Age- and Activity-Related Differences in the Abundance of Myosin Essential and Regulatory Light Chains in Human Muscle
Abstract
:1. Introduction
2. Results
2.1. MyHC Isoform Analysis
2.2. LC-MS-MS Mining of the Myofibrillar Proteome
2.3. Differences in the Myofibrillar Proteome Due to Age or Habitual Levels of Physical Activity
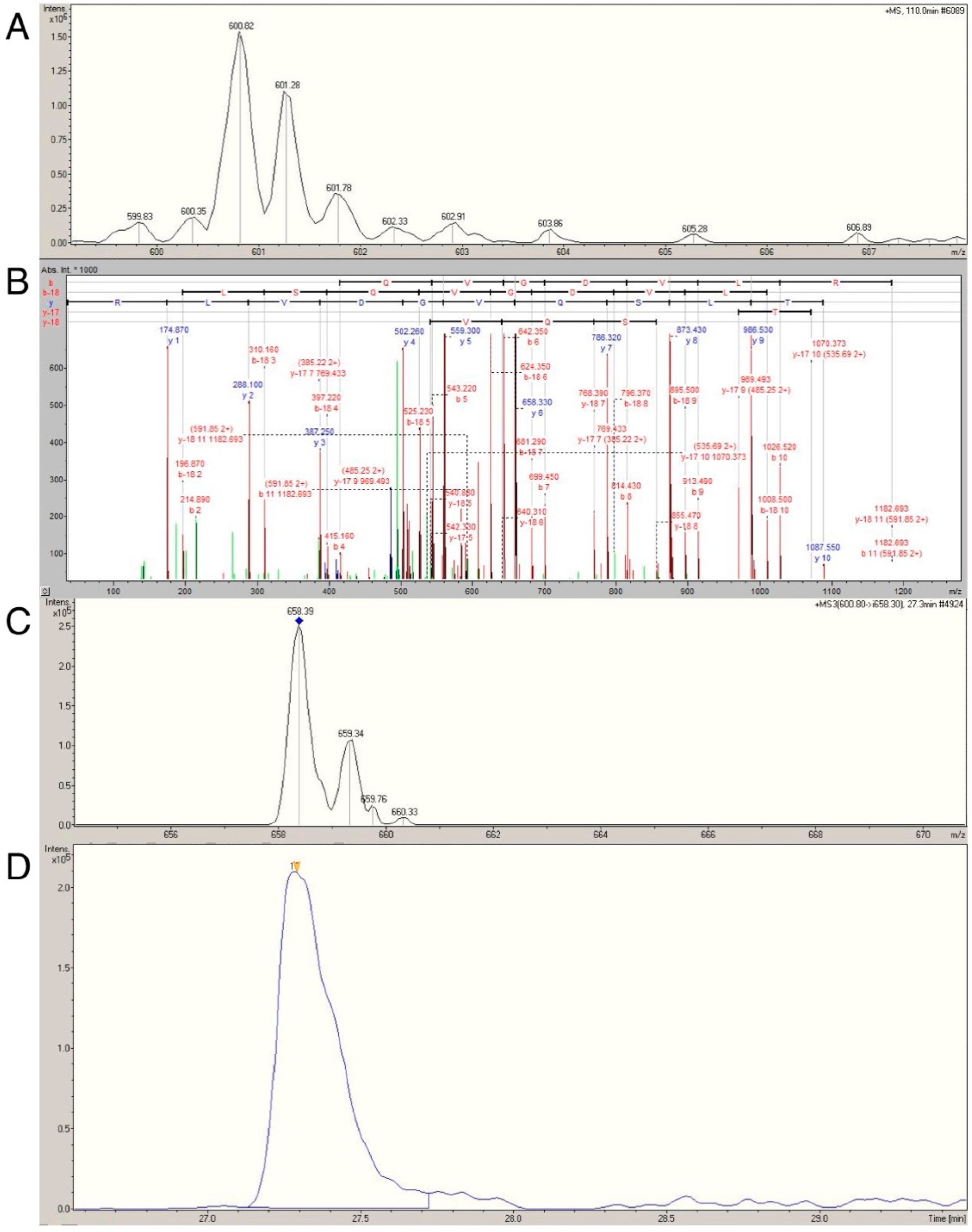
2.4. Confirmation of Age- and Activity-Related Differences in Myosin Light and Regulatory Chain Isoforms Using Selective Reaction Monitoring
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Muscle Biopsies
4.3. MyHC Isoform Staining
4.4. Label Free Liquid Chromatography Coupled Mass Spectrometry (LC-MS)
4.5. Selective Reaction Monitoring (SRM)
4.6. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 2DGE | Two-dimensional gel electrophoresis |
| LC-MS | liquid chromatography mass spectrometry |
| MLRS | Myosin Light Regulatory Chain, fast isoform |
| MRM | Multiple Reaction Monitoring |
| MyHC | Myosin Heavy Chain |
| MYL1 | Myosin Light Chain 1 |
| MYL3 | Myosin Light Chain 3 |
| MU | Motor unit |
| OT | Old Trained |
| OU | Old Untrained |
| SRM | Selective Reaction Monitoring |
| YT | Young Trained |
| YU | Young Untrained |
References
- Cobley, J.N.; Moult, P.R.; Burniston, J.G.; Morton, J.P.; Close, G.L. Exercise improves mitochondrial and redox-regulated stress responses in the elderly: Better late than never! Biogerontology 2015, 16, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Piasecki, M.; Ireland, A.; Jones, D.A.; McPhee, J.S. Age-dependent motor unit remodelling in human limb muscles. Biogerontology 2015. [Google Scholar] [CrossRef] [PubMed]
- Pette, D.; Staron, R.S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 2000, 50, 500–509. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–531. [Google Scholar] [CrossRef] [PubMed]
- Deschenes, M.R. Effects of aging on muscle fibre type and size. Sports Med. 2004, 34, 809–824. [Google Scholar] [CrossRef] [PubMed]
- Zierath, J.R.; Hawley, J.A. Skeletal muscle fiber type: Influence on contractile and metabolic properties. PLoS Biol. 2004, 2, e348. [Google Scholar] [CrossRef] [PubMed]
- Gelfi, C.; Viganò, A.; Ripamonti, M.; Pontoglio, A.; Begum, S.; Pellegrino, M.A.; Grassi, B.; Bottinelli, R.; Wait, R.; Cerretelli, P. The human muscle proteome in aging. J. Proteome Res. 2006, 5, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Kammoun, M.; Cassar-Malek, I.; Meunier, B.; Picard, B. A simplified immunohistochemical classification of skeletal muscle fibres in mouse. Eur. J. Histochem. 2014, 58, 2254. [Google Scholar] [CrossRef] [PubMed]
