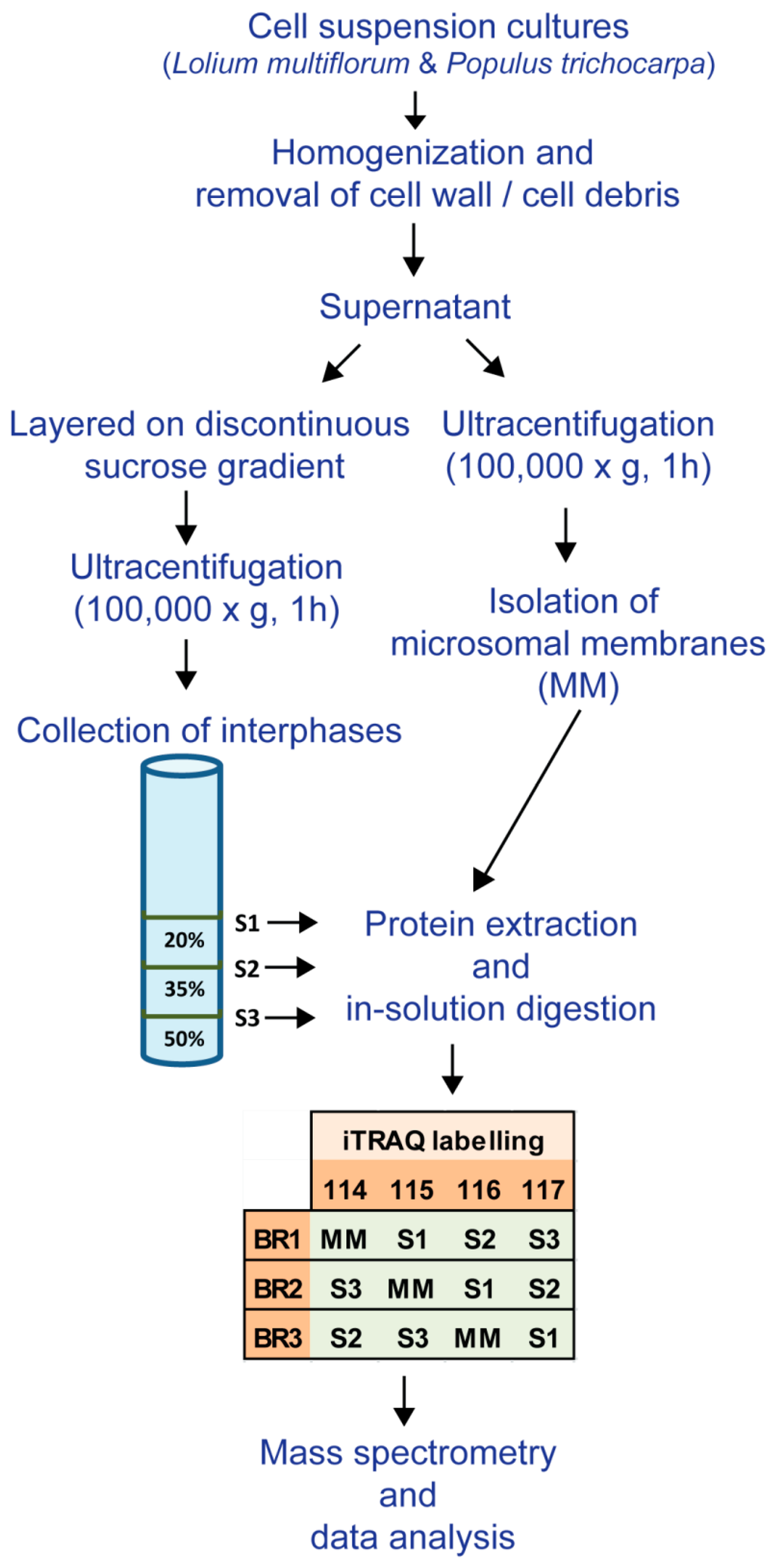
Figure 1.
Experimental workflow used for the enrichment of microsomal membranes (MMs), the subcellular fractions (S1, S2 and S3) and the iTraq labelling of the biological replicates (BR1, BR2 and BR3) for the different membrane fractions from L. multiflorum and P. trichocarpa suspension cell cultures (SCCs).
2.8. Non-labelled Protein Hydrolysis
Three biological replicates of the L. multiflorum S1 fraction and 2 biological replicates of the S1 P. trichocarpa were re-suspended in 6 M urea; 100 mM ammonium bicarbonate and reduced with 10 mM DTT at 60 °C for 1 h. The samples were allowed to cool and then alkylated with 50 mM iodoacetamide for 45 min at RT. The samples were diluted to 1 M urea with 100 mM ammonium bicarbonate and hydrolyzed with trypsin (4 μg; 1:25 w/w, sequencing grade, Promega) for 16 h at 37 °C. Proteolysis was stopped with 1 μL of neat formic acid and the sample desalted using a Sep-Pak C18 Plus Short cartridge (Waters) as described previously [20]. Samples were concentrated under vacuum to approximately 100 μL.
2.9. Peptide Fractionation and Mass Spectrometry
iTRAQ and the L. multiflorum S1 samples were fractionated by strong cation exchange (SCX) chromatography and fractions analyzed by LC-MS/MS as described in Ford et al. [20]. Peptides (1 μg) from the P. trichocarpa S1 fraction were analyzed on a Q Exactive Plus mass spectrometer (Thermo Scientific) coupled to an Ultimate 3000 RSLC nanosystem (Dionex, Sunnyvale, CA, USA). The nanoLC system was equipped with an Acclaim Pepmap nano-trap column (Dionex) and an Acclaim Pepmap analytical column (Dionex), operating at a flow rate of 3 μL·min−1 with a 90 min gradient of 3%–80% (v/v) acetonitrile containing 0.1% formic acid. The Q Exactive Plus mass spectrometer was operated in positive mode, with the spray voltage set to 1800 kV, S-lens RF level at 50 and heated capillary at 250 °C. Peptides were fragmented using normalized collision energy of 35 and activation time of 0.1 ms in the data-dependent mode, whereby the top 10 ions between 400 and 1600 m/z with a charge state between 2+ and 5+ were selected for MS/MS.
2.10. Protein Identification
Mass spectra obtained from the L. multiflorum SCCs were searched against an in-house database for protein predictions from a L. multiflorum SCC transcriptome and the Lolium perenne V1 protein database [21]. Data from the P. trichocarpa samples were searched against the Populus protein database V3 [22] using ProteinPilot version 4.5 (Sciex) with the following parameters: Sample type: iTRAQ 4plex (Peptide labelled); Cys Alkylation: MMTS; Digestion: Trypsin; Search Effort: Thorough ID. Peak lists generated by ProteinPilot were exported and used to search against the same databases using MASCOT version 2.4 (Matrix Science, London, UK). The MASCOT parameters were: Enzyme: Trypsin; Fixed modifications: iTRAQ 4plex (N-term), iTRAQ 4plex (K), Methylthiol (C); MS peptide tolerance: 10 ppm; MS/MS tolerance: 0.15 Da; Number of missed cleavages: up to 1. The peptide list from MASCOT was generated with a p < 0.05. The peptide summary report was exported from ProteinPilot and filtered by confidence level according to the local 5% false discovery rate reported by ProteinPilot. Peptides found using both search algorithms were selected for further analysis. Peptides were used to search the databases (in the case of L. multiflorum the databases were combined into one) using KNIME [23] to find all proteins containing any of the peptides. The list was then reduced to a minimum set of proteins by selecting only those that contained two or more peptides; I and L were treated as the same amino acid. Where 2 or more proteins had the same set of peptides matching, the protein with the longest sequence was selected.
The non-labelled spectra from the S1 protein profiling experiment were analyzed similarly to the iTRAQ spectra except for the following ProteinPilot parameters: Sample type: Identification; Cys Alkylation: Iodoacetamide; Digestion: Trypsin; Search Effort: Thorough ID. Peak lists generated by ProteinPilot were exported and used to search against the same databases using MASCOT version 2.4 (Matrix Science). The MASCOT parameters were: Enzyme: Trypsin; Fixed modifications: carbamidomethyl (C); MS peptide tolerance: 10 ppm; MS/MS tolerance: 0.15 Da; Number of missed cleavages: up to 1.
BLAST analyses were performed on the identified proteins against the TAIR10 [24] database and the Oryza sativa subsp. japonica Uniprot reference proteome (downloaded November 2015) [25]. Arabidopsis proteins with the highest similarity were used to search SUBA3 [26] for protein sub-cellular location based on the experimental data rather than the predicted locations and the Carbohydrate-Active Enzymes database (CAZy) [27] for possible cell wall-associated proteins. Sequences from the P. trichocarpa and O. sativa proteins with the highest similarity to the L. multiflorum sequence were used for analysis with SignalP 4.0 [28] to determine the possible occurrence of a signal peptide.
