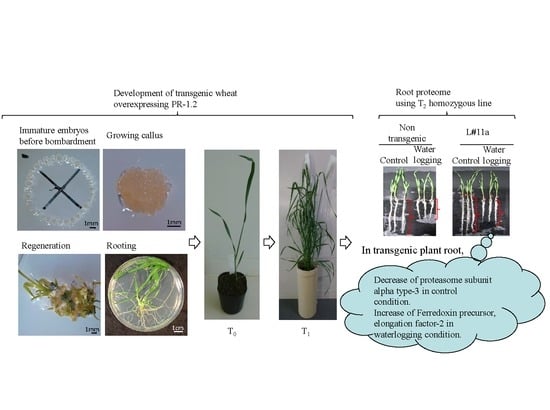
Quantitative Proteomics of the Root of Transgenic Wheat Expressing TaBWPR-1.2 Genes in Response to Waterlogging
Abstract
:1. Introduction
2. Experimental Section
2.1. Construct Preparation
2.2. Plant Material Preparation for Transformation
2.3. Isolation of Scutellar Tissues from Immature Embryos
2.4. Biolistic Transformation
2.5. Tissue Culture and Selection of Transgenic Plants
2.6. PCR Analysis of Transgenic Plants
2.7. Analysis of Gene Expression in Different Organs by RT-PCR
2.8. Gene Expression Analysis by qRT-PCR in Homozygous Transformants under WL
2.9. Protein Extraction and Immunoblot Analysis with Rice Anti-PR-1 Antibody
2.10. Preparation of Proteins for Mass Spectrometry (MS)
2.11. Data Acquisition by Nano-Liquid Chromatography (LC) MS/MS
2.12. Protein Identification
2.13. Analysis of Differential Protein Abundance Using Acquired MS Data
3. Results
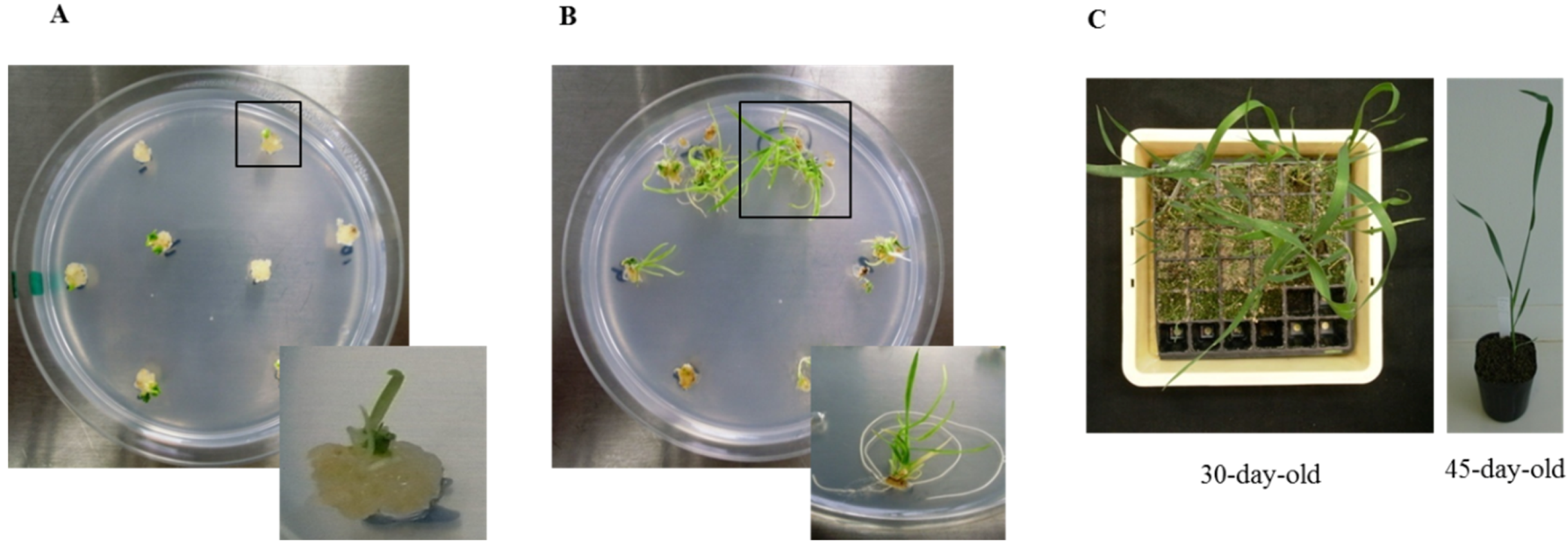
3.1. Regeneration and Establishment of Homozygous Lines
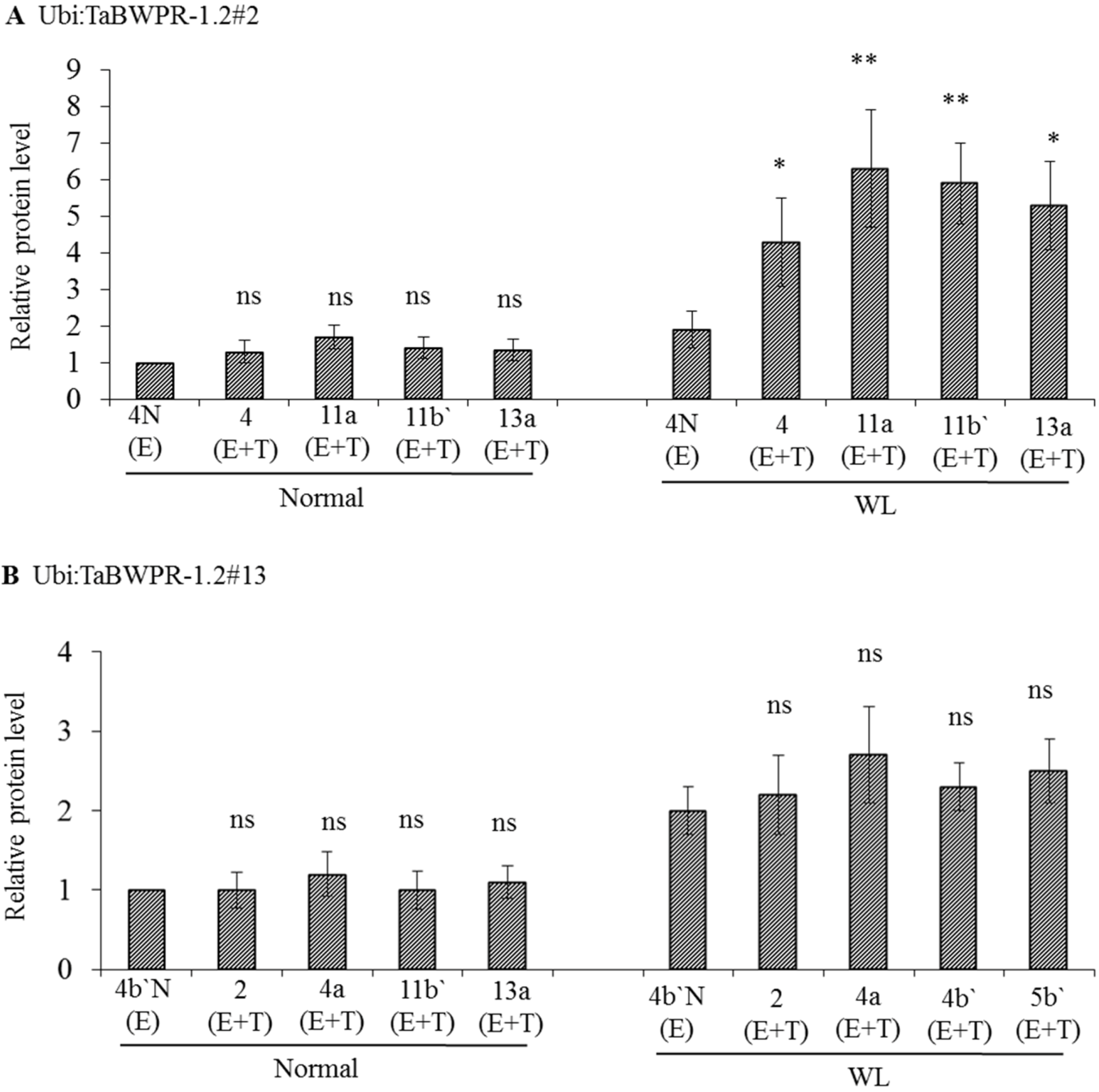
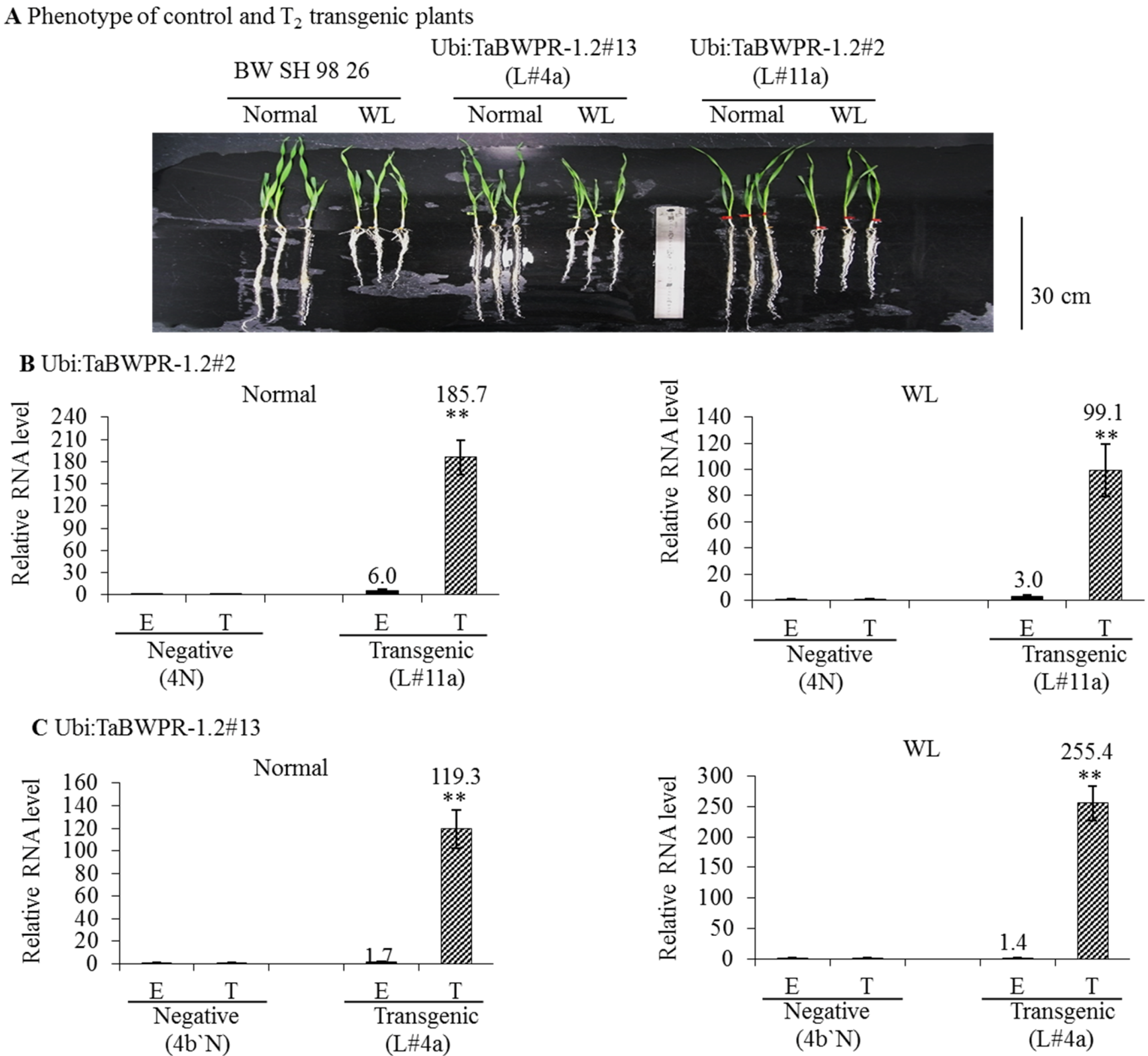
3.2. mRNA and Protein Levels in Ubi:TaBWPR-1.2 Transformants
3.3. Changes in Protein Levels in Seminal Roots of TaBWPR-1.2#2-Overexpressor Transgenic Plants
| Protein Name | Accession No. a | Organism | MP b | Ratio c | SD d | |
|---|---|---|---|---|---|---|
| Normal conditions (11a/4N) | ||||||
| 1 | Pathogenesis-related protein 1_17 | F8S6U7 | T. aestivum | 4 | 3.0 | 0.45 |
| 2 | Pathogenesis-related protein 1_14 | F8S6U4 | T. aestivum | 2 | 1.6 | 0.5 |
| 3 | Pathogenesis-related protein | H2KXF7 | T. aestivum | 4 | 0.44 | 0.02 |
| 4 | Pathogenesis-related protein 10 | B5B3P8 | T. aestivum | 4 | 0.46 | 0.08 |
| 5 | Unknown Proteasome subunit alpha type-3 | AK332255 * | T. aestivum | 5 | 0.58 | 0.05 |