- Bloemberg, D.; Quadrilatero, J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS ONE 2012, 7, e35273. [Google Scholar] [CrossRef] [PubMed]
- Lanza, I.R.; Short, D.K.; Short, K.R.; Raghavakaimal, S.; Basu, R.; Joyner, M.J.; McConnell, J.P.; Nair, K.S. Endurance exercise as a countermeasure for aging. Diabetes 2008, 57, 2933–2942. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A.; Hamadeh, M.J.; Kaczor, J.J.; Raha, S.; DeBeer, J.; Tarnopolsky, M.A. Aberrant mitochondrial homeostasis in the skeletal muscle of sedentary older adults. PLoS ONE 2010, 5, e10778. [Google Scholar] [CrossRef] [PubMed]
- Malik, Z.A.; Cobley, J.N.; Morton, J.P.; Close, G.L.; Edwards, B.J.; Koch, L.G.; Britton, S.L.; Burniston, J.G. Label-Free LC-MS Profiling of Skeletal Muscle Reveals Heart-Type Fatty Acid Binding Protein as a Candidate Biomarker of Aerobic Capacity. Proteomes 2013, 1, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Drexler, H.C.A.; Ruhs, A.; Konzer, A.; Mendler, L.; Bruckskotten, M.; Looso, M.; Gunther, S.; Boettger, T.; Kruger, M.; Braun, T. On Marathons and Sprints: An Integrated Quantitative Proteomics and Transcriptomics Analysis of Differences Between Slow and Fast Muscle Fibers. Mol. Cell. Proteom. 2012, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Burniston, J.G.; Kenyani, J.; Gray, D.; Guadagnin, E.; Jarman, I.H.; Cobley, J.N.; Cuthbertson, D.J.; Chen, Y.W.; Wastling, J.M.; Lisboa, P.J.; et al. Conditional independence mapping of DIGE data reveals PDIA3 protein species as key nodes associated with muscle aerobic capacity. J. Proteom. 2014, 106, 230–245. [Google Scholar] [CrossRef] [PubMed]
- Spangenburg, E.E.; Booth, F.W. Molecular regulation of individual skeletal muscle fibre types. Acta Physiol. Scand. 2003, 178, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Rowan, S.L.; Rygiel, K.; Purves-Smith, F.M.; Solbak, N.M.; Turnbull, D.M.; Hepple, R.T. Denervation causes fiber atrophy and myosin heavy chain co-expression in senescent skeletal muscle. PLoS ONE 2012, 7, e29082. [Google Scholar] [CrossRef] [PubMed]
- Burniston, J.G.; Hoffman, E.P. Proteomic responses of skeletal and cardiac muscle to exercise. Expert Rev. Proteom. 2011, 8, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.C.; Walsh, R.J.; Salajegheh, M.; Amato, A.A.; Krastins, B.; Sarracino, D.A.; Greenberg, S.A. Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J. Proteome Res. 2009, 8, 3265–3277. [Google Scholar] [CrossRef] [PubMed]
