Abstract
Many clinically available anticancer compounds are designed to target DNA. This commonality of action often yields overlapping cellular response mechanisms and can thus detract from drug efficacy. New compounds are required to overcome resistance mechanisms that effectively neutralise compounds like cisplatin and those with similar chemical structures. Studies have shown that 56MESS is a novel compound which, unlike cisplatin, does not covalently bind to DNA, but is more toxic to many cell lines and active against cisplatin-resistant cells. Furthermore, a transcriptional study of 56MESS in yeast has implicated iron and copper metabolism as well as the general yeast stress response following challenge with 56MESS. Beyond this, the cytotoxicity of 56MESS remains largely uncharacterised. Here, yeast was used as a model system to facilitate a systems-level comparison between 56MESS and cisplatin. Preliminary experiments indicated that higher concentrations than seen in similar studies be used. Although a DNA interaction with 56MESS had been theorized, this work indicated that an effect on protein synthesis/ degradation was also implicated in the mechanism(s) of action of this novel anticancer compound. In contrast to cisplatin, the different mechanisms of action that are indicated for 56MESS suggest that this compound could overcome cisplatin resistance either as a stand-alone treatment or a synergistic component of therapeutics.
1. Introduction
Platinum-based drugs are among the most commonly and effectively used compounds in the treatment of solid tumours [1,2]. Cisplatin has been used extensively in the treatment of cancers such as head and neck, ovarian, testicular, and small cell lung variants [2,3]. While cisplatin and its derivatives all covalently bind to DNA at purine bases forming intra- and inter-strand crosslinks [4], the differential activities of these compounds arise from their structural differences. The chloride [5], cyclobutane-1,1-dicarboxylate and oxalate groups influence the anticancer potential, range of activity, capacity for cell recognition, and response to the DNA adducts formed [6]. Most cisplatin derivatives are less toxic than the parent molecule and can be used at higher dosages for longer periods [7]. Nonetheless, these compounds produce severe side effects, the most common being nephrotoxicity, ototoxicity and neurotoxicity [1,6]. Moreover, despite the large number of compounds that are structurally similar to cisplatin (e.g., oxaliplatin, nedaplatin), between 1990 and 2002, only 71 new compounds for the treatment of cancer and symptomatic illnesses were found suitable to be granted marketing approval and cleared for clinical use [8]. Among these new molecules were anti-metabolite, retinoid and anti-mitotic anthracycline compounds. Additional platinum based compounds, satraplatin and picoplatin, have also been under trial but are yet to be approved [9,10].
The disparity between supply and demand of viable cancer therapeutics combined with the increasing prevalence of tumour resistance to treatment, drive novel anti-cancer compound formulation and the necessary characterisation of their molecular mechanisms of action. Given the complexity of biological systems and their interaction with chemotherapeutics, it is necessary to apply methods that can accommodate the intricacies of multifaceted responses but also determine mechanisms of action on a system-wide basis [11]. Systems Biology takes the multiple levels of biological hierarchy into account and capitalises on their interrelationships [12]. It is thus generally held, that using such larger-scale approaches (i.e., the ‘Omics’) provides the best access to the fundamental principles of a biological response, in this case to chemotherapeutics.
An alternative mechanism of action for chemotherapeutics that has been investigated is intercalation [13]. The insertion of a compound between the base pairs of DNA results in a longer, partially unwound, irregular helix and this can alter gene expression, replication, cell growth, and cause apoptosis [14,15]. Intercalators previously implemented in chemotherapy include the anthracyclines, duanomycin and doxorubicin, actinomycin D, and the podophyllotoxin, etoposide [16]. An example of a novel intercalator is [(5,6-dimethyl-1,10-phenanthroline)(1S,2S-diaminocyclohexane) platinum(II)]2+ (56MESS) [13,17], which has an IC50 of 9 ± 2 nM in L1210 murine leukaemia cells, compared to 500–1000 nM for cisplatin [18,19]. Furthermore, 56MESS has also proven active in cisplatin-resistant cell lines [13]. The molecular basis for this cytotoxicity is a prime subject for investigation, given that this represents a rationally designed alternative therapeutic for overcoming cisplatin resistance in tumours.