| ACN10361 * | Salmo salar | |||||
| WL conditions (11a/4N) | ||||||
| 1 | Pathogenesis-related protein 1_6 | F8S6T6 | T. aestivum | 2 | 2.0 | 0.6 |
| 2 | Ferredoxin precursor | Q8S3J5 | T. aestivum | 2 | 2.0 | 1.2 |
| 3 | Elongation factor-2 | Q9M7S5 | T. aestivum | 2 | 1.9 | 1.1 |
| 4 | Unknown (contig 2626) | AK331943 * | T. aestivum | 2 | 1.4 | 0.1 |
| 5 | Pathogenesis-related protein | H2KXF7 | T. aestivum | 4 | 0.6 | 0.09 |
4. Discussion
5. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Abdul, R.; Ma, Z.; Wang, H. Genetic transformation of wheat (Triticum aestivum L): A review. Triticeae Genome Genet. 2010, 1, 1–7. [Google Scholar]
- Bhalla, P.L.; Ottenhof, H.H.; Singh, M.B. Wheat transformation—An update of recent progress. Euphytica 2006, 149, 353–366. [Google Scholar] [CrossRef]
- Jones, H.D. Wheat transformation: Current technology and applications to grain development and composition. J. Cereal Sci. 2005, 41, 137–147. [Google Scholar] [CrossRef]
- Zeller, S.L.; Kalinina, O.; Brunner, S.; Keller, B.; Schmid, B. Transgene environment interactions in genetically modified wheat. PLoS One 2010, 5, e11405. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, H.; Li, Y.; Ren, J.; Wang, X.; Niu, H.; Yin, J. Identification of changes in wheat (Triticum aestivum L.) seeds proteome in response to anti-trxs gene. PLoS One 2011, 6, e22255. [Google Scholar] [CrossRef] [PubMed]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Pellegrineschi, A.; Reynolds, M.; Pacheco, M.; Brito, R.M.; Almeraya, R.; Yamaguchi-Shinozaki, K.; Hoisington, D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 2004, 47, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.H.; Sharrock, R.A.; Quail, P.H. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 1992, 18, 675–689. [Google Scholar] [CrossRef] [PubMed]