- Gelfi, C.; De Palma, S.; Cerretelli, P.; Begum, S.; Wait, R. Two-dimensional protein map of human vastus lateralis muscle. Electrophoresis 2003, 24, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Højlund, K.; Yi, Z.; Hwang, H.; Bowen, B.; Lefort, N.; Flynn, C.R.; Langlais, P.; Weintraub, S.T.; Mandarino, L.J. Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol. Cell. Proteom. 2008, 7, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Doran, P.; Donoghue, P.; O’Connell, K.; Gannon, J.; Ohlendieck, K. Proteomics of skeletal muscle aging. Proteomics 2009, 9, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Gannon, J.; Doran, P.; Kirwan, A.; Ohlendieck, K. Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur. J. Cell Biol. 2009, 88, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Connell, K.O.; Gannon, J.; Doran, P.; Ohlendieck, K.A.Y.; O’Connell, K. Proteomic profiling reveals a severely perturbed protein expression pattern in aged skeletal muscle. Int. J. Mol. Med. 2007, 20, 145–153. [Google Scholar]
- Staunton, L.; Zweyer, M.; Swandulla, D.; Ohlendieck, K. Mass spectrometry-based proteomic analysis of middle-aged vs. aged vastus lateralis reveals increased levels of carbonic anhydrase isoform 3 in senescent human skeletal muscle. Int. J. Mol. Med. 2012, 30, 723–733. [Google Scholar] [PubMed]
- Capitanio, D.; Vasso, M.; de Palma, S.; Fania, C.; Torretta, E.; Cammarata, F.P.; Magnaghi, V.; Procacci, P.; Gelfi, C. Specific protein changes contribute to the differential muscle mass loss during ageing. Proteomics 2016, 16, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Gannon, J.; Doran, P.; Kirwan, A.; Ohlendieck, K. Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur. J. Cell Biol. 2009, 88, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, P.R.; Holzhütter, H.G.; Apweiler, R.; Schlüter, H. The speciation of the proteome. Chem. Cent. J. 2008, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Burniston, J.G.; Connolly, J.; Kainulainen, H.; Britton, S.L.; Koch, L.G. Label-free profiling of skeletal muscle using high-definition mass spectrometry. Proteomics 2014, 14, 2339–2344. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, A.S.; Murgia, M.; Nagaraj, N.; Treebak, J.T. Deep Proteomics of Mouse Skeletal Muscle Enables Quantitation of Protein Isoforms, Metabolic Pathways, and Transcription Factors. Mol. Cell. Proteom. 