2.11. Relative Quantification
The reporter ion peak areas for the peptides identified above were imported into KNIME [23] from the peptide summary report generated in ProteinPilot and used for relative quantitation as applied in Ford et al. [20], with some modifications. Briefly, any peptide with a reporter ion peak area of less than 20 was removed from quantification. Only unique peptides and peptides with a charge state of either 2+ or 3+ were used for quantification. Each peptide signal was then normalized by the sum of the corresponding channel intensities (114, 115, 116 and 117). Peptides were ignored when the normalized peptide value was more than 2 SD units from the calculated mean of the protein the peptide matched to. The mean was then calculated for proteins with two or more peptides that fulfilled the above criteria. A change was considered significant if it was greater than 1.3-fold, based on a 1% FDR calculated from a same-same iTRAQ control experiment (Table S1).
Proteins were clustered using the k-means clustering node in KNIME [23] with k set to 13.
3. Results
3.1. Enrichment of Microsomal Membranes
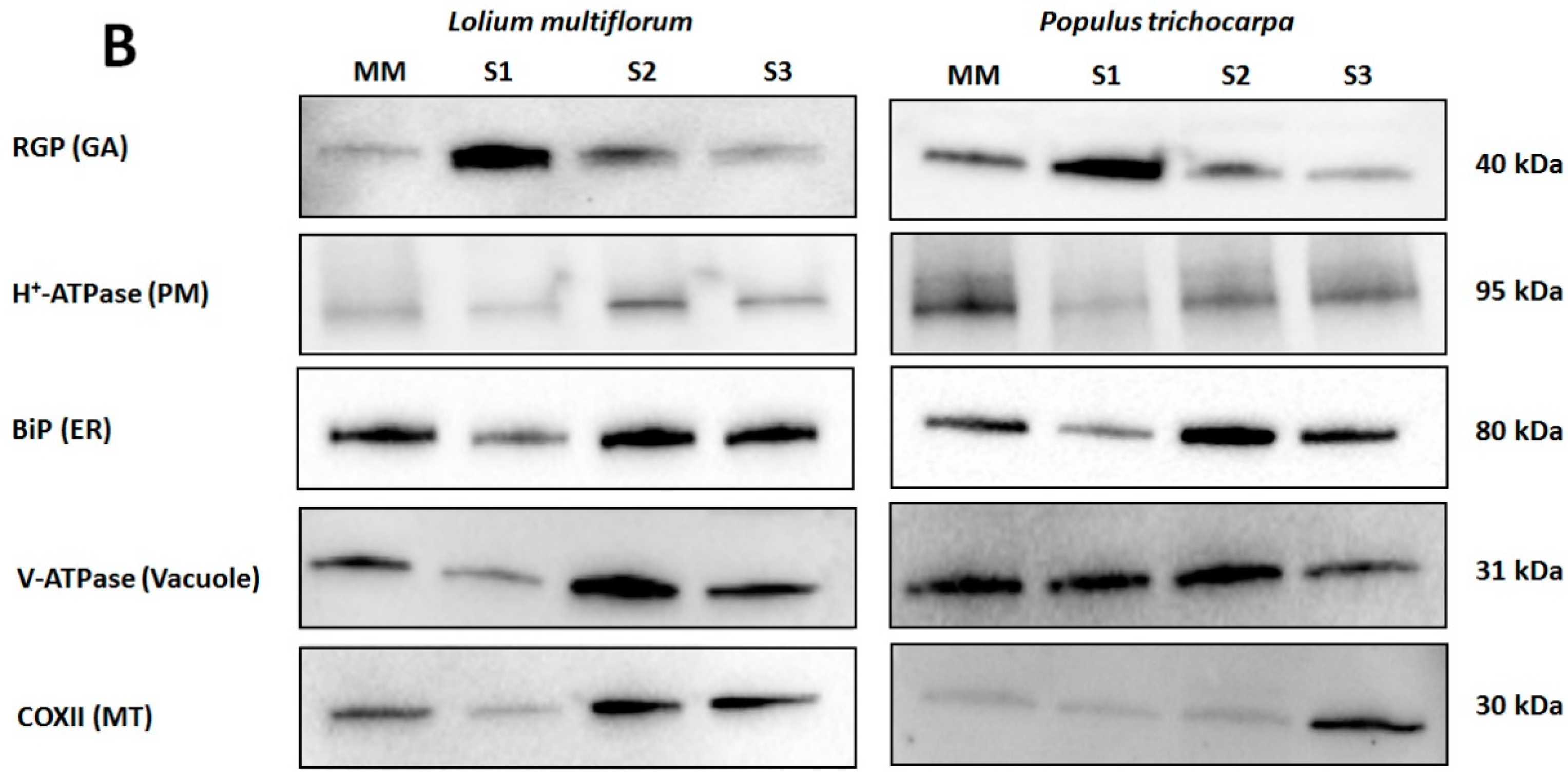
Enrichment of microsomal membranes (MMs) was assessed by enzyme marker assays and Western blot analysis using antibodies to proteins known to be located in particular membrane types. For L. multiflorum the S1 fraction showed an increase in IDPase acitivity (GA) (Table 1) consistent with an increase of α-RGP 1 (GA) signal compared to all other fractions (Figure 2B). Decreases in both the nitrate- and vanadate-sensitive ATPases (vacuole and PM, respectively) as well as the cytochrome-c oxidase assay (MT) in S1 compared to all other fractions (Table 1) are consistent with the decrease in H+-ATPase (PM), COX II (MT) and the V-ATPase (vacuole) markers, respectively. The ER markers, cytochrome-c reductase activity (Table 1) and BiP (Figure 2B) showed a small decrease in S1 compared to all other fractions. Thus, for L. multiflorum the S1 fraction was chosen for proteomic analyses as this was assumed to be the most “GA-enriched” fraction by both the Western blot antibody marker distributions and the marker enzyme activity with a calculated 1.7-fold enrichment for the latter.

Table 1.
Enzymatic marker assays of fractions MM, S1, S2 and S3 from L. multiflorum SCCs.
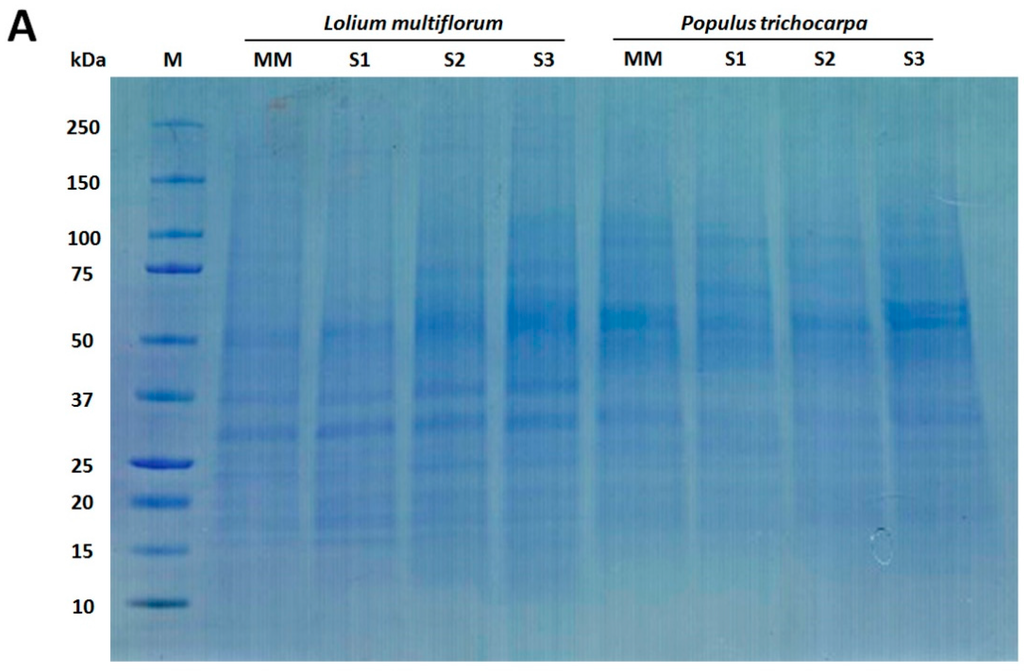
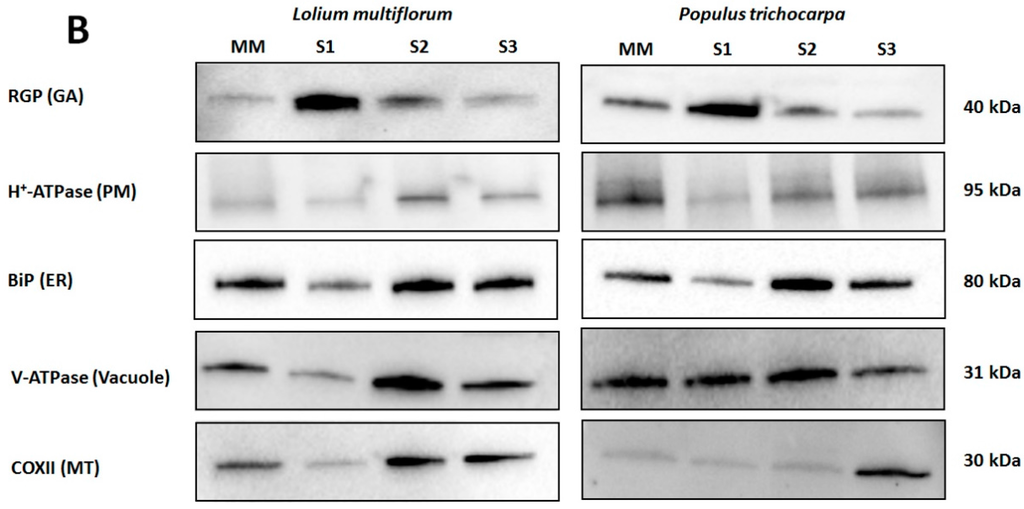
Figure 2.
