The Pollen Coat Proteome: At the Cutting Edge of Plant Reproduction
Abstract
:1. Introduction
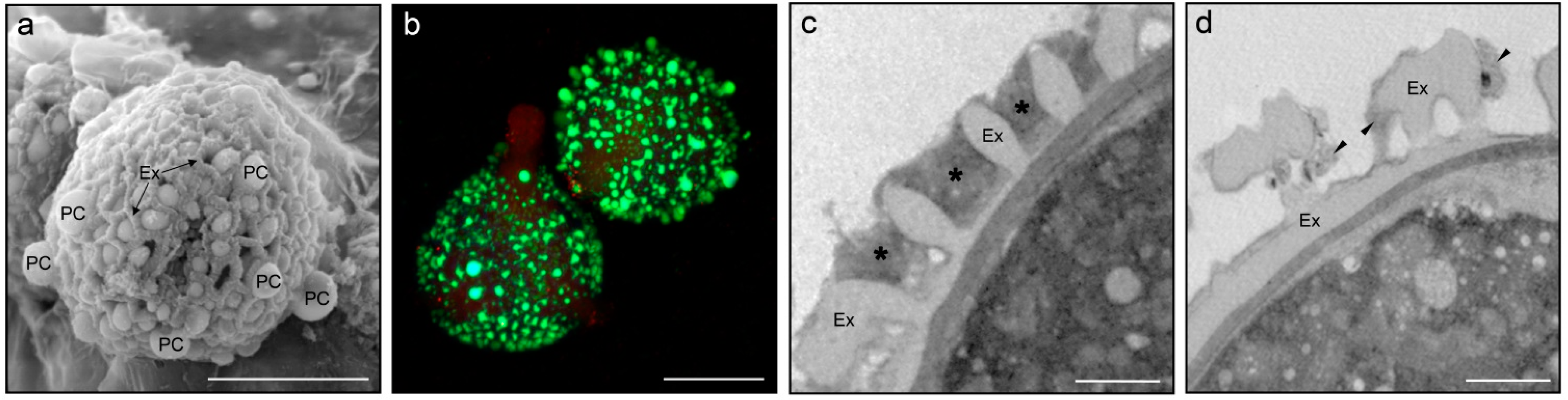
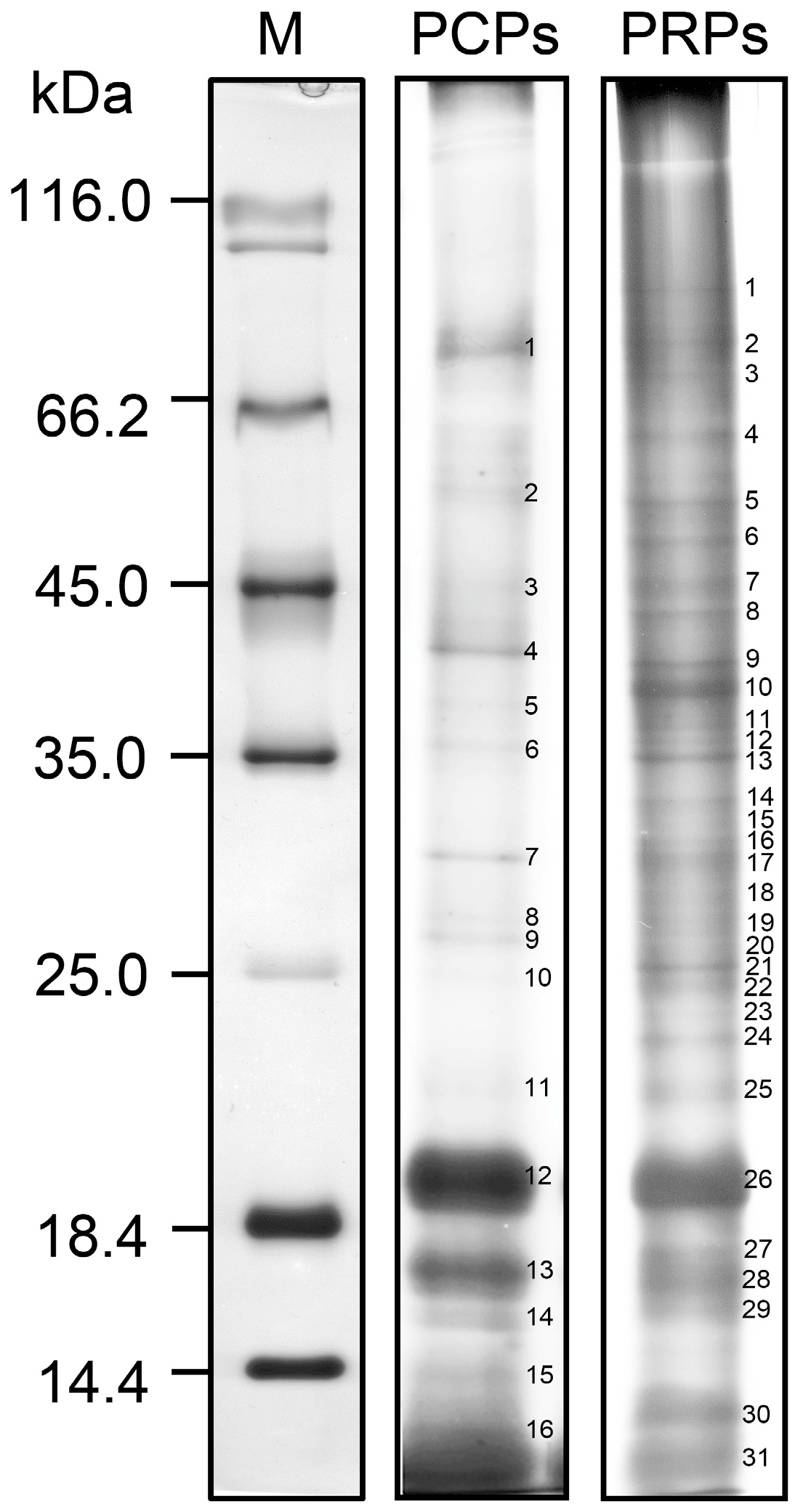
2. The Pollen Coat Proteome
| Protein Name 1 | Species 2 | Putative Function | References |
|---|---|---|---|
| Acetyl cholinesterases (EC 3.1.1.7) | Pollen-stigma communication | ||
| Acetyl cholinesterase | Olea europaea (EC) | [32] | |
| Cholinesterase | Vicia faba (EC) | [33] | |
| Acylesterases (EC 3.1.1.1) | Helianthus annuus (IGEA) | Pollen tube growths | [34] |
| Arabinogalactan proteins (JIM13 epitope) | Olea europaea (IL) | Pollen-stigma adhesion | [35] |
| Β-expansins | Pollen tube growth | ||
| Cyn d 1 * | Cynodon dactylon (WB) | [36] | |
| Phl p 1 * | Phleum pretense (IL) | [37] | |
| EXPB1a (Ory s 1 *) and OsEXPB13 | Oryza sativa (MS) | [26] | |
| Expansins B1 and B4 | xTriticosecale (MS) | [38] | |
| β-expansins-1 and 10 (Zea m 1 *) | Zea mays (IL/MS/WB) | [27,39,40,41,42,43] | |
| Β-glucanases (EC 3.2.1.6) | Pollen tube growth | ||
| Β-1,3-glucanase | Zea mays (SQ/IVEA) | [44] | |
| Ole e 9 * | Olea europaea (MS) | [28] (Table S1) | |
| Β-glucanase | Oryza sativa (MS) | [26] | |
| Endo-β-1,3-glucanase | xTriticosecale (MS) | [38] | |
| Caleosins | Pollen-stigma communication | ||
| EF-hand Ca2+-binding protein | Arabidopsis thaliana (SQ) | [24] | |
| Caleosin | Brassica napus (SQ) | [25] | |
| Caleosin | Olea europaea (IL/WB) | [15,45] | |
| ABA-induced caleosin | Zea mays (MS) | [27] | |
| Calmodulin-like proteins | Pollen tube growth | ||
| Bra r 1 * | Brassica rapa (IL/WB) | [46] | |
| Serine/threonine kinase | Olea europaea (MS) | [28] (Table S1) | |
| Cysteine proteases (EC 3.4.22) | Tapetum PCD/Pollen tube growth | ||
| CEP1 | Arabidopsis thaliana (IL) | [47] | |
| BGP-CP * | Cynodon dactylon (MS/WB) | [36] | |
| Phl p CP * | Phleum pretense (WB) | [36] | |
| Sor h CP * | Sorghum halepense (WB) | [36] | |
| Cysteine protease | Zea Mays (IL/MS/SQ/WB) | [27,48] | |
| GDSL esterases/lipases (EC 3.1.1.-) | Pollen rehydration/Pollen tube growth | ||
| EXL4 and EXL6 lipases | Arabidopsis thaliana (SQ/WB) | [24,49] | |
| GDSL esterase/lipase | Olea europaea (MS) | [28] (Table S1) | |
| Lipases (EC 3.1.1.3) | Helianthus annuus (EC/IVEA) | Unknown | [34] |
| Ole e 1 allergen family | Pollen tube growth | ||
| Ole e 1 * | Olea europaea (IL) | [50,51] (Table S1) | |
| Ole e 1-like * | xTriticosecale (MS) | [38] | |
| Pectinesterases (EC 3.1.1.11) | Pollen tube growth | ||
| Pectin esterase | Brassica napus (SQ) | [25] | |
| Ole e 11 * | Olea europaea (MS) | [28] (Table S1) | |
| Pectate lyases (EC 4.2.2.2) | Pollen tube growth | ||
| Cry j 1 * | Cryptomeria japonica (IL) | [52,53] | |
| Cup a 1 * | Cupressus arizonica (IL) | [54,55] | |
| Cry j 1-like * | Cupressus sempervirens (IL) | [55] | |
| Phl p 4 * | Phleum pretense (IL/MS/WB) | Unknown | [36,56] |
| Polygalaturonases (EC 3.2.1.15) | Pollen tube growth | ||
| Polygalacturonase | Brassica napus (IL) | [57] | |
| Polygalacturonase | Olea europaea (MS) | [28] (Table S1) | |
| Polygalacturonase | xTriticosecale (MS) | [38] | |
| Exopolygalacturonase (Zea m 13 *) | Zea mays (MS) | [27,44] | |
| Pollen coat protein, class A (PCP-A) | Self-incompatibility/Pollen rehydration/Pollen adhesion | ||
| PCP7-like | Brassica napus (IVIA) | [58] | |
| PCP7/PCP-A1 | Brassica oleracea (IVIA) | [29,59,60] | |
| PCP1 | Brassica oleracea (SQ) | [61] | |
| BcPCP-A1 | Brassica rapa (IVIA) | [20] | |
| SLR1-BP1 and SLR1-BP2 | Brassica rapa (IVIA/MS) | [20] | |
| SP11/SCR (male S-determinant) | Brassica rapa (IL/IVIA) | [18,23,62,63,64] | |
| Profilins | Unknown | ||
| Ole e 2 * | Olea europaea (IL/MS) | [65] | |
| Profilin/Ory s 12 * | Oryza sativa (MS) | [26] | |
| Profilin * | xTriticosecale (MS) | [38] | |
| Profilin/Zea m 12 * | Zea mays (MS/SQ) | [27,44] | |
| Receptor-like protein kinases | Unknown | ||
| Kinase | Arabidopsis thaliana (SQ) | [24] | |