2015, 14, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Sakellariou, G.K.; Murray, S.; Waldron, S.; Gregson, W.; Burniston, J.G.; Morton, J.P.; Iwanejko, L.A.; Close, G.L. Lifelong endurance training attenuates age-related genotoxic stress in human skeletal muscle. Longev. Health 2013, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Staron, R.S.; Hagerman, F.C.; Hikida, R.S.; Murray, T.F.; Hostler, D.P.; Crill, M.T.; Ragg, K.E.; Toma, K. Fiber Type Composition of the Vastus Lateralis Muscle of Young Men and Women. J. Histochem. Cytochem. 2000, 48, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, M.; Strehler, E.E.; Garfinkel, L.I.; Gubits, R.M.; Ruiz-Opazo, N.; Nadal-Ginard, B. Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J. Biol. Chem. 1984, 259, 13595–13604. [Google Scholar] [PubMed]
- Cohen-Haguenauer, O.; Barton, P.J.R.; van Cong, N.; Cohen, A.; Masset, M.; Buckingham, M.; Frézal, J. Chromosomal assignment of two myosin alkali light-chain genes encoding the ventricular/slow skeletal muscle isoform and the atrial/fetal muscle isoform (MYL3, MYL4). Hum. Genet. 1989, 81, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Hepple, R.T.; Rice, C.L. Innervation and neuromuscular control in ageing skeletal muscle. J Physiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- De Winter, F.; Vo, T.; Stam, F.J.; Wisman, L.A.B.; Bär, P.R.; Niclou, S.P.; van Muiswinkel, F.L.; Verhaagen, J. The expression of the chemorepellent Semaphorin 3A is selectively induced in terminal Schwann cells of a subset of neuromuscular synapses that display limited anatomical plasticity and enhanced vulnerability in motor neuron disease. Mol. Cell. Neurosci. 2006, 32, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Pasterkamp, R.J. Getting neural circuits into shape with semaphorins. Nat. Rev. Neurosci. 2012, 13, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Picotti, P.; Aebersold, R. Selected reaction monitoring–based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Torgerud, W.S.; Mosser, K.H.H.; Hirai, H.; Watanabe, S.; Asakura, A.; Thompson, L.V. Myosin light chain 3f attenuates age-induced decline in contractile velocity in MHC type II single muscle fibers. Aging Cell 2012, 11, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Billeter, R.; Heizmann, C.W.; Howald, H.; Jenny, E. Analysis of myosin light and heavy chain types in single human skeletal muscle fibers. Eur. J. Biochem. 