- McElroy, D.; Blowers, A.D.; Jenes, B.; Wu, R. Construction of expression vectors based on the rice actin-1 (Act1) 5' region for use in monocot transformation. Mol. Gen. Genet. 1991, 231, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Rooke, L.; Bekes, F.; Fido, R.; Barro, F.; Gras, P.; Tatham, A.S.; Barcelo, P.; Lazzeri, P.; Shewry, P.R. Overexpression of a gluten protein in transgenic wheat results in greatly increased dough strength. J. Cereal Sci. 2000, 30, 115–120. [Google Scholar] [CrossRef]
- Stoger, E.; Williams, S.; Keen, D.; Christou, P. Constitutive versus seed specific expression in transgenic wheat: Temporal and spatial control. Transgenic Res. 1999, 8, 73–82. [Google Scholar] [CrossRef]
- Kovalchuk, N.; Jia, W.; Eini, O.; Morran, S.; Pyvovarenko, T.; Fletcher, S.; Bazanova, N.; Harris, J.; Beck-Oldach, K.; Shavrukov, Y.; Langridge, P.; Lopato, S. Optimization of TaDREB3 gene expression in transgenic barley using cold-inducible promoters. Plant Biotechnol. J. 2013, 11, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, M.; Gao, S.; Zhang, Z.; Zhao, X.; Zhao, C.; Zhang, F.; Chen, X. Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol. Plant. 2012, 144, 210–224. [Google Scholar] [CrossRef] [PubMed]
- Eldakak, M.; Milad, S.I.M.; Nawar, A.; Rohila, J.S. Proteomics: A biotechnology tool for crop improvement. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, D.; Forshey, K.L.; Grimsrud, P.A.; Ane, J.M. Leveraging proteomics to understand plant-microbe interactions. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Redillas, M.C.F.R.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 2012, 10, 792–805. [Google Scholar] [CrossRef]