| Protein kinase | Brassica napus (SQ) | [25] | |
| Subtilisin-like Ser proteases (EC 3.4.21.-) | Unknown | ||
| Subtilisin-like Ser protease | Oryza sativa (MS) | [26] | |
| Putative subtilase | Zea mays (MS) | [27] | |
| Tapetal oleosins (T-oleosins) | Pollen rehydration/Tapetosome formation/Pollen dehydration tolerance | ||
| GRP17 | Arabidopsis thaliana (MU/PAGE/WB) | [22] | |
| GRP14 & GRP16−19 | Arabidopsis thaliana (PAGE/SQ) | [24] | |
| T3, T5 & T6 oleosins | Arabidopsis thaliana (AMT/MU) | [14] | |
| BnOlnB;4 | Arabidopsis thaliana (AMT/IL/WB) | [66] | |
| BnOlnB;4 | Brassica carinata (AMT/IL/WB) | [67] | |
| BnOlnB;3, BnOlnB;4 & BnOlnB;6 | Brassica napus (PAGE/SQ) | [68] | |
| BnOlnB1−6 & 11/Pollenins 1−6 & 11 | Brassica napus (IL/SQ/WB) | [6,25] | |
| BnOlnB;3 & BnOlnB;4 | Brassica napus (IL/WB) | [8,9] | |
| 39-kDa oleosin fragment | Brassica oleracea (SQ/WB) | [9] | |
| BOPC3, BOPC4 & BOPC5 | Brassica oleracea (IS/PAGE/WB) | [69] | |
| 37-kDa oleosin fragment | Brassica rapa (SQ/WB) | [8,9] | |
| Xylanases (EC 3.2.1.8) | Pollen tube growth | ||
| EXY * | Cynodon dactylon (MS/WB) | [36] | |
| 30-kDa endoxylanase * | Phleum pretense (MS/WB) | [36] | |
| 1,4-β-xylanase | Oryza sativa (MS/WB) | [26] | |
| Endoxylanase | Zea Mays (MS/SQ/WB/IVEA) | [27,30,70,71] |
2.1. The Pollen Coat Proteome of Brassicaceae
2.2. The Pollen Coat Proteome of Grasses
2.3. The Pollen Coat Proteome of the Olive Tree
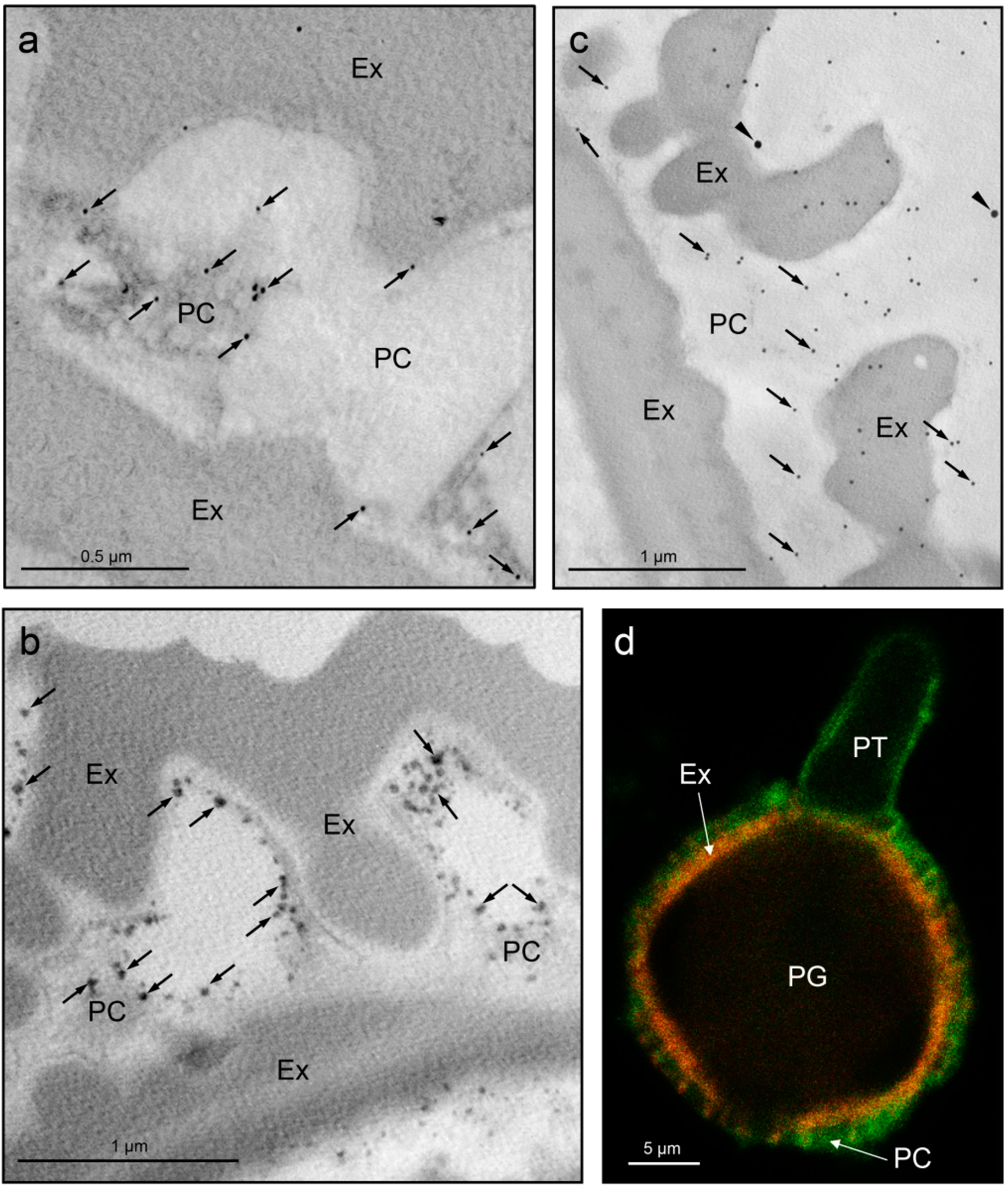
3. Molecular and Biological Functions of the Pollen Coat-Associated Proteins
3.1. Pollen Adhesion and Hydration
3.2. Pollen–Stigma Communication
3.3. Pollen Germination
3.4. Expanding the List of Pollen Coat Proteins and Functions
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lord, E.M.; Russell, S.D. The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 2002, 18, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef] [PubMed]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.C.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Pacini, E.; Hesse, M. Pollenkitt—Its composition, forms and functions. Flora 2005, 200, 399–415. [Google Scholar] [CrossRef]
- Piffanelli, P.; Ross, J.H.E.; Murphy, D.J. Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 1998, 11, 65–80. [Google Scholar] [CrossRef]
- Murphy, D.J.; Ross, J.H.E. Biosynthesis, targeting and processing of oleosin-like proteins, which are major pollen coat components in Brassica napus. Plant J. 1998, 13, 1–16. [Google Scholar] [PubMed]
- Wu, S.S.H.; Platt, K.A.; Ratnayake, C.; Wang, T.W.; Ting, J.T.L.; Huang, A.H.C. Isolation and characterization of neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc. Natl. Acad. Sci. USA 1997, 94, 12711–12716. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, K.; Huang, A.H.C. Lipid-rich tapetosomes in Brassica tapetum are composed of oleosin-coated oil droplets and vesicles, both assembled in and then detached from the endoplasmic reticulum. Plant J. 2005, 43, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Ting, J.T.L.; Wu, S.S.H.; Ratnayake, C.; Huang, A.H.C. Constituents of the tapetosomes and elaioplasts in Brassica campestris tapetum and their degradation and retention during microsporogenesis. Plant J. 1998, 16, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.S.H.; Moreau, R.A.; Whitaker, B.D.; Huang, A.H.C. Steryl esters in the elaioplasts of the tapetum in developing Brassica anthers and their recovery on the pollen surface. Lipids 1999, 34, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, K.; Huang, A.H.C. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 2007, 19, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, H.G. Role of plastids in formation of pollen grain coatings. Cytobios 1973, 8, 25–40. [Google Scholar] [PubMed]
- Pacini, E.; Casadoro, G. Tapetum plastids of Olea europaea L. Protoplasma 1981, 106, 289–296. [Google Scholar] [CrossRef]