1981, 116, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S.; Bedrin, N.G.; Callahan, D.M.; Previs, M.J.; Jennings, M.E.; Ades, P.A.; Maughan, D.W.; Palmer, B.M.; Toth, M.J. Age-related slowing of myosin actin cross-bridge kinetics is sex specific and predicts decrements in whole skeletal muscle performance in humans. J. Appl. Physiol. 2013, 115, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.J.; Selby, A.; Atherton, P.; Smith, K.; Kumar, V.; Glover, E.L.; Philips, S.M. Facts, noise and wishful thinking: Muscle protein turnover in aging and human disuse atrophy. Scand. J. Med. Sci. Sport 2010, 20, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Sakellariou, G.K.; Owens, D.J.; Murray, S.; Waldron, S.; Gregson, W.; Fraser, W.D.; Burniston, J.G.; Iwanejko, L.A.; McArdle, A.; et al. Lifelong training preserves some redox-regulated adaptive responses after an acute exercise stimulus in aged human skeletal muscle. Free Radic. Biol. Med. 2014, 70, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Reidy, P.T.; Hinkley, J.M.; Trappe, T.A.; Trappe, S.W.; Harber, M.P. Protein composition of endurance trained human skeletal muscle. Int. J. Sports Med. 2014, 35, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Bartlett, J.D.; Kayani, A.; Murray, S.W.; Louhelainen, J.; Donovan, T.; Waldron, S.; Gregson, W.; Burniston, J.G.; Morton, J.P.; et al. PGC-1α transcriptional response and mitochondrial adaptation to acute exercise is maintained in skeletal muscle of sedentary elderly males. Biogerontology 2012, 13, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Talmadge, R.J.; Roy, R.R. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J. Appl. Physiol. 1993, 75, 2337–2340. [Google Scholar] [PubMed]
- Bamman, M.M.; Clarke, M.S.; Talmadge, R.J.; Feedback, D.L. Enhanced protein electrophoresis technique for separating human skeletal muscle myosin heavy chain isoforms General. Electrophoresis 1999, 20, 466–468. [Google Scholar] [CrossRef]
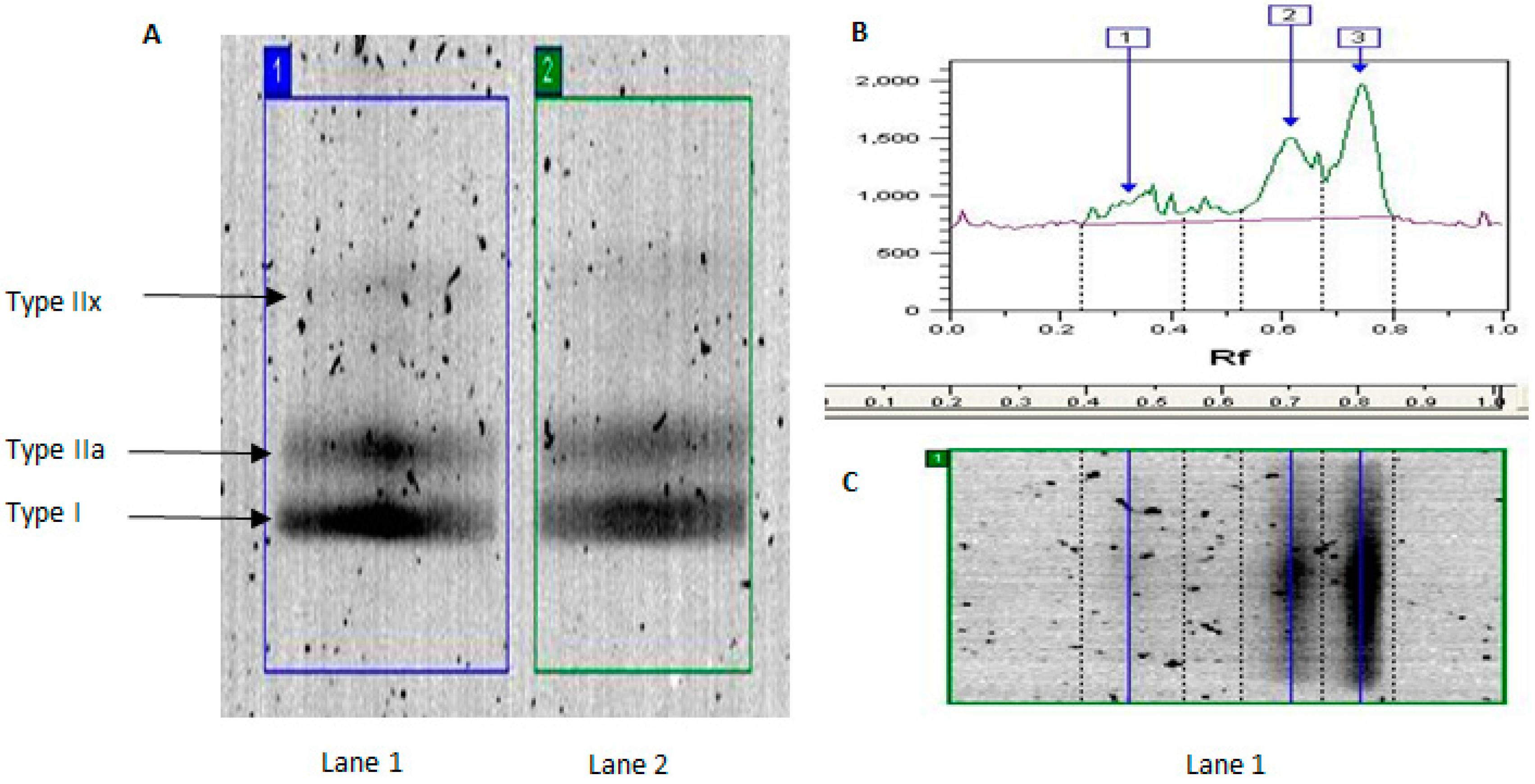
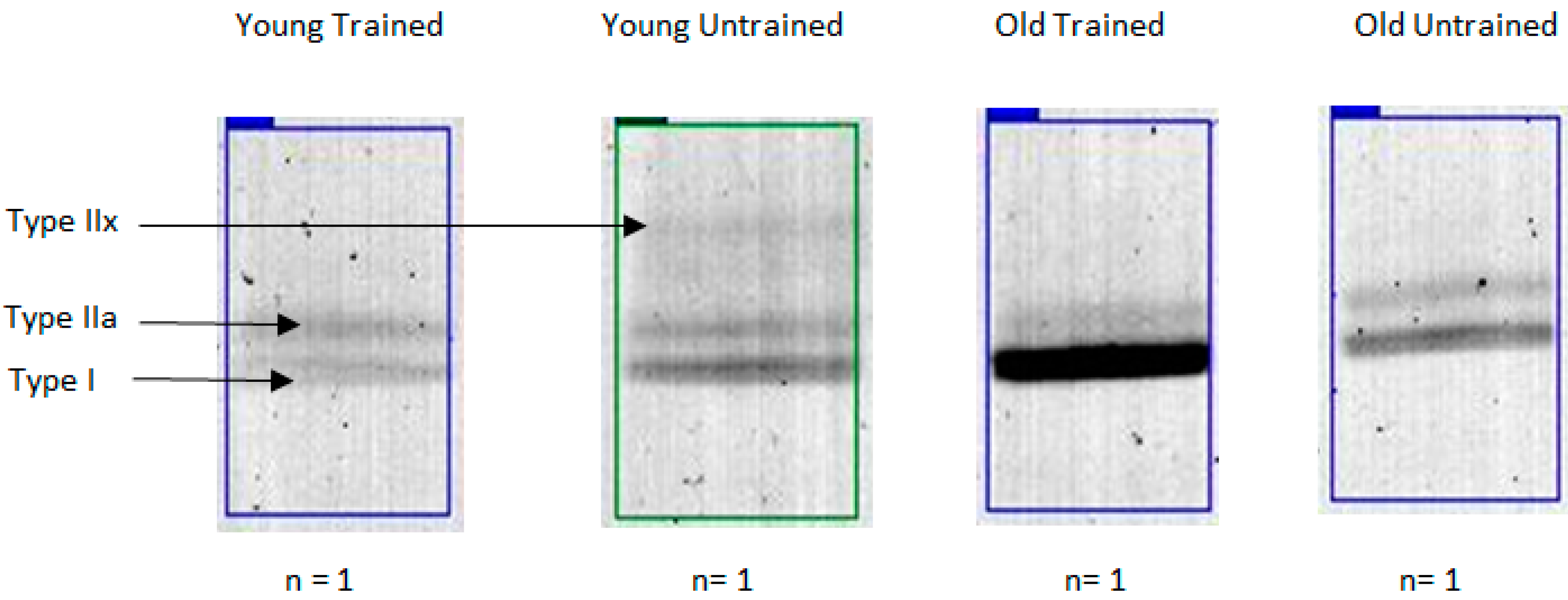
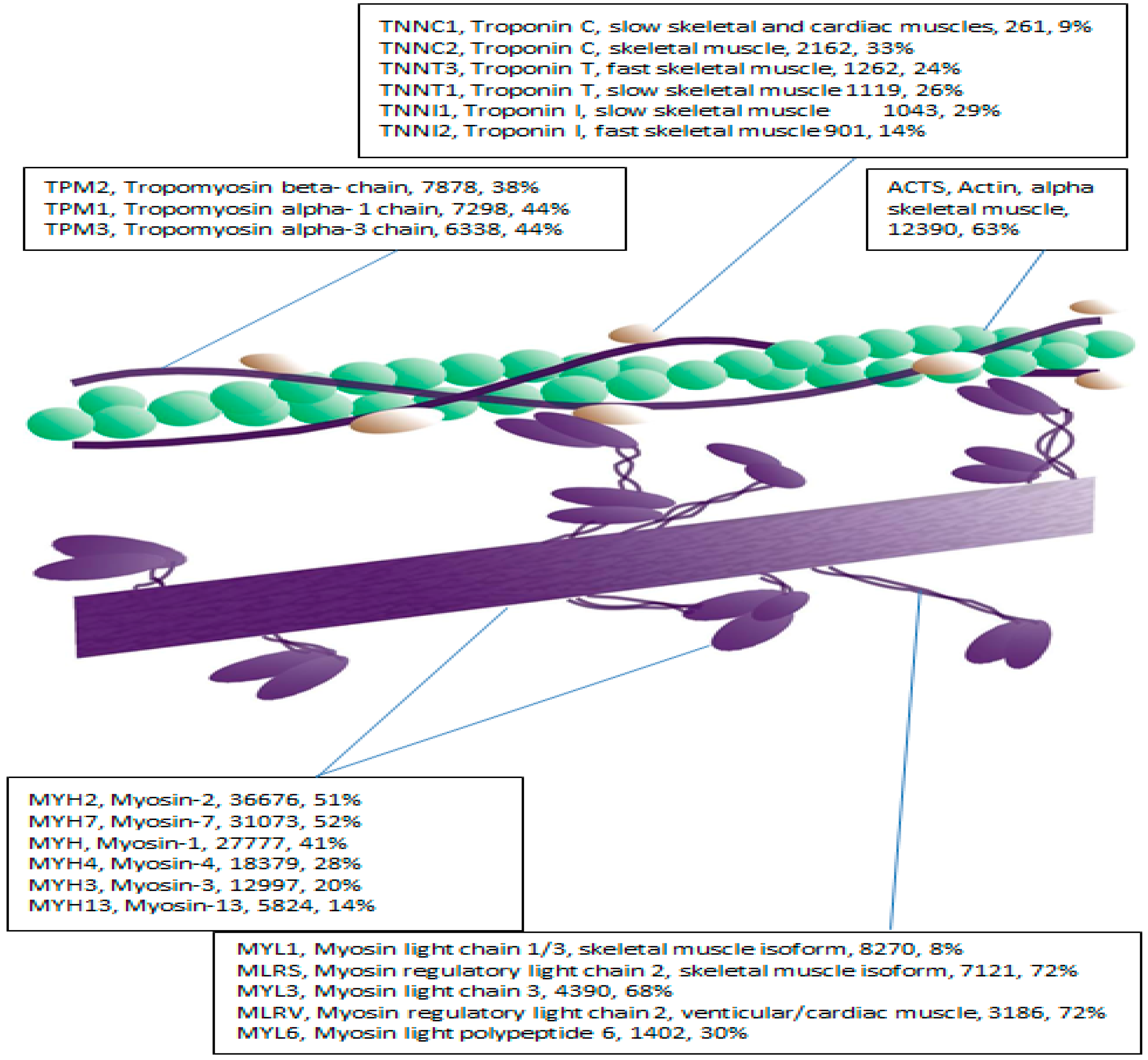
| Band | YT | YU | OT | OU | Effect of Age in Trained | Effect of Age in Untrained |
|---|---|---|---|---|---|---|
| IIx (%) | 9.0 ± 8.3 | 13.8 ± 13.5 | 0.2 ± 0.4 | 9.5 ± 13.1 | 98% lower | 31% lower |
| Iia (%) | 35.6 ± 14.2 | 50.2 ± 15.1 | 21.7 ± 13.8 * | 30.0 ± 20.7 | 40% lower | 40% lower |
| I (%) | 55.4 ± 27.8 | 36.0 ± 12.1 | 78.1 ± 14.2 * | 60.5 ± 30.1 | 41% higher | 68% higher |