- Lu, S.; Friesen, T.L.; Faris, J.D. Molecular characterization and genomic mapping of the pathogenesis-related protein 1 (PR-1) gene family in hexaploid wheat (Triticum aestivum L.). Mol. Genet. Genomics 2011, 285, 485–503. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.; Gorlach, J.; Volrath, S.; Ryals, J. Wheat genes encoding two types of PR-1 proteins are pathogen inducible, but do not respond to activators of systemic acquired resistance. Mol. Plant Microbe Interact. 1999, 12, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Milla, M.A.R.; Butler, E.D.; Huete, A.R.; Wilson, C.F.; Anderson, O.; Gustafson, J.P. Expressed sequence tag-based gene expression analysis under aluminum stress in rye. Plant Physiol. 2002, 130, 1706–1716. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Kisseleva, L.; Sawa, S.; Furukawa, T.; Komatsu, S.; Koshiba, T. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 2004, 45, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.R.; Chen, Z.Y.; Brown, R.L.; Bhatnagar, D. Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J. Plant. Physiol. 2010, 167, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Gyohda, A.; Tominaga, M.; Kawakats, M.; Atakeyama, A.; Ishii, N.; Shimaya, K.; Nishimura, T.; Riemann, M.; Nick, P.; Hashimoto, M.; et al. RSOsPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol. 2011, 52, 1686–1696. [Google Scholar] [CrossRef]
- Haque, M.E.; Abe, F.; Kawaguchi, K. Formation and extension of lysigenous aerenchyma in seminal root cortex of spring wheat (Triticum aestivum cv. Bobwhite line SH 98 26) seedlings under different strengths of waterlogging. Plant Root 2010, 4, 31–39. [Google Scholar] [CrossRef]