- Huang, C.Y.; Chen, P.Y.; Huang, M.D.; Tsou, C.H.; Jane, W.N.; Huang, A.H.C. Tandem oleosin genes in a cluster acquired in Brassicaceae created tapetosomes and conferred additive benefit of pollen vigor. Proc. Natl. Acad. Sci. USA 2013, 110, 14480–14485. [Google Scholar] [CrossRef] [PubMed]
- Zienkiewicz, K.; Zienkiewicz, A.; Rodríguez-García, M.I.; Castro, A.J. Characterization of a caleosin expressed during olive (Olea europaea L.) pollen ontogeny. BMC Plant Biol. 2011, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Keijzer, C.J. The processes of anther dehiscence and pollen dispersal. II. The formation and the transfer mechanism of pollenkitt, cell-wall development of the loculus tissues and a function of orbicules in pollen dispersal. New Phytol. 1987, 105, 499–507. [Google Scholar] [CrossRef]
- Gong, F.; Wu, X.; Wang, W. Diversity and function of maize pollen coat proteins: From biochemistry to proteomics. Front. Plant Sci. 2015, 6, 199. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Shiba, H.; Iwano, M.; Shimosato, H.; Che, F.S.; Kai, N.; Watanabe, M.; Suzuki, G.; Hinata, K.; Isogai, A. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 2000, 97, 1920–1925. [Google Scholar] [CrossRef] [PubMed]
- Wolters-Arts, M.; Lush, W.M.; Mariani, C. Lipids are required for directional pollen-tube growth. Nature 1998, 392, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Shiba, H.; Iwano, M.; Asano, K.; Hara, M.; Che, F.S.; Watanabe, M.; Hinata, K.; Isogai, A. Isolation and characterization of pollen coat proteins of Brassica campestris that interact with S locus-related glycoprotein 1 involved in pollen-stigma adhesion. Proc. Natl. Acad. Sci. USA 2000, 97, 3765–3770. [Google Scholar] [CrossRef] [PubMed]
- Preuss, D.; Lemieux, B.; Yen, G.; Davis, R.W. A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 1993, 7, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, J.A.; Preuss, D. Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2000, 2, 128–130. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. The male determinant of self-incompatibility in Brassica. Science 1999, 286, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, J.A.; Fiebig, A.; Johnstone, S.E.; Preuss, D. Gene families from the Arabidopsis thaliana pollen coat proteome. Science 2001, 292, 2482–2485. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.J. The extracellular pollen coat in members of the Brassicaceae: Composition, biosynthesis, and functions in pollination. Protoplasma 2006, 228, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Li, L.; Chen, T.; Chong, K.; Xue, Y.; Wang, T. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 2006, 6, 2504–2529. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cai, G.; Gong, F.; An, S.; Cresti, M.; Wang, W. Proteome profiling of maize pollen coats reveals novel protein components. Plant Mol. Biol. Rep. 2015, 33, 975–986. [Google Scholar] [CrossRef]
- Rejón, J.D. Characterization of esterase enzymes involved in pollen-pistil interaction in Olea europaea L. Ph.D. Thesis, University of Granada, Granada, Spain, 2012. [Google Scholar]
- Doughty, J.; Hedderson, F.; McCubbin, A.; Dickinson, H.G. Interaction between a coating-borne peptide of the Brassica pollen grain and stigmatic-S (self-incompatibility)-locus-specific glycoproteins. Proc. Natl. Acad. Sci. USA 1993, 90, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Bih, F.Y.; Wu, S.S.H.; Ratnayake, C.; Walling, L.L.; Nothnagel, E.A.; Huang, A.H.C. The predominant protein on the surface of maize pollen is an endoxylanase synthesized by a tapetum mRNA with a long 5′ leader. J. Biol. Chem. 1999, 274, 22884–22894. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Scali, M.; Vignani, R.; Spadafora, A.; Sensi, E.; Mazzuca, S.; Cresti, M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 2003, 24, 2369–2375. [Google Scholar] [CrossRef] [PubMed]