| Accession | Protein Name | Score | SC (%) | Peptides | MW | pI |
|---|---|---|---|---|---|---|
| MYH3 | Myosin-3 | 12,997 | 20 | 1 | 223.8 | 5.5 |
| MYH13 | Myosin-13 | 5824 | 14 | 1 | 223.5 | 5.4 |
| MYH7 | Myosin-7 | 31,073 | 52 | 71 | 223.0 | 5.5 |
| MYH1 | Myosin-1 | 27,777 | 41 | 11 | 223.0 | 5.5 |
| MYH2 | Myosin-2 | 36,676 | 51 | 94 | 222.9 | 5.5 |
| MYH4 | Myosin-4 | 18,379 | 28 | 1 | 222.9 | 5.6 |
| MYOM2 | Myomesin-2 | 116 | 9 | 5 | 164.8 | 5.8 |
| MYPC1 | Myosin-binding protein C, slow type | 763 | 17 | 14 | 128.2 | 5.7 |
| ACTN2 | Alpha-actinin-2 | 5705 | 45 | 31 | 103.8 | 5.2 |
| LDB3 | LIM domain-binding protein 3 | 1259 | 18 | 9 | 77.1 | 9.4 |
| ALBU | Serum albumin precursor | 1689 | 41 | 20 | 69.3 | 5.9 |
| K2C1 | Keratin, type II cytoskeletal 1 | 171 | 10 | 6 | 66.0 | 8.8 |
| PDLI5 | PDZ and LIM domain protein 5 | 135 | 15 | 4 | 63.9 | 9.6 |
| K1C10 | Keratin, type I cytoskeletal | 116 | 6 | 3 | 58.8 | 5.0 |
| ATPB | ATP synthase subunit beta, mitochondrial | 261 | 21 | 6 | 56.5 | 5.1 |
| DESM | Desmin | 1778 | 39 | 15 | 53.5 | 5.1 |
| VIME | Vimentin | 424 | 14 | 2 | 52.6 | 4.9 |
| ENOB | Beta-enolase | 324 | 47 | 4 | 46.9 | 8.6 |
| CASQ1 | Calsequestrin-1 | 1718 | 11 | 2 | 44.5 | 3.9 |
| KCRM | Creatine kinase M-type | 5182 | 43 | 1 | 43.1 | 6.9 |
| ACTS | Actin, alpha skeletal muscle | 12,390 | 63 | 23 | 42.0 | 5.1 |
| ACTB | Actin, cytoplasmic 1 | 6353 | 30 | 1 | 41.7 | 5.2 |
| ALDOA | Fructose-bisphosphate aldolase A | 1159 | 41.8 | 9 | 39.4 | 9.2 |
| ALDOC | Fructose-biphosphate aldolase C | 102 | 18 | 1 | 39.4 | 6.5 |
| FHL1 | Four and a half LIM domains protein 1 | 398 | 14 | 3 | 36.2 | 10.5 |
| G3P | Glyceraldehyde-3-phosphate dehydrogenase | 599 | 38 | 7 | 36.0 | 9.3 |
| TNNT1 | Troponin T, slow skeletal muscle | 1119 | 26 | 6 | 32.9 | 5.8 |
| TPM2 | Tropomyosin beta- chain | 7878 | 38 | 13 | 32.8 | 4.5 |
| TPM3 | Tropomyosin alpha-3 chain | 6338 | 44 | 7 | 32.8 | 4.5 |
| TPM1 | Tropomyosin alpha- 1 chain | 7298 | 44 | 11 | 32.7 | 4.5 |
| TNNT3 | Troponin T, fast skeletal muscle | 1262 | 24 | 6 | 31.8 | 5.6 |
| MYOZ1 | Myozenin-1 | 599 | 51 | 7 | 31.7 | 9.3 |
| CAH3 | Carbonic anhydrase 3 | 760 | 38 | 6 | 29.5 | 7.1 |
| MYL6B | Myosin light chain 6B | 152 | 15 | 1 | 22.7 | 5.5 |
| MYL3 | Myosin light chain 3 | 4390 | 68 | 10 | 21.9 | 4.9 |
| TNNI1 | Troponin I, slow skeletal muscle | 1043 | 29 | 5 | 21.7 | 10.3 |
| TNNI2 | Troponin I, fast skeletal muscle | 901 | 14 | 3 | 21.3 | 9.6 |