Western blot analysis of Lolium multiflorum and Populus trichocarpa suspension cell culture (SCC) membrane fractions. Fractions (10 μg of total protein each) from MM, S1, S2, and S3 were separated by SDS-PAGE. (A) Coomassie blue staining of the total proteins separated on SDS-PAGE demonstrating the equal protein loading across the membrane fractions, (B) Western blots performed with anti-RGP (GA marker), anti-H+-ATPase (PM marker), anti-BiP (ER marker), anti-V-ATPase (vacuole marker), and anti-COX II (MT marker) antibodies. The molecular masses of the respective marker proteins are indicated on the right.
In P. trichocarpa the S1 fraction showed an increased α-RGP 1 (GA) signal compared to the MM, S2 and S3 fractions as well as decreases in H+-ATPase (PM), COX II (MT) and the BiP2 (ER) markers (Figure 2B), consistent with an enrichment of GA. There appears to be considerable vacuolar contamination with little variation in the V-ATPase (vacuole) marker (Figure 2B) in the S1 compared to the S2 and MM fractions.
3.2. Mass Spectrometric Analyses
3.2.1. Protein Profiling of the Non-labelled GA-Enriched Fractions from L. multiflorum and P. trichocarpa
The L. multiflorum S1 fraction contained 835 proteins that were identified with at least 2 peptides, in at least 2 biological replicates (Table S2) whereas the P. trichocarpa counterpart contained 1236 proteins identified with at least 2 peptides, in 2 biological replicates (Table S3). BLAST analysis of the L. multiflorum proteins against the Arabidopsis database revealed that 79 of these proteins are known to reside in the GA, 127 proteins have been identified as GA-localized in previous proteomic analysis [6,7,8,9,10,11,29] and 35 proteins present significant levels of similarity to proteins in the CAZy database [27]. The O. sativa sequence with the highest similarity to each L. multiflorum sequence was used for signal peptide prediction using SignalP as more complete sequences were needed. There were 79 proteins identified with a predicted signal peptide, indicating that they are destined for delivery to post-Golgi membrane compartments downstream in the secretion pathway. In total, this provided a list of 211 (25%) of the identified proteins conceivably either residing or transiting through the GA. The same analysis of the P. trichocarpa proteins revealed 69 proteins known to reside within the GA, 104 proteins identified as GA-localized in previous proteomic analysis [6,7,8,9,10,11,29], 33 proteins with similarities to proteins in the CAZy database and 96 proteins with a predicted signal peptide. In total, 227 (18%) proteins were identified that likely reside or transit through the GA. Of the remaining L. multiflorum proteins there were 81 (10%) known MT proteins, 34 (4%) PM proteins, 37 (4%) ER proteins, 42 (5%) cytoplasmic proteins (CP), 29 (3%) nuclear proteins, 27 (3%) plastid proteins, 8 (1%) vacuolar proteins and 375 (45%) proteins without a known sub-cellular location. Comparatively for the remaining P. trichocarpa proteins, 80 (6%) were known MT proteins, 68 (6%) PM proteins, 11 (1%) ER proteins, 137 (11%) cytoplasmic proteins (CP), 26 (2%) nuclear proteins, 47 (4%) plastid proteins, 23 (2%) vacuolar proteins and 628 (51%) proteins without a known location.
Amongst the GA proteins identified in the L. multiflorum S1 fraction was the GA marker protein RGP1 (Table S2). Even though RGP1 was not identified in P. trichocarpa, RGP2 and RGP3, two proteins from the same family, were identified in this species (Table S3). In both the grass and tree cell samples, the more abundant GA proteins were the endomembrane 70 (EMP70) proteins and S-adenosyl-l-methionine-dependent methyltransferases (SAM). Enzymes active on nucleotides or involved in nucleotide-sugar interconversion that were reported in other GA proteomes were also present in both species, such as apyrase 1, UDP-d-glucose/UDP-d-galactose 4 epimerase (UGE1) and UDP-d-glucose pyrophosphorylase 2 (UGP2) in L. multiflorum and UGP2 and an uncharacterized nucleotide-diphospho-sugar transferase in P. trichocarpa. Proteins expected to reside in the trans-Golgi network (TGN) were well represented, with the presence of RAB GTPases and vacuolar sorting receptors (VSR) in both species, with the addition of the YIP1, syntaxins and transport protein particle (TRAPP) proteins in L. multiflorum (Table S2) and the N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) and vesicle-associated membrane proteins (VAMP) in P. trichocarpa (Table S3). Proteins expected to be more loosely associated with the GA and identified within the S1 fraction in both species were the coatomer complex proteins, with the addition of tether proteins in L. multiflorum, such as the conserved oligomeric Golgi complex (COG) and golgin candidate 6 (GC6).
A more noticeable difference between the two species was observed in the glycosyltransferases (GTs) identified in the GA-enriched fractions. In L. multiflorum there were 5 proteins identified from the GT2 family involved in cell wall biosynthesis, i.e., cellulose synthase-like F6 (CSLF6) involved in the synthesis of MLG [30], cellulose synthases involved in primary cell wall biosynthesis (CESAs 1, 3 and 6) and a cellulose synthase-like D (CSLD) possibly involved in (1-4)-β-glucan [31] or mannan [32] activity. Five glucan synthase-like (GSL) proteins predicted to be responsible for callose biosynthesis [33] were identified from the GT48 family. Other GTs present in the L. multiforum samples were 3 GTs from the GT61 family and 1 from the GT47 family, both families have been implicated in xylan biosynthesis [34,35], a GT29 protein, possibly involved in pectin biosynthesis [36], the UDP-glucose: glycoprotein glycosyltransferase (EBS1) involved in protein folding and an uncharacterized GT with a domain of unknown function (DUF707) [27]. In P. trichocarpa neither CESA nor GSL proteins were observed, or any other GT involved in cell wall biosynthesis, possibly due to the lower GA-enrichment in the samples from this species combined with an expected low abundance of GTs. The only similarity between the lists of GTs observed in the samples from the two species was the presence of the dolichyl-diphospho-oligosaccharide protein glycosyltransferase (DGL1) involved in protein N-glycosylation [37] from the GT2 family and trehalose phosphatase/synthase 7 (ATTPS7) involved in secondary metabolism [38] from the GT20 family, both contaminants from the ER and the CP, respectively. In addition, a different set of GTs was observed in the samples from P. trichocarpa, which contained 3 GT1 family members, UDP-glycosyltransferase 71B1 (UGT71B1), UDP-glycosyltransferase 73B4 (UGT73B4) and an uncharacterized UDP-glycosyltransferase, as well as the UDP-glycosyltransferase/trehalose phosphatase (ATTPS6) from the GT20 family. These 4 GTs are most likely involved in secondary metabolism rather than cell wall biosynthesis.