Previous studies have indicated that 56MESS does indeed interact with cells via a different mechanism to cisplatin. Microprobe synchrotron radiation X-ray fluorescence (SRXRF) imaging was used to map cellular targets of cisplatin and 56MESS in A549 human lung cancer cells and it was observed that the platinum in 56MESS was primarily localised to the nucleus while cisplatin exposure resulted in more widespread cellular distribution [20]. Moreover, platinum from 56MESS accumulated in the nucleus after only 4 h of exposure. Co-localisation with phosphorus indicated that this DNA-based interaction was targeted to heterochromatin and involved the intact metallointercalator molecule [20]. Additionally, a yeast transcriptomic study using cDNA microarrays identified glutathione, iron and copper related pathways in the cellular response to 56MESS [21]. Specifically, the up-regulated genes were involved in the homeostasis of iron and copper, and the transport of these metal ions across the plasma membrane. This is notable as copper transport has been implicated in the transport of cisplatin [22,23] and may be part of a more generalised response to heavy metals. Down-regulated genes were associated with amino acid metabolism, the yeast stress response, and cellular respiration [21]. Previous studies suggest that other heavy metals affect cell division and when in excess will be stored in the vacuole for use during times of scarcity and as protection from cytotoxicity [24,25]. The varied mechanisms of 56MESS activity suggested by this evidence imply that there are multiple factors at play in a cellular response to 56MESS. In this work, we have used a well-characterized eukaryotic model and a Systems Biology approach that includes both genomic and proteomic analyses to refine our understanding of mechanisms underlying the cellular response to 56MESS relative to cisplatin.
Saccharomyces cerevisiae is a single celled eukaryote that is extremely amenable to genetic and other experimental manipulations [26]. As such, S. cerevisiae has been used extensively as a model for human cells, with many highly conserved genes, and thus proteins, molecular mechanisms, as well as cellular structures and processes [27]; this has facilitated its use in the screening of potential anticancer agents [28]. In light of the conservation of fundamental mechanisms, the usefulness of this model has been further facilitated by the construction of a commercial yeast deletion mutant library (EUROSCARF deletion library [29]). This library can be used in conjunction with the Affymetrix TAG3 microarray system [28], enabling the identification of gene deletions that alter cellular response to a specific environmental perturbation [26]. The library has previously been used with TAG microarrays, to identify mechanisms potentially involved in the biological response to cisplatin [30]. Here, these arrays enabled assessment of genomic information that could then be more effectively consolidated with proteomic data.
While it is not surprising that 56MESS, a compound designed to interact with DNA is, in part, localised to the nucleus, additional mechanisms also appear to be at work [20,21]. Thus, we hypothesized that the increased efficacy of 56MESS was due to a mode of action potentially quite different to cisplatin. Screening for mutants with increased sensitivity to 56MESS implicated protein trafficking and degradation functions and, as such, the proteome was subsequently examined using high resolution, top-down proteomics (i.e., one- and two-dimensional gel electrophoresis) in direct comparison with cisplatin treated cells. This integrated Systems Biology approach has thus provided insight into the anticancer potential of 56MESS, particularly with regard to its likely alternate mechanisms of cytotoxicity relative to the current therapeutic standard, cisplatin.
2. Experimental Section
2.1. Reagents and Suppliers
All reagents were of analytical grade or higher, and supplied by Amresco unless otherwise stated. The manufacture of 56MESS was undertaken using published methods [17]. Cisplatin (99.9% purity) was obtained from Sigma (Sydney, Australia).
2.2. Media, Yeast Strains and Growth Conditions
Yeast growth was carried out in liquid yeast extract/peptone/dextrose (YEPD) media prepared as described previously [31], with orbital shaking (120 rpm). Stationary incubation of yeast on YEPD agar (2% (w/v)) plates was carried out at 30 °C.