- Haque, M.E.; Kawaguchi, K.; Komatsu, S. Analysis of proteins in aerenchymatous seminal roots of wheat grown in hypoxic soils under waterlogged conditions. Protein Pept. Lett. 2011, 18, 912–924. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.E.; Abe, F.; Mori, M.; Oyanagi, A.; Komatsu, S.; Kawaguchi, K. Characterization of a wheat pathogenesis related protein-1.2, TaBWPR-1.2, expressed in seminal roots in response to waterlogging. J. Plant Physiol. 2014, 171, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.H.; Quail, P.H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Pellegrineschi, A.; Noguera, L.M.; Skovmand, B.; Brito, R.M.; Velazquez, L.; Salgado, M.M.; Hernandez, R.; Warburton, M.; Hoisington, D. Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome 2002, 45, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Toki, S.; Takamatsu, S.; Nojiri, C.; Ooba, S.; Anzai, H.; Iwata, M.; Christensen, A.H.; Quail, P.H.; Uchimiya, H. Expression of a maize ubiquitin gene promoter-bar chimeric gene in transgenic rice plants. Plant Physiol. 1992, 100, 1503–1507. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Kawata, M.; Ando, I.; Shimizu, T.; Ohshima, M. Selecting genetic transformants of indica and indica-derived rice cultivars using bispyribac sodium and a mutated ALS gene. Plant Cell Rep. 2010, 29, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Abe, F.; Kawahigashi, H.; Nakazono, K.; Tagiri, A.; Matsumoto, T.; Utsugi, S.; Ogawa, T.; Handa, H.; Ishida, H.; et al. A Wheat Homolog of MOTHER OF FT AND TFL1 Acts in the Regulation of Germination. Plant Cell 2011, 23, 3215–3229. [Google Scholar] [CrossRef]
- Rakwal, R.; Komatsu, S. Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis 2000, 21, 2492–2500. [Google Scholar] [CrossRef] [PubMed]
- TriFLDB: Triticeae Full-Length CDS DataBase. Available online: http://trifldb.psc.riken.jp/download.pl (accessed on 20 February 2013).