| MYL1 | Myosin light chain 1/3, skeletal muscle | 8270 | 8 | 15 | 21.1 | 4.8 |
| MLRS | Myosin regulatory light chain 2, skeletal muscle | 7121 | 72 | 2 | 19 | 4.7 |
| MLRV | Myosin regulatory light chain 2, venticular/cardiac | 3186 | 72 | 11 | 18.8 | 4.7 |
| TNNC1 | Troponin C, slow skeletal and cardiac | 261 | 9 | 2 | 18.4 | 3.9 |
| TNNC2 | Troponin C, skeletal muscle | 2162 | 33 | 4 | 18.1 | 3.9 |
| MYG | Myoglobin | 3163 | 55 | 8 | 17.2 | 7.9 |
| MYL6 | Myosin light polypeptide 6 | 1402 | 30 | 2 | 16.9 | 4.4 |
| HBB | Hemoglobin subunit beta | 11,403 | 88 | 12 | 16 | 6.9 |
| HBD | Hemoglobin subunit delta | 4206 | 69 | 2 | 16 | 9.1 |
| HBA | Hemoglobin subunit alpha | 2586 | 53 | 6 | 15.2 | 9.4 |
| Accession | Retention Time: Mass/Charge Ratio | p-Value | |||
|---|---|---|---|---|---|
| Age | Training | Age × Training | Sequence | ||
| ACTS | 40.1 min: 644.31 m/z | 0.552 | 0.289 | 0.02 | GILTLK |
| 98.9 min: 895.86 m/z | 0.024 | 0.315 | 0.336 | SYELPDGQVITIGNER | |
| 44.7 min: 1162.47 m/z | 0.043 | 0.537 | 0.678 | EITALAPSTMK | |
| MLRS | 83.4 min: 652.64 m/z | 0.159 | 0.029 | 0.05 | VAPEEHPTLLTEAPLNPK |
| 103.8 min: 1524.61 m/z | 0.117 | 0.03 | 0.2 | FLEELLTTQCDR | |
| 32.1 min: 1173.43 m/z | 0.06 | 0.026 | 0.399 | DGIIDKEDLR | |
| MYH | 104.4 min: 1319.51 m/z | 0.045 | 0.046 | 0.465 | GADPEDVITGAFK |
| 117.6 min: 895.91 m/z | 0.026 | 0.096 | 0.211 | NAYEESLDQLETLK | |
| 21.9 min: 1004.37 m/z | 0.268 | 0.348 | 0.041 | ELEEISER | |
| 144.6 min: 1893.88 m/z | 0.004 | 0.051 | 0.051 | LLSTLFANYAGADAPIEK | |
| 67.1 min: 1507.59 m/z | 0.015 | 0.275 | 0.302 | LTYTQQLEDLKR | |
| 21.5 min: 1272.36 m/z | 0.015 | 0.51 | 0.87 | TLEDQMNEHR | |
| MYL1 | 20.0 min: 1411.48 m/z | 0.876 | 0.832 | 0.027 | ELTYQTEEDRK |
| 17.8 min: 1482.51 m/z | 0.108 | 0.555 | 0.033 | KVQHELDEAEER | |
| 119.4 min: 1010.42 m/z | 0.508 | 0.022 | 0.444 | EAFLLFDR | |
| MYL3 | 116.4 min: 1542.60 m/z | 0.001 | 0.841 | 0.798 | DQATYEDFVEGLR |
| 22.9 min: 1524.60 m/z | 0.03 | 0.917 | 0.662 | AAPAPAPPPEPERPK | |
| TPM | 68.8 min: 1243.50 m/z | 0.04 | 0.574 | 0.63 | IQLVEEELDR |
| Transition | Young | Old | Age (p Value) | Trained | Untrained | Activity (p-Value) |
|---|---|---|---|---|---|---|
| MYL1 (771.7 → 835.4) | 1372 ± 669 | 796 ± 280 | 0.06 | 868 ± 388 | 1343 ± 684 | 0.024 |
| MYL1 (600.8 → 658.3) | 2790 ± 1309 | 1682 ± 640 | 0.007 | 1761 ± 848 | 2806 ± 1252 | 0.013 |
| MYL1 (505.7 → 550.2) | 3829 ± 2133 | 1764 ± 1187 | 0.007 | 2014 ± 1515 | 3744 ± 2149 | 0.025 |