3.2.2. iTRAQ Analysis of the L. multiflorum and P. trichocarpa GA-enriched Fractions
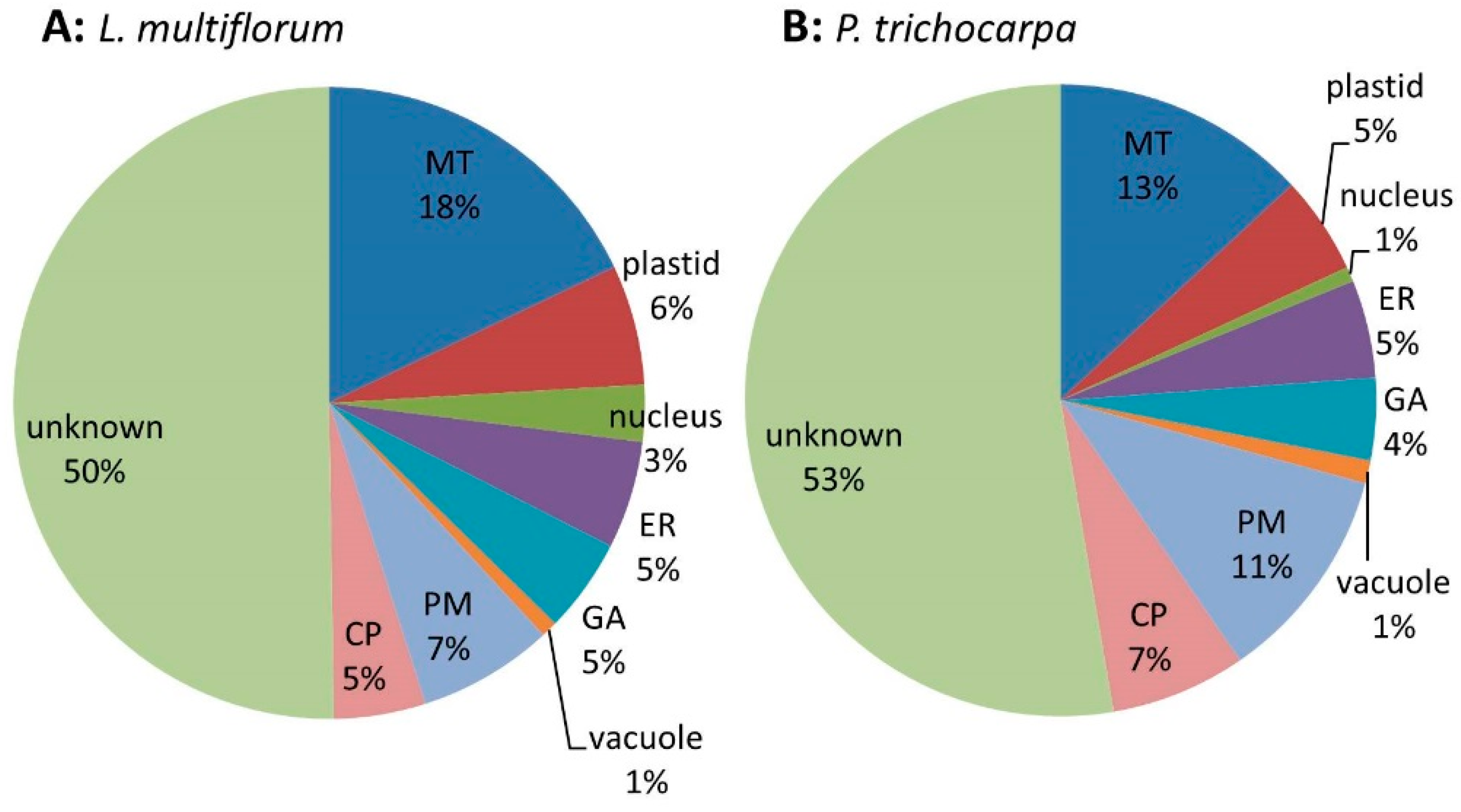
Samples prepared from three biological replicates were fractionated into MM, S1, S2 and S3 and their protein composition was compared using the iTRAQ technology. Quantitative data were obtained for a total of 619 proteins (Table S4) across all 3 biological replicates from L. multiflorum. These proteins were assigned putative sub-cellular locations based on experimental data of Arabidopsis proteins (SUBA3) with the highest similarity to the Lolium sequences. Proteins that have been shown to reside in multiple locations were assigned as ‘unknown’. Of the 619 proteins 18% were predicted to be located in MT, 6% in plastids, 3% in nucleus, 5% in the ER, 5% in GA, 1% in the vacuole, 7% in the PM, 5% in the cytosoluble phase (CP) and the remaining 50% as unknown (Figure 3A). Quantitative data were obtained from 260 proteins in the P. trichocarpa samples (Table S5) with similar proportions as for L. multiflorum. These proteins are predicted to belong to the various subcellular organelles, with 13% assigned to MT, 5% to plastids, 1% to nucleus, 5% to ER, 4% to GA, 1% to vacuole, 11% to PM, 7% to CP and the remaining 53% as unknown (Figure 3B).
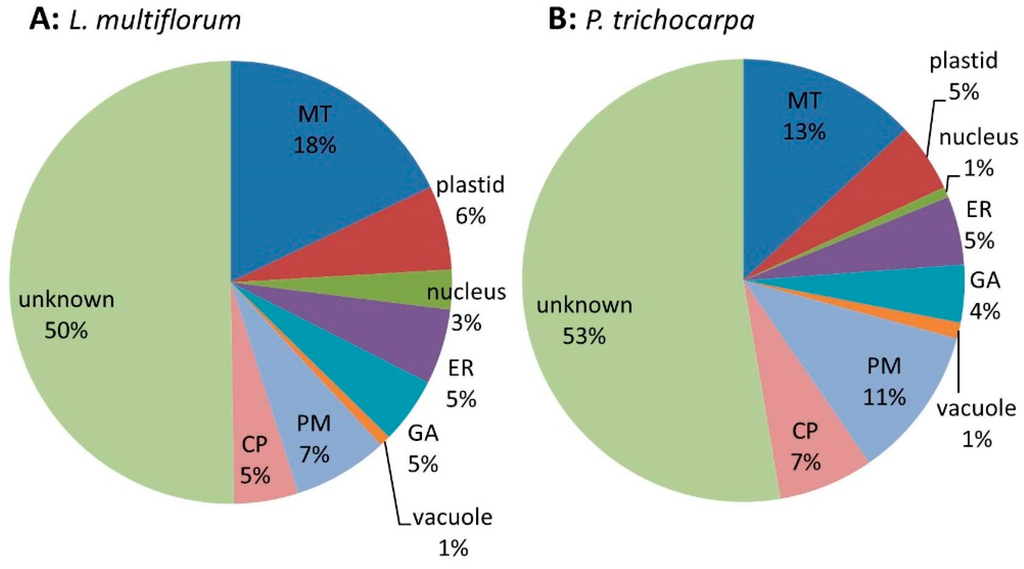
Figure 3.
Pie chart of the expected sub-cellular locations of (A) the 619 proteins from L. multiflorum and (B) the 260 proteins from P. trichocarpa from which quantitative data were obtained. The pie charts show a similar distribution of proteins in the different sub-cellular compartments, i.e., mitochondria (MT), endoplasmic reticulum (ER), Golgi apparatus (GA), plasma membrane (PM), vacuole and cytoplasmic (CP).
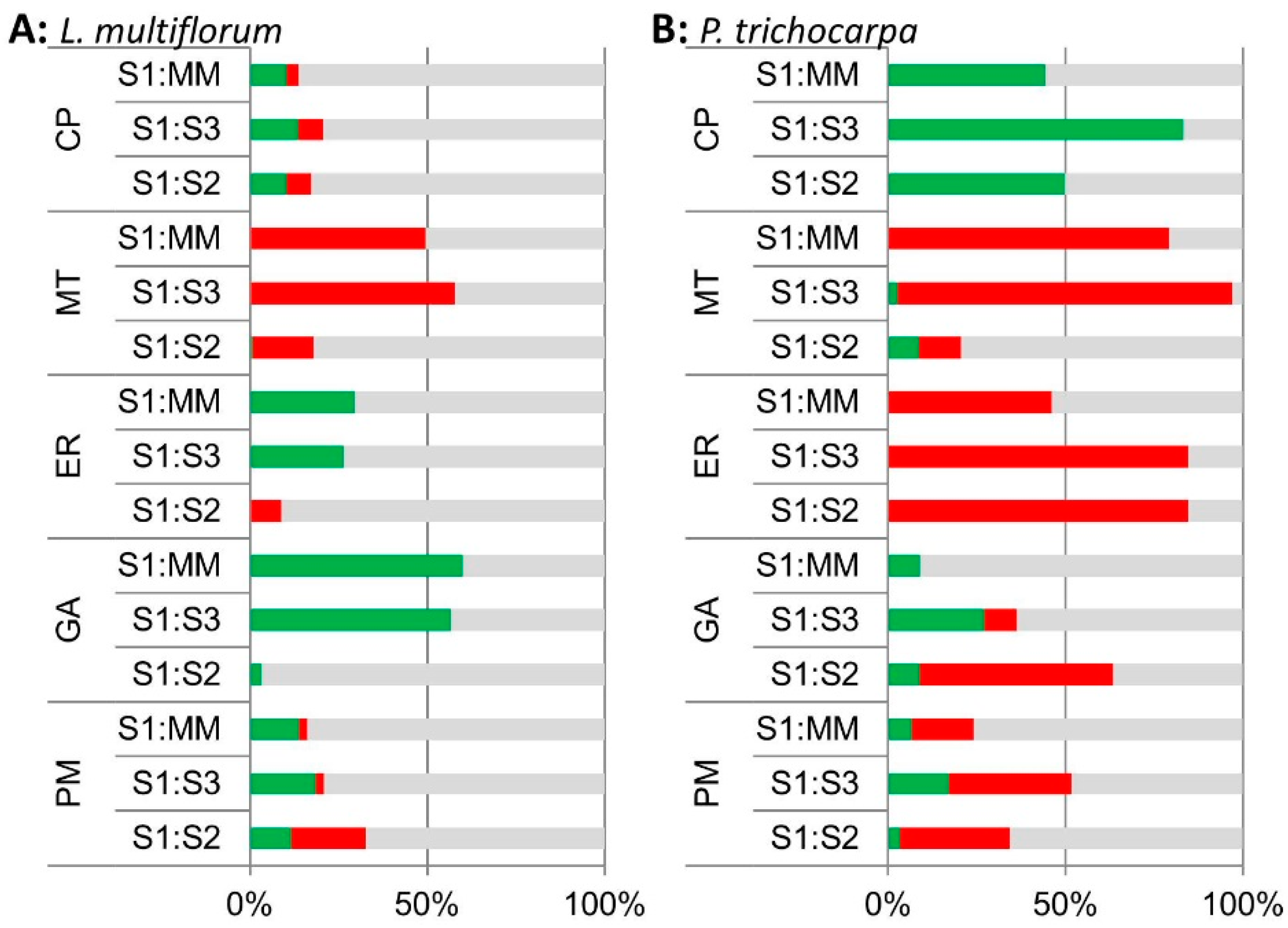
A comparison of the percentage of proteins changing in their iTRAQ ratios in the different predicted sub-cellular locations identified significant increases in GA and ER proteins in S1 compared to MM (S1:MM) and S1:S3 in the L. multiflorum samples (Figure 4A). Surprisingly, the P. trichocarpa samples had a different pattern, with most significant increases occurring for CP proteins in S1:S3, S1:S2 and S1:MM, and only some increases in GA proteins in S1:MM and S1:S3, most of which decreased in S1:S2 (Figure 4B). Samples from both plant species showed a decrease in MT protein in S1 relative to MM, S2 and S3.