- Rejón, J.D.; Zienkiewicz, A.; Rodríguez-García, M.I.; Castro, A.J. Profiling and functional classification of esterases in olive (Olea europaea) pollen during germination. Ann. Bot. 2012, 110, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Bednarska, E. The localization of nonspecific esterase and cholinesterase activity in germinating pollen and in pollen tube of Vicia faba. I. The effect of actinomycin-D and cycloheximide. Biol. Plant. 1992, 34, 229–240. [Google Scholar] [CrossRef]
- Shakya, R.; Bhatla, S.C. A comparative analysis of the distribution and composition of lipidic constituents and associated enzymes in pollen and stigma of sunflower. Sex. Plant Reprod. 2010, 23, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.J.; Suarez, C.; Zienkiewicz, K.; Alché, J.D.; Zienkiewicz, A.; Rodríguez-García, M.I. Electrophoretic profiling and immunocytochemical detection of pectins and arabinogalactan proteins in olive pollen during germination and pollen tube growth. Ann. Bot. 2013, 112, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Bashir, M.E.H.; Ward, J.M.; Cummings, M.; Karrar, E.E.; Root, M.; Mohamed, A.B.A.; Naclerio, R.M.; Preuss, D. Dual function of novel pollen coat (surface) proteins: IgE-binding capacity and proteolytic activity disrupting the airway epithelial barrier. PLoS ONE 2013, 8, e53337. [Google Scholar]
- Staff, I.A.; Taylor, P.E.; Smith, P.; Singh, M.B.; Knox, R.B. Cellular localization of water-soluble, allergenic proteins in rye-grass (Lolium perenne) pollen using monoclonal and specific IgE antibodies with immunogold probes. Histochem. J. 1990, 22, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, M.A.; O’Leary, S.; Wu, S.; Gleddie, S.; Eudes, F.; Laroche, A.; Robert, L.S. A molecular and proteomic investigation of proteins rapidly released from triticale pollen upon hydration. Plant Mol. Biol. 2012, 79, 101–121. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Bedinger, P.A.; Volk, C.; Jones, A.D.; Cosgrove, D.J. Purification and characterization of four β-expansins (Zea m 1 isoforms) from maize pollen. Plant Physiol. 2003, 132, 2073–2085. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Milanesi, C.; Faleri, C.; Cresti, M. Localization of group-1 allergen Zea m 1 in the coat and wall of maize pollen. Acta Histochem. 2006, 108, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Yennawar, N.H.; Li, L.C.; Dudzinski, D.M.; Tabuchi, A.; Cosgrove, D.J. Crystal structure and activities of EXPB1 (Zea m 1), α,β-expansin and group-1 pollen allergen from maize. Proc. Natl. Acad. Sci. USA 2006, 103, 14664–14671. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, E.R.; Stephenson, A.G.; Durachko, D.M.; Cosgrove, D.J. Class B β-expansins are needed for pollen separation and stigma penetration. Sex. Plant Reprod. 2009, 22, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, A.; Li, L.C.; Cosgrove, D.J. Matrix solubilization and cell wall weakening by β-expansin (group-1 allergen) from maize pollen. Plant J. 2011, 68, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Suen, D.F.; Wu, S.S.H.; Chang, H.C.; Dhugga, K.S.; Huang, A.H.C. Cell wall reactive proteins in the coat and wall of maize pollen—Potential role in pollen tube growth on the stigma and through the style. J. Biol. Chem. 2003, 278, 43672–43681. [Google Scholar] [CrossRef] [PubMed]
- Zienkiewicz, K.; Castro, A.J.; Alché, J.D.; Zienkiewicz, A.; Suarez, C.; Rodríguez-García, M.I. Identification and localization of a caleosin in olive (Olea europaea L.) pollen during in vitro germination. J. Exp. Bot. 2010, 61, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Zhang, Z.J.; Russell, S.D.; Toriyama, K. Localization of the Ca2+-binding protein, Bra r 1, in anthers and pollen tubes. Plant Cell Physiol. 1999, 40, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Suen, D.F.; Huang, C.Y.; Kung, S.Y.; Huang, A.H.C. The maize tapetum employs diverse mechanisms to synthesize and store proteins and flavonoids and transfer them to the pollen surface. Plant Physiol. 2012, 158, 1548–1561. [Google Scholar] [CrossRef] [PubMed]