Wildtype BY4743 S. cerevisiae (mating type: MATa/MATα, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, met15Δ0/+, lys2Δ0/+, ura3Δ0/ura3Δ0) was grown to log phase (OD600 = 1.0) and exposed to 56MESS to determine the optimal concentration and time span for selecting sensitive mutants. This culture was divided into subcultures and exposed to 56MESS concentrations ranging from 0–8 mM. Aliquots of the exposed cultures were taken every 15 min for the first hour, every 30 min for the second hour, and then hourly for the following 4 h and grown on YEPD plates to establish the loss of cell viability caused by each concentration. The percentage of cell death was measured as the loss in total number of colony forming units present in the culture.
The homozygous diploid deletion mutant pool in the BY4743 genetic background (Invitrogen, Melbourne, Australia) was used in the TAG microarray experiments [26,32]. Master cultures of the mutant library pool were grown to log phase (OD600 = 1.0). Cultures were then exposed to 2 mM 56MESS for 3 h and aliquots from each were tested as described above, to yield a dose response curve [31].
2.3. Tag Microarray
Duplicate 50 µL aliquots were taken from cultures treated with 0, 4 and 8 mM 56MESS at 0.25, 0.5, 0.75, 1, 1.5, 2 and 3 h. All aliquots were inoculated into separate 100 mL volumes of fresh YEPD and supplemented daily with fresh YEPD for 3 days (i.e., yielding OD600 = 6.0). Cells were then harvested by centrifugation at 600× g for 5 min and cell pellets were frozen in liquid nitrogen and stored at −80 °C until DNA extraction was carried out as described previously [33].
The UP- and DOWNTAG molecular barcode sequences were amplified separately from 1.5 µg of extracted DNA using previously identified primers (Invitrogen, Melbourne, Australia) [32]. These primers were designed specifically for amplification of the molecular barcodes of the deletion mutants. The PCR protocol used 20 mM tris-HCl (pH 8.4) / 50 mM KCl (PCR buffer from Invitrogen), 0.4 mM of each deoxynucleotide triphosphate, Taq polymerase (20 U mL−1), forward and reverse primer (1 µg mL−1) and chromosomal DNA (20–30 µg mL−1).
The UP- and DOWNTAG PCR products for each sample were combined and prepared for hybridization as described previously [32]. Briefly, UP- and DOWNTAG PCR products were combined with 1 M NaCl / 66 mM NaH2PO4 / 6.6 mM EDTA (pH 7.4) / 0.005% (w/v) Triton X-100 (6 × SSPE-T) and 400 pmol each of the UP- and DOWNTAG amplified DNA samples. Sample hybridization cocktails were heated at 100 °C for 5 min, cooled on ice for 2 min, injected into an Affymetrix Tag3 GeneChip® high-density oligonucleotide microarray, and hybridized for 16 h at 42 °C with orbital rotation (60 rpm). Washing and staining of chips was carried out as previously described [32] and chips were scanned at 570 nm using an Affymetrix GeneChip® scanner [26,34].
Resultant scanner images of each GeneChip® microarray were quantified for analysis using the 75th percentile of each separate TAG element on the microarray [26]. Mutants were then linked to their corresponding molecular UP- and DOWNTAG marker by GeneChip® Operating Software (Version 1.4) with reference to a GeneFlex® library of TAG sequences.
Raw data were then sorted for perfect match (PM) values. PM values are signal readings associated with the hybridization of each TAG to its exact sequence complement on the microarray. A ratio of solvent signal over treatment signal was calculated; all mutants with a 1.5-fold change (p < 0.05) in their complement of UP- and DOWNTAG signals were retained for further testing (Table S1).