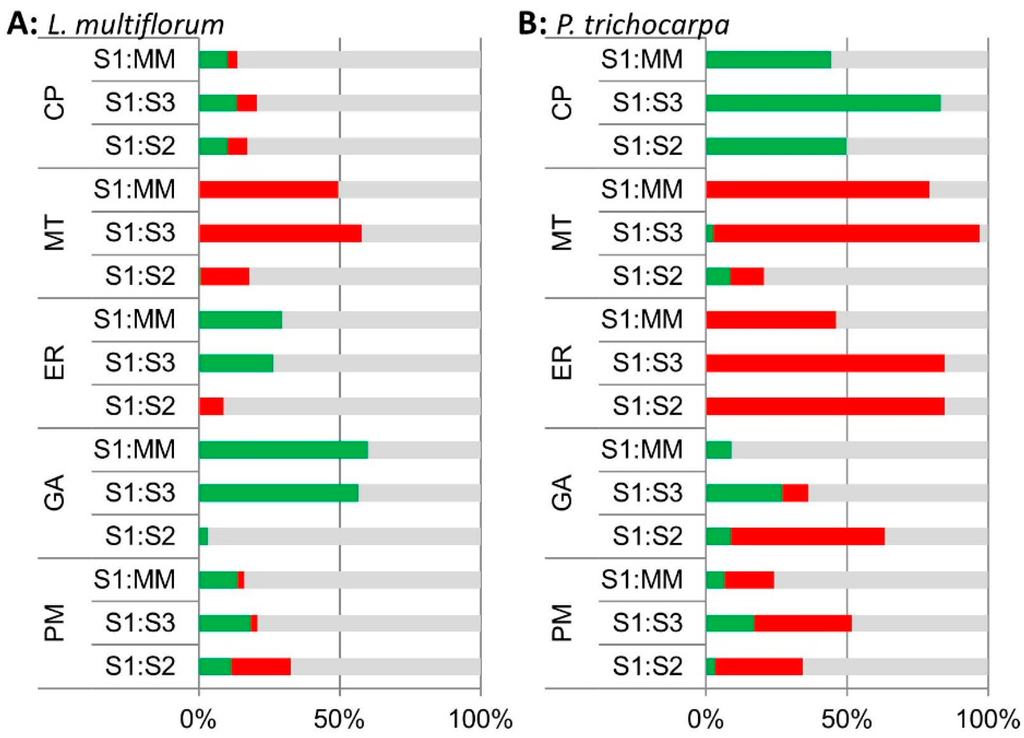
Figure 4.
Bar chart of the percentage of proteins in the S1:MM, S1:S3 and S1:S2 iTRAQ ratios that are significantly enriched (green), depleted (red) and unchanged (grey) across all 3 biological replicates based on their expected sub-cellular locations; cytoplasmic proteins (CP), mitochondria (MT), endoplasmic reticulum (ER), Golgi (GA) and plasma membrane (PM) locations in (A) L. multiflorum and (B) P. trichocarpa samples based on iTRAQ analyses.
Of the thirty proteins that were assigned as GA-localized in L. multiflorum, only one, the vacuolar sorting receptor homolog 1, was significantly increased in S1:S2 (Table S4). The GA proteins are mostly enriched in S1:S3 and S1:MM (Figure 4A). There were a total of 79 proteins significantly enriched across all 3 biological replicates in S1:S3 and S1:MM. Over half (17) of the GA proteins were significantly enriched in both S1:S3 and S1:MM, together with 3 CP proteins, 8 ER proteins, 6 PM proteins, 2 nuclear proteins, 2 vacuolar proteins, 1 plastid protein and 40 proteins with an unclear, hence unassigned, sub-cellular location. Collectively, approximately 30% of these proteins (24) have previously been observed as GA-localized in other proteomics experiments [6,7,8,9,10,29]. These included GA proteins identified in the non-labelled protein profiling of the S1 fraction, EMP70, VSR, apyrase and nucleotide-sugar interconversion proteins.
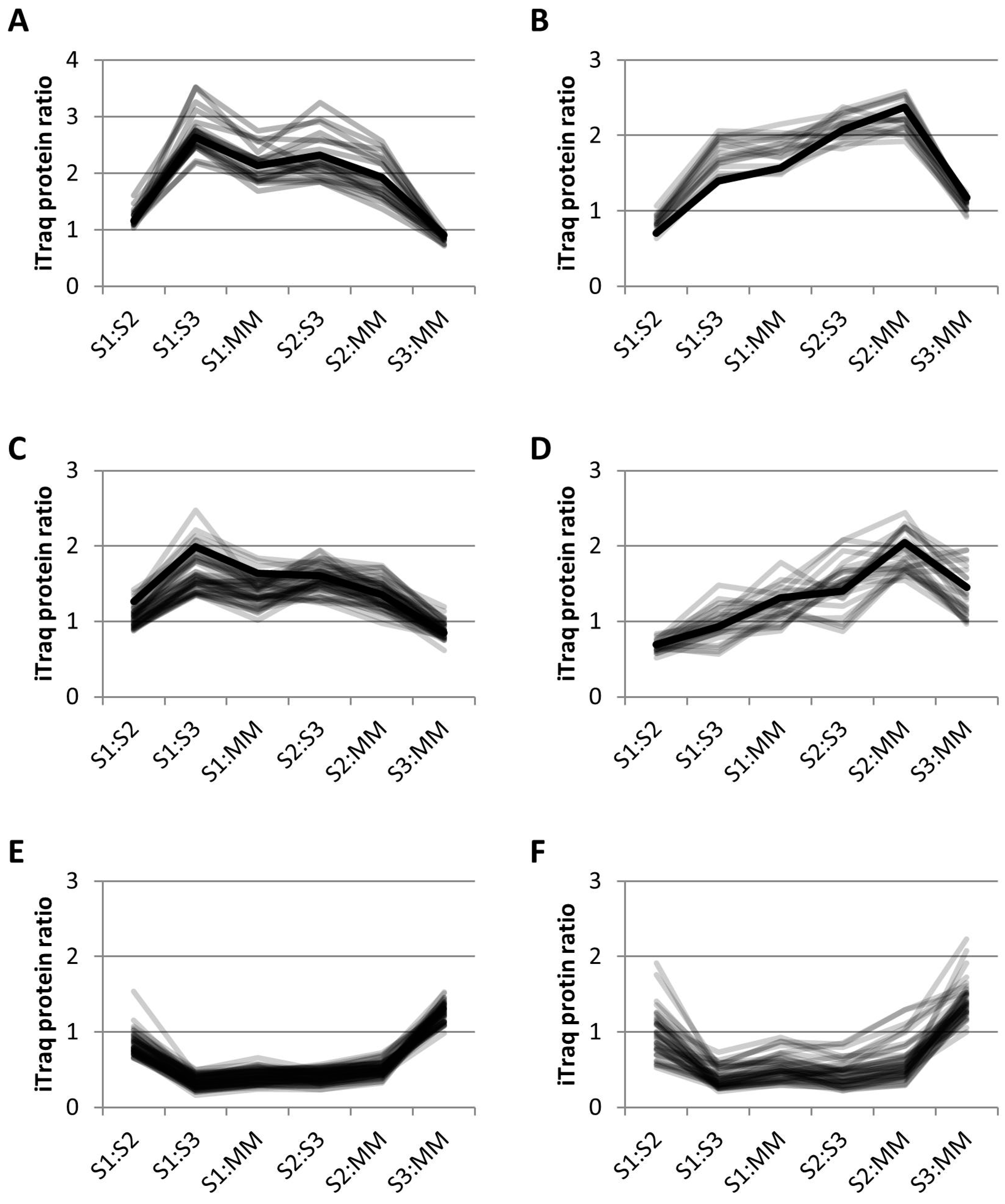
Clustering using k-means (see Materials and Methods) separated the iTRAQ data into 13 different clusters. Of the 24 proteins previously identified as GA-localized, 13 clustered together using k-means clustering, along with 9 other proteins in cluster 13 (Figure 5A). Sixty-eight percent of the proteins in this group have been identified in previous proteomics studies (Table 2). In this cluster, there were 22 proteins that included 13 GA proteins, 2 vacuolar proteins and 7 unassigned proteins. There were 4 EMP70 family proteins in this cluster that are reportedly the most abundant GA proteins [8]. Although the monosaccharide transporter is labelled as a tonoplast protein in the TAIR10 Arabidopsis database, it contains a KXD/E motif at the C-terminus, indicative of GA localization [39], as observed previously [9]. There were 2 nucleotide-sugar biosynthetic proteins, i.e., a UDP-glucuronic acid decarboxylase and the UDP-arabinose 4-epimerase 1, as well as a putative sugar phosphate/phosphate translocator. The L. multiflorum protein p6229 contains an O-fucosyltransferase (FucT)-like domain and has some homology to an unclassified Arabidopsis GT. The apyrase 1 protein is within this cluster and has previously been localized to the GA both by proteomics and microscopy [9]. Thus, the 22 proteins in this cluster could all conceivably be GA proteins.