- NCBI: National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov (accessed on 20 February 2013).
- UniProt. Available online: http://www.uniprot.org (accessed on 20 February 2013).
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.E.; Abe, F.; Mori, M.; Oyanagi, A.; Komatsu, S.; Kawaguchi, K.; NARO Institute of Crop Science, Tsukuba, Japan. Unpublished data. 2012.
- Rivière, M.P.; Marais, A.; Ponchet, M.; Willats, W.; Galiana, E. Silencing of acidic pathogenesis-related PR-1 genes increases extracellular b-(1/3)-glucanase activity at the onset of tobacco defence reactions. J. Exp. Bot. 2008, 59, 1225–1239. [Google Scholar]
- Altpeter, F.; Vasil, V.; Srivastava, V.; Stoger, E.; Vasil, I.K. Accelerated production of transgenic wheat (Triticum aestivum L.) plants. Plant Cell Rep. 1996, 16, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S. Position effects and variegation enhancers in an autosomal region of Drosophila melanogaster. Genetics 1979, 93, 105–115. [Google Scholar] [PubMed]
- Gottschling, D.E.; Aparicio, O.M.; Billington, B.L.; Zakian, V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 1990, 63, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Wang, S.; Li, Y.; Smalle, J. Proteasome regulation, plant growth and stress tolerance. Plant Signal Behav. 2009, 4, 924–927. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Komatsu, S. Ubiquitin/proteasome-mediated proteolysis is involved in the response to flooding stress in soybean roots, independent of oxygen limitation. Plant Sci. 2012, 185–186, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Chamarro, J.; Granell, A. A nonphotosynthetic ferredoxin gene is induced by ethylene in citrus organs. Plant. Mol. Biol. 1995, 29, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Onda, Y.; Matsumura, T.; Kimata-Ariga, Y.; Sakakibara, H.; Sugiyama, T.; Hase, T. Differential interaction of maize root Ferredoxin: NADP1 oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol. 2000, 123, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, S.W.; Redinbaugh, M.G.; Shiraishi, N.; Vrba, J.M.; Campbell, W.H. Identification of a maize root transcript expressed in the primary response to nitrate: Characterization of a cDNA with homology to ferredoxin-NADP1 oxidoreductase. Plant Mol. Biol. 1994, 26, 678–690. [Google Scholar] [CrossRef]
- Justice, M.C.; Hsu, M.J.; Tse, B.; Ku, T.; Balkovec, J.; Schmatz, D.; Nielsen, J. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 1998, 273, 3148–3151. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Nanjo, Y.; Nishimura, M. Proteomic analysis of the flooding tolerance mechanism in mutant soybean. J. Proteomics 2013, 79, 231–250. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haque, E.; Abe, F.; Mori, M.; Nanjo, Y.; Komatsu, S.; Oyanagi, A.; Kawaguchi, K. Quantitative Proteomics of the Root of Transgenic Wheat Expressing TaBWPR-1.2 Genes in Response to Waterlogging. Proteomes 2014, 2, 485-500. https://doi.org/10.3390/proteomes2040485
Haque E, Abe F, Mori M, Nanjo Y, Komatsu S, Oyanagi A, Kawaguchi K. Quantitative Proteomics of the Root of Transgenic Wheat Expressing TaBWPR-1.2 Genes in Response to Waterlogging. Proteomes. 2014; 2(4):485-500. https://doi.org/10.3390/proteomes2040485
Chicago/Turabian StyleHaque, Emdadul, Fumitaka Abe, Masahiko Mori, Yohei Nanjo, Setsuko Komatsu, Atsushi Oyanagi, and Kentaro Kawaguchi. 2014. "Quantitative Proteomics of the Root of Transgenic Wheat Expressing TaBWPR-1.2 Genes in Response to Waterlogging" Proteomes 2, no. 4: 485-500. https://doi.org/10.3390/proteomes2040485
APA StyleHaque, E., Abe, F., Mori, M., Nanjo, Y., Komatsu, S., Oyanagi, A., & Kawaguchi, K. (2014). Quantitative Proteomics of the Root of Transgenic Wheat Expressing TaBWPR-1.2 Genes in Response to Waterlogging. Proteomes, 2(4), 485-500. https://doi.org/10.3390/proteomes2040485