- Updegraff, E.P.; Zhao, F.; Preuss, D. The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex. Plant Reprod. 2009, 22, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Alché, J.D.; Castro, A.J.; Olmedilla, A.; Fernández, M.C.; Rodríguez, R.; Villalba, M.; Rodríguez-García, M.I. The major olive pollen allergen (Ole e I) shows both gametophytic and sporophytic expression during anther development, and its synthesis and storage takes place in the RER. J. Cell Sci. 1999, 112, 2501–2509. [Google Scholar]
- Alché, J.D.; M´rani-Alaoui, M.; Castro, A.J.; Rodríguez-García, M.I. Ole e 1, the major allergen from olive (Olea europaea L.) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant Cell Physiol. 2004, 45, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Miki-Hirosige, H.; Nakamura, S.; Yasueda, H.; Shida, T.; Takahashi, Y. Immunocytochemical localization of the allergenic proteins in the pollen of Cryptomeria japonica. Sex. Plant Reprod. 1994, 7, 95–100. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Ono, A.; Sawatani, M.; Nanba, M.; Kohno, K.; Usui, H.; Kurimoto, M.; Matuhasi, T. Cry j I, a major allergen of Japanese cedar pollen, has pectate lyase enzyme activity. Allergy 1995, 50, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Aceituno, E.; Del Pozo, V.; Mínguez, A.; Arrieta, I.; Cortegano, I.; Cárdaba, B.; Gallardo, S.; Rojo, M.; Palomino, P.; Lahoz, C. Molecular cloning of major allergen from Cupressus arizonica pollen: Cup a 1. Clin. Exp. Allergy 2000, 30, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Cervera, M.; Takahashi, Y.; Vega-Maray, A.; Seoane-Camba, J.A. Immunocytochemical localization of Cry j 1, the major allergen of Cryptomeria japonica (Taxodiaceae) in Cupressus arizonica and Cupressus sempervirens (Cupressaceae) pollen grains. Sex. Plant Reprod. 2003, 16, 9–15. [Google Scholar]
- Fischer, S.; Grote, M.; Fahlbusch, B.; Muller, W.D.; Kraft, D.; Valenta, R. Characterization of Phl p 4, a major timothy grass (Phleum pratense) pollen allergen. J. Allergy Clin. Immunol. 1996, 98, 189–198. [Google Scholar] [CrossRef]
- Dearnaley, J.D.W.; Daggard, G.A. Expression of a polygalacturonase enzyme in germinating pollen of Brassica napus. Sex. Plant Reprod. 2001, 13, 265–271. [Google Scholar] [CrossRef]
- Hiscock, S.J.; Doughty, J.; Willis, A.C.; Dickinson, H.G. A 7-kDa pollen coating-borne peptide from Brassica napus interacts with S-locus glycoprotein and S-locus-related glycoprotein. Planta 1995, 196, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Doughty, J.; Dixon, S.; Hiscock, S.J.; Willis, A.C.; Parkin, I.A.P.; Dickinson, H.G. PCP-A1, a defensin-like Brassica pollen coat protein that binds the S locus glycoprotein, is the product of gametophytic gene expression. Plant Cell 1998, 10, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, A.G.; Doughty, J.; Dixon, S.; Elleman, C.; Hiscock, S.J.; Dickinson, H.G. The male determinant of self-incompatibility in Brassica oleracea is located in the pollen coating. Plant J. 1997, 12, 1351–1359. [Google Scholar] [CrossRef]
- Stanchev, B.S.; Doughty, J.; Scutt, C.P.; Dickinson, H.G.; Croy, R.R.D. Cloning of PCP1, a member of a family of pollen coat protein (PCP) genes from Brassica oleracea encoding novel cysteine-rich proteins involved in pollen-stigma interactions. Plant J. 1996, 10, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Kai, N.; Hirose, T.; Fukui, K.; Nishio, T.; Takayama, S.; Isogai, A.; Watanabe, M.; Hinata, K. Genomic organization of the S locus: Identification and characterization of genes in SLG/SRK region of S-9 haplotype of Brassica campestris (syn. rapa). Genetics 1999, 153, 391–400. [Google Scholar] [PubMed]