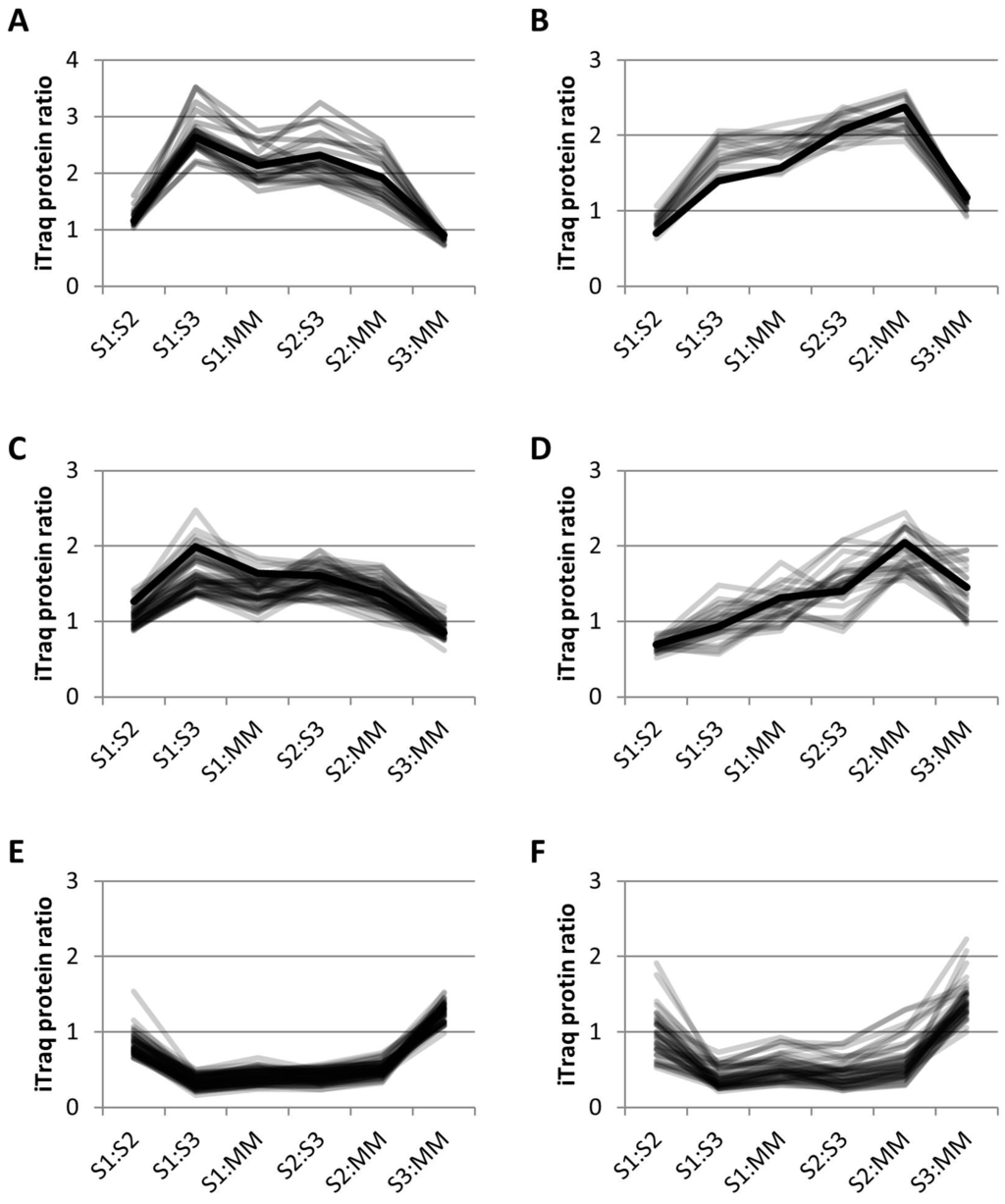
Figure 5.
Relative abundance profiles using k-means clustering of iTRAQ data. (A) L. multiflorum cluster 13; 22 proposed GA proteins in grey and the profile of apyrase, a well characterized GA protein in black; (B) L. multiflorum cluster 12; 13 proposed ER proteins in grey and the profile of the ER dolichyl-diphosphooligosaccharide glycosyltransferase subunit 2 (DGL2) in black; (C) L. multiflorum cluster 9; 49 proposed TGN proteins in grey and known TGN protein, vacuolar sorting protein 45 (VSPS45) in black, (D) P. trichocarpa cluster 9; 25 proposed TGN proteins in grey and known TGN protein, vacuolar ATPase subunit D (VATD) in black, (E) L. multiflorum cluster 11; 61 proposed MT proteins, and (F) P. trichocarpa cluster 11; 44 proposed MT proteins.

Table 2.
Proteins from cluster 13 (representing GA proteins) based on the iTRAQ analysis of L. multiflorum fractions.
Thirty-one percent of the proteins in cluster 12 (Figure 5B, Table S6) have previously been identified in GA proteomes. This cluster contained 7 out of the 8 ER proteins that are significantly enriched in S1:S3 and S1:MM (Figure 4A), as well as 6 other proteins expected to be ER-localized, together with 1 plastid and 5 unassigned proteins. Included in this cluster was DGL2, an essential subunit of the N-oligosaccharyltransferase (OST) complex which catalyses the transfer of a high mannose oligosaccharide from a lipid-linked oligosaccharide donor to a nascent polypeptide chain; an evolutionarily conserved and a well characterized process that occurs in the ER [40]. The plastid protein has been observed in previous proteomics studies in various locations such as the PM, plastid, MT and GA (SUBA database [26]). This cluster could be conceivably labeled as ER-localized, with 72% of the proteins known to localize to the ER.
Cluster 9 (Figure 5C, Table S7) from L. multiflorum contained 12 GA proteins as well as 5 ER, 2 plastid, 1 nuclear, 1 vacuolar, 5 PM and 24 unassigned proteins. The 12 GA proteins in this cluster are 6 vacuolar ATPases, 2 vacuolar sorting proteins, 3 Rab GTPases and an alpha SNAP protein, all of which are expected to reside in the TGN. The vacuolar sorting protein VSP45, a regulatory protein of the SNARE complex, is present in this cluster and has been shown to be localized in the GA using proteomics as well as immunocytochemistry [29]. Four of these proteins were also identified in the P. trichocarpa cluster 9 (Figure 5D, Table S8). This cluster contained the greatest number of GA proteins (4) compared to the other 12 clusters from P. trichocarpa, along with 2 PM proteins and 1 each from ER, MT, plastid and vacuole; the remaining 15 proteins were not assigned. Cluster 9 from both species appears to contain either TGN proteins or cargo proteins, although the profiles are slightly different (Figure 5C,D), with a higher enrichment in S1:S3 for L. multiflorum samples and S2:MM for P. trichocarpa.
In L. multiflorum the majority of the proteins depleted in S1:S3 and S1:MM are proteins expected to reside in the MT and plastid. The cluster with the most significant decreases, i.e., cluster 11 (Figure 5E), contained 42 MT, 17 plastid and 2 unknown proteins. The P. trichocarpa sample produced a cluster with a similar profile (Figure 5F) which contained 31 MT, 6 ER, 1 PM, 1 plastid, 1 vacuolar and 4 unknown proteins. In both species, cluster 11 could be considered as representing MT proteins.