| MYL1 total | 2081 ± 977 | 1239 ± 450 | 0.006 | 1315 ± 614 | 2075 ± 953 | 0.014 |
| MLY3 (498.2 → 759.4) | 3579 ± 1682 | 6353 ± 3347 | 0.021 | 5881 ± 3140 | 3867 ± 2403 | 0.083 |
| MYL3:MYL1 ratio | 1.72 | 5.13 | 4.47 | 1.86 | ||
| MLRS (660.2 → 735.4) | 1029 ± 436 | 740 ± 399 | 0.108 | 701 ± 379 | 1097 ± 410 | 0.029 |
| MLRS (762.8 → 780.2) | 2101 ± 1135 | 1236 ± 834 | 0.048 | 1251 ± 904 | 2164 ± 1080 | 0.039 |
| MLRS total | 1565 ± 778 | 988 ± 591 | 0.055 | 976 ± 630 | 1630 ± 725 | 0.031 |
| Isform Specific Transition | YT | YU | OT | OU |
|---|---|---|---|---|
| MYL1 (771.7 → 835.4) | 1034 ± 482 | 1779 ± 672 | 703 ± 181 * | 907 ± 356 * |
| MYL1 (600.8 → 658.3) | 5777 ± 2532 | 5707 ± 7583 | 5457 ± 5430 * | 8436 ± 8908 |
| MYL1 (505.7 → 550.2) | 2829 ± 1822 | 4829 ± 2072 | 1199 ± 373 * | 2441 ± 1515 |
| MYL1 Average | 1568 ± 801 | 2698 ± 459 | 1062 ± 186 * | 1451 ± 600 * |
| MYL3 (498.2 → 759.4) | 4529 ± 1478 | 2438 ± 1166 | 7233 ± 3889 * | 5296 ± 2556 |
| MYL3:MYL1 ratio | 2.89 | 0.90 | 6.81 | 3.65 |
| MLRS (762.8 → 780.2) | 1632 ± 116 | 2571 ± 980 | 869 ± 321 * | 1675 ± 1081 |
| MLRS (660.2 → 735.4) | 829 ± 450 | 1229 ± 345 | 573 ± 273 * | 939 ± 46 |
| MLRS Average | 1231 ± 794 | 1900 ± 658 | 721 ± 295 * | 1307 ± 730 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cobley, J.N.; Ab. Malik, Z.; Morton, J.P.; Close, G.L.; Edwards, B.J.; Burniston, J.G. Age- and Activity-Related Differences in the Abundance of Myosin Essential and Regulatory Light Chains in Human Muscle. Proteomes 2016, 4, 15. https://doi.org/10.3390/proteomes4020015
Cobley JN, Ab. Malik Z, Morton JP, Close GL, Edwards BJ, Burniston JG. Age- and Activity-Related Differences in the Abundance of Myosin Essential and Regulatory Light Chains in Human Muscle. Proteomes. 2016; 4(2):15. https://doi.org/10.3390/proteomes4020015
Chicago/Turabian StyleCobley, James N., Zulezwan Ab. Malik, James P. Morton, Graeme L. Close, Ben J. Edwards, and Jatin G. Burniston. 2016. "Age- and Activity-Related Differences in the Abundance of Myosin Essential and Regulatory Light Chains in Human Muscle" Proteomes 4, no. 2: 15. https://doi.org/10.3390/proteomes4020015
APA StyleCobley, J. N., Ab. Malik, Z., Morton, J. P., Close, G. L., Edwards, B. J., & Burniston, J. G. (2016). Age- and Activity-Related Differences in the Abundance of Myosin Essential and Regulatory Light Chains in Human Muscle. Proteomes, 4(2), 15. https://doi.org/10.3390/proteomes4020015