2.4. Sensitivity Confirmation
A library of individually stored mutants obtained from the European S. cerevisiae Archive for Functional Analysis (EUROSCARF) was used for secondary analysis of the 133 mutants identified by the microarray experiments. Mutants identified as sensitive to 56MESS were each grown in a separate well on a microtitre plate, spiked with glycerol (15% w/v) and stored at −80 °C. Mutants were thawed and replicated onto a fresh YEPD-filled microtitre plate, grown to confluence and incubated an additional 4 h following addition of fresh media. All mutants were then exposed to a solvent control or 2 mM of either 56MESS or cisplatin over 24 h. Well contents were plated onto YEPD agar at times 0, 1, 2, 4, 8 and 24 h. Empty wells were distributed randomly across each mutant plate to indicate potential levels of cross contamination. BY4743 was used as a reference strain with five randomly assigned wells on each microtitre plate. This process was repeated for all mutants.
2.5. Protein Sample Preparation and Extraction
Triplicate BY4743 cultures were grown to OD600 = 1.0 and split into three subcultures for treatment with solvent, cisplatin, or 56MESS. Prior to administration of treatments, initial samples were taken from each culture (i.e., 0 h), and then another at 4 h for protein extraction (as well as an aliquot for plating onto YEPD to generate a concurrent kill curve). Samples were centrifuged at 600× g for 5 min and the cell pellet was snap frozen in liquid nitrogen before being stored at −80 °C (i.e., AFD; automated frozen disruption). Frozen yeast cell pellets were pulverised to a fine powder using a Mikro-Dismembranator S (Sartorius AG, Goettingen, Germany) before solubilisation (i.e., AFD; automated frozen disruption) [35,36]. All protein fractions were quantified using the EZQ Protein quantification assay (BioRad, Gladesville, Australia) and then stored at −80 °C until needed [37].
2.6. 1D SDS Polyacrylamide Gel Electrophoresis
Protein samples were diluted with 2 × sample buffer (0.125 M tris (pH 8.8) / 5% (w/v) SDS / 25% glycerol / 0.0625 M dithiothreitol (DTT) / 0.001% (w/v) bromophenol blue) with a protease inhibitor cocktail (2 µg mL−1 each of aprotinin, pepstatin and leupeptin) [35,36]. All samples were diluted to enable uniform total protein loads in every analysis. Gel electrophoresis was carried out as previously described with minor modifications [38,39,40,41]. Once complete, gels were fixed for a minimum of 1 h.
2.7. Tris-Tricine Polyacrylamide Gel Electrophoresis
Samples were prepared for tris-tricine PAGE as described above for 1D electrophoresis. Tricine PAGE was carried out as described previously with minor alterations [42,43,44,45]. Resolving gels (17% T, 5.88% C / 1 M tris / 0.1% (w/v) SDS / 13.5% (w/v) glycerol / 0.033% (v/v) TEMED / 0.033% (w/v) APS) were cast and allowed to polymerize overnight. A stacking gel prepared as described above was then overlaid on the resolving gel and formed into wells. Anodic buffer consisted of 0.2 M tris (pH 8.9) and the cathodic buffer of 0.1 M tris, 0.1 M tricine and 0.1% (w/v) SDS. Gel electrophoresis was carried out at 4 °C for 4 h at a constant 50 mA and a maximum of 120 V. Gels were then fixed overnight (min 18 h).
2.8. 2D SDS Polyacrylamide Gel Electrophoresis
2D SDS PAGE (2DE) was carried out as described previously [35,36,41,44,45,46,47,48]. Briefly, following AFD, the resulting ‘powdered’ cell pellets were solubilised in 8 M urea, 2 M thiourea and 4% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPS) with a protease inhibitor cocktail (2 µg mL−1 each of aprotinin, pepstatin and leupeptin) [35,36]. Each sample was reduced and alkylated as described previously and a volume equivalent to 100 µg of protein/sample was passively hydrated into 7 cm 3–10 non-linear immobilised pH gradient (IPG) strips (Bio-Rad) for 16 h at room temperature (RT) [35,36,41,44,45,46,47,48].