4. Discussion
Although there have been considerable efforts made in defining the GA proteome in Arabidopsis, there are only two published studies in cereals (rice [11,12]) and 2 in conifers [13,14]). Here we provide a comparative analysis of the GA proteomes of a dicot (P. trichocarpa) and a commelinid monocot (L. multiflorum). For this purpose, we have used classical membrane fractionation procedures available in most laboratories (sucrose density gradient centrifugation). Profiling of the GA-enriched fractions (S1) in both species revealed the occurrence of known resident GA proteins and TGN proteins. The list of identified proteins comprised loosely associated and/or tethered proteins and transient or cargo proteins, all of which confirmed significant enrichment of intact GA. Interestingly, the GA-associated proteins that are present in the samples from both species, such as the coatomer complex proteins and RGP proteins, have not been identified in the Arabidopsis GA proteome [2].
Most noticeably absent from the P. trichocarpa S1 fraction were GTs involved in cell wall biosynthesis, possibly due to the lower level of GA enrichment in the samples prepared from this species. Although many expected GA proteins were present in the S1 fraction from both species, only 25% and 18% of the proteins in this fraction were assigned as GA proteins in L. multiflorum and P. trichocarpa, respectively. Similar limited levels of enrichments have been reported for other species [13]. Approximately 30% of the proteins in the GA-enriched fractions from L. multiflorum and P. trichocarpa were assigned to other organelles. As these data did not represent sufficient evidence for any biological conclusions, a relative quantitation approach was taken using the iTRAQ technology.
Although the marker enzyme assays and the immunological characterization of the different fractions suggested that the S1 fraction was enriched in GA compared to all other fractions, this was not reflected in the S1:S2 ratio following sucrose density gradient fractionation. The cut-off chosen for enrichment in the iTRAQ analysis was 1.3, i.e., very similar to the fold observed (1.4) in the enzyme assays. When combined with the inherent problem of ratio suppression in the iTRAQ method [41], this lead to virtually no observed changes in GA proteins in S1 compared to S2. Thirty known GA proteins were identified in the L. multiflorum sample using the iTRAQ approach, with 17 of these proteins enriched in S1:S3 and S1:MM. In addition, enriched in both S1:S3 and S1:MM were proteins known to reside in other sub-cellular locations, i.e., 3 CP proteins, 8 ER proteins, 6 PM proteins, 2 nuclear proteins, 2 vacuolar proteins and 1 plastid protein. As the ER is closely associated to the GA and therefore difficult to separate, it is a common contaminant in GA preparations prepared by density centrifugation. This is reflected in the enrichment of 8 ER proteins in S1:S3 and S1:MM in L. multiflorum. To discriminate the GA proteins from others, k-means clustering was applied to the iTRAQ data, producing 13 clusters from the samples from both species. A potential GA cluster (cluster 13) and a potential ER cluster (cluster 12) was observed in L. multiflorum. No similar clusters were observed in P. trichocarpa, although potential TGN (cluster 9) and MT (cluster 11) clusters were observed.
Although the immunological analyses of the P. trichocarpa profile was similar to that of L. multiflorum, the “enrichment” was not reflected in the proteomic analysis. Thus, the iTRAQ analysis did give some insights into the GA proteome of L. multiflorum but not for that of P. trichocarpa. Whilst a similar distribution of sub-cellular locations of the identified proteins was observed the trends in iTRAQ ratios were not and neither were members of the most abundant GA protein family, the EMP70 detected.
5. Conclusions
Protein profiling of the GA-enriched fractions allowed the identification of approximately 220 GA proteins from each species, but 30% of the proteins found in the GA-“enriched” fractions were considered to be contaminating proteins. The iTRAQ analyses only identified 83 potential GA proteins when cluster 13 (GA) and cluster 9 (TGN) are considered in L. multiflorum and 26 proteins in P. tricocarpa from cluster 9 (TGN). Although iTRAQ analysis is quantitative and hence gives more definitive localization information it is not as comprehensive as qualitative protein profiling due to the inherent limitations of the methodology. Development of methods allowing greater enrichment of the GA is needed in order to gain meaningful data from a protein profiling experiment. There is evidence to suggest that GA morphology can change during active secretion [42] and although we chose SCCs as a source of homogenous single cell type, it is known that the number of GA stacks and their size can vary within a single cell type [43]. For example, it is not clear whether all GA make and/or transport the same cargo proteins simultaneously [1] and as a consequence, the components in each individual GA could vary, contributing to different GA morphology/density/properties. Despite the evidence that GA is a highly dynamic organelle the enrichment techniques employed thus far, such as density gradients and FFE, have relied on the GA being morphologically homogeneous. Thus, a greater understanding of GA morphology, only possible through immuno-imaging approaches, would greatly enhance our ability to consider alternate techniques to purify GA. For example, using a series of antibodies to isolate various compartments of the GA, such as recently reported by Drakakaki et al. [29] to analyze the TGN, should be considered.
Supplementary Materials
The following are available online at www.mdpi.com/2227-7382/4/3/23/s1, Table S1: iTRAQ data of same-same trial to determine iTRAQ ratio cut-offs, Table S2: L. multiflorum protein profiling results of fraction S1, Table S3: P. trichocarpa protein profiling results of fraction S1, Table S4: L. multiflorum iTRAQ results, Table S5: P. trichocarpa iTRAQ results, Table S6: L. multiflorum cluster 12 (ER proteins), Table S7: L. multiflorum cluster 9 (TGN proteins), Table S8: P. trichocarpa cluster 9 (TGN proteins).
Acknowledgments
The authors thank Tingting Guo for advice on antibody and western analysis. This work was supported by a grant to the ARC Centre of Excellence in Plant Cell Walls (M.S.D., V.B., A.B.; (CE110001007)) and by a grant from the Swedish Research Council Vetenskapsrådet (V.B. (2011-4150)). The ARC Centre of Excellence in Plant Cell Walls acknowledges the support of funds from the Australian Government’s National Collaborative Research Infrastructure Program (NCRIS) administered through Bioplatforms Australia (BPA) Ltd.
Author Contributions
K.L.F., T.C., V.S., M.S.D., V.B. and A.B. conceived and designed the experiments; K.F., T.C., V.S. and W.Z. performed the experiments; K.F., V.B. and A.B. wrote the paper. All co-authors read and contributed to the editing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| ATTPS | trehalose phosphatase/synthase |
| BiP | luminal-binding protein |
| COG | oligomeric Golgi complex |
| COX | cytochrome-c oxidase |
| CP | cytoplasmic |
| CSL | cellulose synthase-like |
| DTT | dithiothreitol |
| EMP70 | endomembrane 70 protein |
| ER | endoplasmic reticulum |
| FFE | Free Flow Electrophoresis |
| GA | Golgi apparatus |
| GC6 | golgin candidate 6 |
| GSL | glucan synthase-like |
| GT | glycosyltransferase |
| IDPase | inosine diphosphatase |
| iTRAQ | Isobaric tags for relative and absolute quantitation |
| LOPIT | Localization of Organelle Proteins by Isotope Tagging |
| MLG | β-1,3;1,4-glucans |
| MM | microsomal membrane |
| MMTS | methylmethane thiosulphate |
| MT | mitochondria |
| PM | plasma membrane |
| RGP | reversibly glycosylated protein |
| SAM | S-adenosyl-l-methionine-dependent methyltransferase |
| SCCs | suspension cell cultures |
| SNARE | N-ethylmaleimide-sensitive factor attachment protein receptors |
| TCEP | tris-(2-carboxyethyl) phosphine |
| TGN | trans-Golgi network |
| TRAPP | transport protein particle |
| UGE | UDP-d-glucose/UDP-d-galactose 4 epimerase |
| UGP | UDP-d-glucose pyrophosphorylase |
| VAMP | vesicle-associated membrane proteins |
| VSR | vacuolar sorting receptors |
References
- Van de Meene, A.M.; Doblin, M.S.; Bacic, A. The plant secretory pathway seen through the lens of the cell wall. Protoplasma 2016. [Google Scholar] [CrossRef] [PubMed]
- Parsons, H.T.; Drakakaki, G.; Heazlewood, J.L. Proteomic dissection of the Arabidopsis golgi and trans-golgi network. Front. Plant Sci. 2013, 3, 298. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.G.; Brandizzi, F.; Hawes, C.; Nakano, A. Vesicles versus tubes: Is endoplasmic reticulum-golgi transport in plants fundamentally different from other eukaryotes? Plant Physiol. 2015, 168, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Gibeaut, D.M.; Carpita, N.C. Separation of membranes by flotation centrifugation for in vitro synthesis of plant cell wall polysaccharides. Protoplasma 1990, 156, 82–93. [Google Scholar] [CrossRef]