- Shiba, H.; Takayama, S.; Iwano, M.; Shimosato, H.; Funato, M.; Nakagawa, T.; Che, F.S.; Suzuki, G.; Watanabe, M.; Hinata, K.; et al. A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica species. Plant Physiol. 2001, 125, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Iwano, M.; Shiba, H.; Funato, M.; Shimosato, H.; Takayama, S.; Isogai, A. Immunohistochemical studies on translocation of pollen S-haplotype determinant in self-incompatibility of Brassica rapa. Plant Cell Physiol. 2003, 44, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Morales, S.; Jiménez-López, J.C.; Castro, A.J.; Rodríguez-García, M.I.; Alché, J.D. Olive pollen profilin (Ole e 2 allergen) co-localizes with highly active areas of the actin cytoskeleton and is released to the culture medium during in vitro pollen germination. J. Microsc. 2008, 231, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Lévesque-Lemay, M.; Chabot, D.; Hubbard, K.; Chan, J.K.; Miller, S.; Robert, L.S. Tapetal oleosins play an essential role in tapetosome formation and protein relocation to the pollen coat. New Phytol. 2016, 209, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Foster, E.; Schneiderman, D.; Cloutier, M.; Gleddie, S.; Robert, L.S. Modifying the pollen coat protein composition in Brassica. Plant J. 2002, 31, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.H.E.; Murphy, D.J. Characterization of anther-expressed genes encoding a major class of extracellular oleosin-like proteins in the pollen coat of Brassicaceae. Plant J. 1996, 9, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Ruiter, R.K.; van Eldik, G.J.; van Herpen, R.M.A.; Schrauwen, J.A.M.; Wullems, G.J. Characterization of oleosins in the pollen coat of Brassica oleracea. Plant Cell 1997, 9, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.S.H.; Suen, D.F.; Chang, H.C.; Huang, A.H.C. Maize tapetum xylanase is synthesized as a precursor, processed and activated by a serine protease, and deposited on the pollen. J. Biol. Chem. 2002, 277, 49055–49064. [Google Scholar] [CrossRef] [PubMed]
- Suen, D.F.; Huang, A.H.C. Maize pollen coat xylanase facilitates pollen tube penetration into silk during sexual reproduction. J. Biol. Chem. 2007, 282, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Kimport, R.; Preuss, D. Comparisons of pollen coat genes across Brassicaceae species reveal rapid evolution by repeat expansion and diversification. Proc. Natl. Acad. Sci. USA 2004, 101, 3286–3291. [Google Scholar] [CrossRef] [PubMed]
- Schein, M.; Yang, Z.H.; Mitchell-Olds, T.; Schmid, K.J. Rapid evolution of a pollen-specific oleosin-like gene family from Arabidopsis thaliana and closely related species. Mol. Biol. Evol. 2004, 21, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Robert, L.S.; Gerster, J.; Allard, S.; Cass, L.; Simmonds, J. Molecular characterization of two Brassica napus genes related to oleosins which are highly expressed in the tapetum. Plant J. 1994, 6, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Hsieh, K.; Ratnayake, C.; Huang, A.H.C. A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 2002, 277, 22677–22684. [Google Scholar] [CrossRef] [PubMed]
- Akoh, C.C.; Lee, G.C.; Liaw, Y.C.; Huang, T.H.; Shaw, J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, E.R.; Sampedro, J.; Lamb, J.C.; Chopra, S.; Cosgrove, D.J. Recent proliferation and translocation of pollen group-1 allergen genes in the maize genome. Plant Physiol. 2007, 143, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Broadwater, A.H.; Rubinstein, A.L.; Chay, C.H.; Klapper, D.G.; Bedinger, P.A. Zea m I, the maize homolog of the allergen-encoding Lol p I gene of rye grass. Gene 1993, 131, 227–230. [Google Scholar] [CrossRef]
- Wu, Y.J.; Meeley, R.B.; Cosgrove, D.J. Analysis and expression of the α-expansin and β-expansin gene families in maize. Plant Physiol. 2001, 126, 222–232. [Google Scholar] [CrossRef] [PubMed]
- SecretomeP 2.0 Server. Available online: http://www.webcitation.org/6ej7Au1nK (accessed on 22 January 2016).
- Kalinowski, A.; Winiarezyk, K.; Radlowski, M. Pollen coat proteins after two-dimensional gel electrophoresis and pollen wall ultrastructure of Secale cereale and Festuca pratensis. Sex. Plant Reprod. 2002, 15, 75–83. [Google Scholar]
- Kalinowski, A.; Radlowski, M.; Bocian, A. Effects of interaction between pollen coat eluates and pistil at the molecular level in self-compatible and self-incompatible plants of Lolium multiflorum Lam. J. Appl. Genet. 2006, 47, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, A.; Klimko, M.; Wojciechowska, B. Pollen morphology and two-dimensional patterns of pollen coat and protoplast proteins in Aegilops kotschyi x Secale cereale amphiploids. Acta Biol. Crac. Ser. Bot. 2005, 47, 97–110. [Google Scholar]
- Rejón, J.D.; Delalande, F.; Schaeffer-Reiss, C.; Carapito, C.; Zienkiewicz, K.; Alché, J.D.; Rodríguez-García, M.I.; van Dorsselaer, A.; Castro, A.J. Proteomics profiling reveals novel proteins and functions of the plant stigma exudate. J. Exp. Bot. 2013, 64, 5695–5705. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.; Batanero, E.; Monsalve, R.I.; De la Peña, M.A.G.; Lahoz, C.; Rodríguez, R. Cloning and expression of Ole e I, the major allergen from olive tree pollen—Polymorphism analysis and tissue-specificity. J. Biol. Chem. 1994, 269, 15217–15222. [Google Scholar] [PubMed]
- Hamman-Khalifa, A.; Castro, A.J.; Jiménez-López, J.C.; Rodríguez-García, M.I.; Alché, J.D. Olive cultivar origin is a major cause of polymorphism for Ole e 1 pollen allergen. BMC Plant Biol. 2008, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.J.; Bednarczyk, A.; Schaeffer-Reiss, C.; Rodríguez-García, M.I.; van Dorsselaer, A.; Alché, J.D. Screening of Ole e 1 polymorphism among olive cultivars by peptide mapping and N-glycopeptide analysis. Proteomics 2010, 10, 953–962. [Google Scholar] [CrossRef] [PubMed]
- Edlund, A.F.; Swanson, R.; Preuss, D. Pollen and stigma structure and function: The role of diversity in pollination. Plant Cell 2004, 16, S84–S97. [Google Scholar] [CrossRef] [PubMed]
- Zinkl, G.M.; Zwiebel, B.I.; Grier, D.G.; Preuss, D. Pollen-stigma adhesion in Arabidopsis: A species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 1999, 126, 5431–5440. [Google Scholar] [PubMed]
- Luu, D.T.; Heizmann, P.; Dumas, C. Pollen-stigma adhesion in kale is not dependent on the self-(in)compatibility genotype. Plant Physiol. 1997, 115, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Luu, D.T.; Marty-Mazars, D.; Trick, M.; Dumas, C.; Heizmann, P. Pollen-stigma adhesion in Brassica spp involves SLG and SLR1 glycoproteins. Plant Cell 1999, 11, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.; Bosch, M.; Bots, M.; Nieuwland, J.; Feron, R.; Mariani, C. Pistil factors controlling pollination. Plant Cell 2004, 16, S98–S106. [Google Scholar] [CrossRef] [PubMed]
- Elleman, C.J.; Franklin-Tong, V.; Dickinson, H.G. Pollination in species with dry stigmas—The nature of the early stigmatic response and the pathway taken by pollen tubes. New Phytol. 1992, 121, 413–424. [Google Scholar] [CrossRef]
- Hulskamp, M.; Kopczak, S.D.; Horejsi, T.F.; Kihl, B.K.; Pruitt, R.E. Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 1995, 8, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Mayfield, J.A.; Miley, N.L.; Chau, S.; Fischer, R.L.; Preuss, D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 2000, 12, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, H.G. Dry stigmas, water and self-incompatibility in Brassica. Sex. Plant Reprod. 1995, 8, 1–10. [Google Scholar] [CrossRef]
- Suárez, C.; Castro, A.J.; Rapoport, H.F.; Rodríguez-García, M.I. Morphological, histological and ultrastructural changes in the olive pistil during flowering. Sex. Plant Reprod. 2012, 25, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Wolters-Arts, M.; van der Weerd, L.; van Aelst, A.C.; van der Weerd, J.; van As, H.; Mariani, C. Water-conducting properties of lipids during pollen hydration. Plant Cell Environ. 2002, 25, 513–519. [Google Scholar] [CrossRef]
- Goldman, M.H.S.; Goldberg, R.B.; Mariani, C. Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J. 1994, 13, 2976–2984. [Google Scholar] [PubMed]
- Bots, M.; Mariani, C. Oleosin-like proteins are not present on the surface of tobacco pollen. Sex. Plant Reprod. 2004, 16, 223–226. [Google Scholar] [CrossRef]
- Alché, J.D.; Castro, A.J.; Rodríguez-García, M.I. Expression of oleosin genes in the olive (Olea europaea) anther. In Anther and Pollen: From Biology to Biotechnology; Clément, C., Pacini, E., Audran, J.C., Eds.; Springer-Verlag: Berlin, Germany, 1999; pp. 91–99. [Google Scholar]