- Van der Woude, W.J.; Lembi, C.A.; Morré, D.J.; Kindinger, J.I.; Ordin, L. Β-glucan synthetases of plasma membrane and golgi apparatus from onion stem. Plant Physiol. 1974, 54, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Dunkley, T.P.J.; Hester, S.; Shadforth, I.P.; Runions, J.; Weimar, T.; Hanton, S.L.; Griffin, J.L.; Bessant, C.; Brandizzi, F.; Hawes, C.; et al. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 2006, 103, 6518–6523. [Google Scholar] [CrossRef] [PubMed]
- Nikolovski, N.; Rubtsov, D.; Segura, M.P.; Miles, G.P.; Stevens, T.J.; Dunkley, T.P.J.; Munro, S.; Lilley, K.S.; Dupree, P. Putative glycosyltransferases and other plant golgi apparatus proteins are revealed by lopit proteomics. Plant Physiol. 2012, 160, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Nikolovski, N.; Shliaha, P.V.; Gatto, L.; Dupree, P.; Lilley, K.S. Label-free protein quantification for plant golgi protein localization and abundance. Plant Physiol. 2014, 166, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Parsons, H.T.; Christiansen, K.; Knierim, B.; Carroll, A.; Ito, J.; Batth, T.S.; Smith-Moritz, A.M.; Morrison, S.; McInerney, P.; Hadi, M.Z.; et al. Isolation and proteomic characterization of the Arabidopsis golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 2012, 159, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Heard, W.; Sklenář, J.; Tomé, D.F.A.; Robatzek, S.; Jones, A.M.E. Identification of regulatory and cargo proteins of endosomal and secretory pathways in Arabidopsis thaliana by proteomic dissection. Mol. Cell. Proteomics 2015, 14, 1796–1813. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Hirose, S.; Katamine, H.; Kitajima, A.; Hori, H.; Sato, M.H.; Fujiwara, M.; Shimamoto, K.; Mitsui, T. Isolation and proteomic analysis of rice golgi membranes: Cis-golgi membranes labeled with GFP-SYP31. Plant Biotechnol. 2006, 23, 475–485. [Google Scholar] [CrossRef]
- Tanaka, N.; Fujita, M.; Handa, H.; Murayama, S.; Uemura, M.; Kawamura, Y.; Mitsui, T.; Mikami, S.; Tozawa, Y.; Yoshinaga, T.; et al. Proteomics of the rice cell: Systematic identification of the protein populations in subcellular compartments. Mol. Genet. Genomics 2004, 271, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Parsons, H.T.; Weinberg, C.S.; Macdonald, L.J.; Adams, P.D.; Petzold, C.J.; Strabala, T.J.; Wagner, A.; Heazlewood, J.L. Golgi enrichment and proteomic analysis of developing Pinus radiata xylem by free-flow electrophoresis. PLoS ONE 2013, 8, e84669. [Google Scholar] [CrossRef] [PubMed]
- Mast, S.; Peng, L.; Jordan, T.W.; Flint, H.; Phillips, L.; Donaldson, L.; Strabala, T.J.; Wagner, A. Proteomic analysis of membrane preparations from developing Pinus radiata compression wood. Tree Physiol. 2010, 30, 1456–1468. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.M.; Stone, B.A. Chemical composition of the cell walls of lolium multiflorum endosperm. Phytochemistry 1973, 12, 1361–1367. [Google Scholar] [CrossRef]
- Smith, M.M.; Stone, B.A. Studies on lolium multiflorum endosperm in tissue-culture. Aust. J. Biol. Sci. 1973, 26, 123–133. [Google Scholar]
- Wilson, S.M.; Ho, Y.Y.; Lampugnani, E.R.; van de Meene, A.M.L.; Bain, M.P.; Bacic, A.; Doblin, M.S. Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3; 1,4)-β-d-glucan in grasses. Plant Cell 2015, 27, 754–771. [Google Scholar] [CrossRef] [PubMed]
- Briskin, D.P.; Leonard, R.T.; Hodges, T.K. Isolation of plasma membrane: Membrane markers and general principles. Methods Enzymol. 1987, 148, 542–558. [Google Scholar]
- Natera, S.H.A.; Ford, K.L.; Cassin, A.M.; Patterson, J.H.; Newbigin, E.J.; Bacic, A. Analysis of the oryza sativa plasma membrane proteome using combined protein and peptide fractionation approaches in conjunction with mass spectrometry. J. Proteome Res. 2008, 7, 1159–1187. [Google Scholar] [CrossRef] [PubMed]
- Ford, K.L.; Cassin, A.; Bacic, A. Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2011, 2, 44. [Google Scholar] [CrossRef] [PubMed]
- Byrne, S.L.; Nagy, I.; Pfeifer, M.; Armstead, I.; Swain, S.; Studer, B.; Mayer, K.; Campbell, J.D.; Czaban, A.; Hentrup, S.; et al. A synteny-based draft genome sequence of the forage grass lolium perenne. Plant J. 2015, 84, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (torr. & gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Sieb, C.; Thiel, K.; Wiswedel, B. Knime: The konstanz information miner. In Data Analysis, Machine Learning and Applications; Preisach, C., Burkhardt, H., Schmidt-Thieme, L., Decker, R., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, Germany, 2008; pp. 319–326. [Google Scholar]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis information resource (tair): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. Uniprot: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Tanz, S.K.; Castleden, I.; Hooper, C.M.; Vacher, M.; Small, I.; Millar, H.A. Suba3: A database for integrating experimentation and prediction to define the subcellular location of proteins in Arabidopsis. Nucleic Acids Res. 2013, 41, D1185–D1191. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (cazy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. Signalp 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Drakakaki, G.; van de Ven, W.; Pan, S.; Miao, Y.; Wang, J.; Keinath, N.F.; Weatherly, B.; Jiang, L.; Schumacher, K.; Hicks, G.; et al. Isolation and proteomic analysis of the syp61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 2012, 22, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Wilson, S.M.; Hrmova, M.; Harvey, A.J.; Shirley, N.J.; Medhurst, A.; Stone, B.A.; Newbigin, E.J.; Bacic, A.; Fincher, G.B. Cellulose synthase-like cslf genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science 2006, 311, 1940–1942. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Szumlanski, A.L.; Gu, F.; Guo, F.; Nielsen, E. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 2011, 13, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Verhertbruggen, Y.; Oikawa, A.; Manisseri, C.; Knierim, B.; Prak, L.; Jensen, J.K.; Knox, J.P.; Auer, M.; Willats, W.G.; et al. The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol. Plant 2011, 4, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, D.; Voigt, C.A. Callose biosynthesis in Arabidopsis with a focus on pathogen response: What we have learned within the last decade. Ann. Bot. 2014, 114, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Jiang, N.; Nadella, R.; Killen, T.L.; Nadella, V.; Faik, A. A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol. 2010, 154, 78–97. [Google Scholar] [CrossRef] [PubMed]
- Anders, N.; Wilkinson, M.D.; Lovegrove, A.; Freeman, J.; Tryfona, T.; Pellny, T.K.; Weimar, T.; Mortimer, J.C.; Stott, K.; Baker, J.M.; et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. USA 2012, 109, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.; Lehner, A.; Bouton, S.; Kiefer-Meyer, M.C.; Voxeur, A.; Pelloux, J.; Lerouge, P.; Mollet, J.C. The cell wall pectic polymer rhamnogalacturonan-II is required for proper pollen tube elongation: Implications of a putative sialyltransferase-like protein. Ann. Bot. 2014, 114, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Lerouxel, O.; Mouille, G.; Andeme-Onzighi, C.; Bruyant, M.P.; Seveno, M.; Loutelier-Bourhis, C.; Driouich, A.; Hofte, H.; Lerouge, P. Mutants in defective glycosylation, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 2005, 42, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.H.; Gao, C.; Yu, P.; Tu, L.; Meng, Z.; Banfield, D.K.; Yao, X.; Jiang, L. Conserved function of the lysine-based KXD/E motif in golgi retention for endomembrane proteins among different organisms. Mol. Biol. Cell 2015, 26, 4280–4293. [Google Scholar] [CrossRef] [PubMed]
- Dell, A.; Galadari, A.; Sastre, F.; Hitchen, P. Similarities and differences in the glycosylation mechanisms in prokaryotes and eukaryotes. Int. J. Microbiol. 2010, 2010, 148178. [Google Scholar] [CrossRef] [PubMed]
- Ow, S.Y.; Salim, M.; Noirel, J.; Evans, C.; Rehman, I.; Wright, P.C. Itraq underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J. Proteome Res. 2009, 8, 5347–5355. [Google Scholar] [CrossRef] [PubMed]
- Philippe, S.; Saulnier, L.; Guillon, F. Arabinoxylan and (1→3),(1→4)-β-glucan deposition in cell walls during wheat endosperm development. Planta 2006, 224, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.F.; Staehelin, L.A. Functional compartmentation of the golgi apparatus of plant cells: Immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol. 1992, 99, 1070–1083. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).