- Takasaki, T.; Hatakeyama, K.; Suzuki, G.; Watanabe, M.; Isogai, A.; Hinata, K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 2000, 403, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Shimosato, H.; Shiba, H.; Funato, M.; Che, F.S.; Watanabe, M.; Iwano, M.; Isogai, A. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 2001, 413, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Shimosato, H.; Yokota, N.; Shiba, H.; Iwano, M.; Entani, T.; Che, F.S.; Watanabe, M.; Isogai, A.; Takayama, S. Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in Brassica self-incompatibility. Plant Cell 2007, 19, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.F.; Tsai, C.C.Y.; Tzen, J.T.C. Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol. 1999, 40, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Sagane, Y.; Nakagawa, T.; Yamamoto, K.; Michikawa, S.; Oguri, S.; Momonoki, Y.S. Molecular characterization of maize acetylcholinesterase. A novel enzyme family in the plant kingdom. Plant Physiol. 2005, 138, 1359–1371. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, T.; Akita, I.; Yoshino, N.; Suzuki, Y. Regulation of self-incompatibility by acetylcholine and cAMP in Lilium longiflorum. J. Plant Physiol. 2007, 164, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D.A. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Upton, C.; Buckley, J.T. A new family of lipolytic enzymes. Trends Biochem. Sci. 1995, 20, 178–179. [Google Scholar] [CrossRef]
- Hiscock, S.J.; Dewey, F.M.; Coleman, J.O.D.; Dickinson, H.G. Identification and localization of an active cutinase in the pollen of Brassica napus L. Planta 1994, 193, 377–384. [Google Scholar] [CrossRef]
- Valdivia, E.R.; Wu, Y.; Li, L.C.; Cosgrove, D.J.; Stephenson, A.G. A group-1 grass pollen allergen influences the outcome of pollen competition in maize. PLoS ONE 2007, 2, e154. [Google Scholar] [CrossRef] [PubMed]
- Shpigel, E.; Roiz, L.; Goren, R.; Shoseyov, O. Bacterial cellulose-binding domain modulates in vitro elongation of different plant cells. Plant Physiol. 1998, 117, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 445–476. [Google Scholar] [CrossRef] [PubMed]
- Huecas, S.; Villalba, M.; Rodríguez, R. Ole e 9, a major olive pollen allergen is a 1,3-β-glucanase. J. Biol. Chem. 2001, 276, 27959–27966. [Google Scholar] [CrossRef] [PubMed]
- Roggen, H.P.J.; Stanley, R.G. Cell-wall-hydrolyzing enzymes in wall formation as measured by pollen-tube extension. Planta 1969, 84, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Yoshikawa, T.; Liu, X.Z.; Nakagawa, N.; Li, Y.Q.; Sakurai, N. Molecular cloning of two exo-β-glucanases and their in vivo substrates in the cell walls of lily pollen tubes. Plant Cell Physiol. 2004, 45, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.J.; Alché, J.D.; Cuevas, J.; Romero, P.J.; Alché, V.; Rodríguez-García, M.I. Pollen from different olive tree cultivars contains varying amounts of the major allergen Ole e 1. Int. Arch. Allergy Immunol. 2003, 131, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Muschietti, J.; Dircks, L.; vancanneyt, G.; McCormick, S. LAT52 protein is essential for tomato pollen development—Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 1994, 6, 321–338. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Ezcurra, I.; Muschietti, J.; McCormick, S. A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 2002, 14, 2277–2287. [Google Scholar] [CrossRef] [PubMed]
- Wengier, D.; Valsecchi, I.; Cabanas, M.L.; Tang, W.H.; McCormick, S.; Muschietti, J. The receptor kinases LePRK1 and LePRK2 associate in pollen and when expressed in yeast, but dissociate in the presence of style extract. Proc. Natl. Acad. Sci. USA 2003, 100, 6860–6865. [Google Scholar] [CrossRef] [PubMed]
- Coffeen, W.C.; Wolpert, T.J. Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 2004, 16, 857–873. [Google Scholar] [CrossRef] [PubMed]
- Coimbra, S.; Almeida, J.; Junqueira, V.; Costa, M.L.; Pereira, L.G. Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. J. Exp. Bot. 2007, 58, 4027–4035. [Google Scholar] [CrossRef] [PubMed]
- Suárez, C.; Zienkiewicz, A.; Castro, A.J.; Zienkiewicz, K.; Majewska-Sawka, A.; Rodríguez-García, M.I. Cellular localization and levels of pectins and arabinogalactan proteins in olive (Olea europaea L.) pistil tissues during development: Implications for pollen-pistil interaction. Planta 2013, 237, 305–319. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rejón, J.D.; Delalande, F.; Schaeffer-Reiss, C.; Alché, J.D.D.; Rodríguez-García, M.I.; Van Dorsselaer, A.; Castro, A.J. The Pollen Coat Proteome: At the Cutting Edge of Plant Reproduction. Proteomes 2016, 4, 5. https://doi.org/10.3390/proteomes4010005
Rejón JD, Delalande F, Schaeffer-Reiss C, Alché JDD, Rodríguez-García MI, Van Dorsselaer A, Castro AJ. The Pollen Coat Proteome: At the Cutting Edge of Plant Reproduction. Proteomes. 2016; 4(1):5. https://doi.org/10.3390/proteomes4010005
Chicago/Turabian StyleRejón, Juan David, François Delalande, Christine Schaeffer-Reiss, Juan De Dios Alché, María Isabel Rodríguez-García, Alain Van Dorsselaer, and Antonio Jesús Castro. 2016. "The Pollen Coat Proteome: At the Cutting Edge of Plant Reproduction" Proteomes 4, no. 1: 5. https://doi.org/10.3390/proteomes4010005
APA StyleRejón, J. D., Delalande, F., Schaeffer-Reiss, C., Alché, J. D. D., Rodríguez-García, M. I., Van Dorsselaer, A., & Castro, A. J. (2016). The Pollen Coat Proteome: At the Cutting Edge of Plant Reproduction. Proteomes, 4(1), 5. https://doi.org/10.3390/proteomes4010005