Isoelectric focusing (IEF) of the hydrated IPG strips was carried out as described previously at 17 °C with an initial desalting step at 250 V for 15 min [35,36,41,44,45,46,47,48]. Voltage was linearly ramped for 2 h to 4000 V with wick changes as required to maintain ramping (i.e., five in this instance). IPG strips were then focused at 4000 V for 37,500 Vh and stored at −30 °C until required. Focused strips were equilibrated first in 2% DTT, then 2.5% (w/v) acrylamide and thoroughly drained of buffer [35,36,41,44,45,46,47,48]. Proteins in the IPG strips were then resolved using stacking and resolving gels prepared as described above, and overlaid with agarose (375 mM tris buffer (pH 8.8) / 0.5% (w/v) low melting agarose / 0.1% (w/v) SDS / 0.01% (v/v) bromophenol blue). Electrophoresis and fixation were carried out as described for 1D SDS PAGE.
2.9. Staining and Quantitative Analysis
Following fixation, gels were washed in distilled water for 3 × 20 min, stained overnight with SYPRO Ruby (SR) and washed with fixative for 3 × 20 min. Gels were then washed for 3 × 20 min in distilled water and imaged using a FLA-9000 fluorescence imager (Ex: 473 nm; Em: 510 nm) (Fujifilm). Fluorescence imaging and quantitative image analysis were carried out as described previously [35,36,41,44,46,47,48]; Delta 2D (Decodon, Greifswald, Germany) was used to analyse the 2D gel images.
2.10. Mass Spectrometric Analysis
Proteins differing substantially in abundance, as identified by Delta 2D (Decodon), were assessed as to their experimental isoelectric points and molecular weights based on the parallel use of calibration standards during 2DE (2-D SDS-PAGE Standards (Bio-Rad, Gladesville, Australia) and PageRuler Unstained Protein Ladder (Thermo Scientific, Scoresby, Australia)). Protein spots that were uniquely present or absent (i.e., above or below the minimal level of detection, respectively) in one experimental condition over the others (by definition, ‘all or none’ changes) were selected for mass spectrometric (MS) analysis and identification. Protein spots were excised and prepared for tryptic digest and peptide extraction as described previously [41] with minor alterations [45,49]. Following peptide extraction, 15 µL was transferred to an autosampler vial for LC/MS/MS analysis. The MS/MS data files produced by the QSTAR were queried using Mascot Daemon (version 2.4, provided by the Walter and Elisa Hall Institute [50]) and searched against the LudwigNR database (comprised of the UniProt, plasmoDB and Ensembl databases (vQ111. 16,818,973 sequences; 5,891,363,821 residues) with the parameter settings described previously [41] and the additional variable modifications: Carbamidomethyl. The results of the search were then filtered by including only protein hits with at least one unique peptide and excluding peptide hits with a p-value greater than 0.05. Peptides identified by Mascot were further validated by manual inspection of the MS/MS spectra for the peptide to ensure the b- and y-ion series were sufficiently extensive for an accurate identification.
2.11. Bioinformatics
Genes identified and confirmed as contributing to 56MESS sensitivity when deleted were analysed for function, localization and molecular interactions using the Yeast Proteome Database (YPD) [51] and Saccharomyces Genome Database (SGB) [52]. The molecular weight and isoelectric points of the products of these genes were also sourced. Based on these classifications and descriptions, genes were categorised according to common function. The SGD Go Slim mapping tool was also used to categorise genes from this and a previously published study [21] for comparison.
3. Results
Drug treatment concentrations were determined by kill curve analysis of wild type yeast and the deletion library was then exposed to these concentrations (Figure 1 and Figure 2). The DNA containing the molecular TAG3 labels of the sensitive mutants was then extracted from culture, amplified by PCR and hybridised to a TAG3 microarray. This data were used to produce a list of potentially sensitive deletion mutants having a statistically significant ≥1.5-fold change in signal between control and treated samples. These genes were categorised according to predicted function and localisation of the proteins they encode (Figure S1). Following verification of sensitivity via microtitre plate assay and individual exposure to 56MESS, a subtotal of 48 gene deletion mutants was confirmed as 56MESS-sensitive.
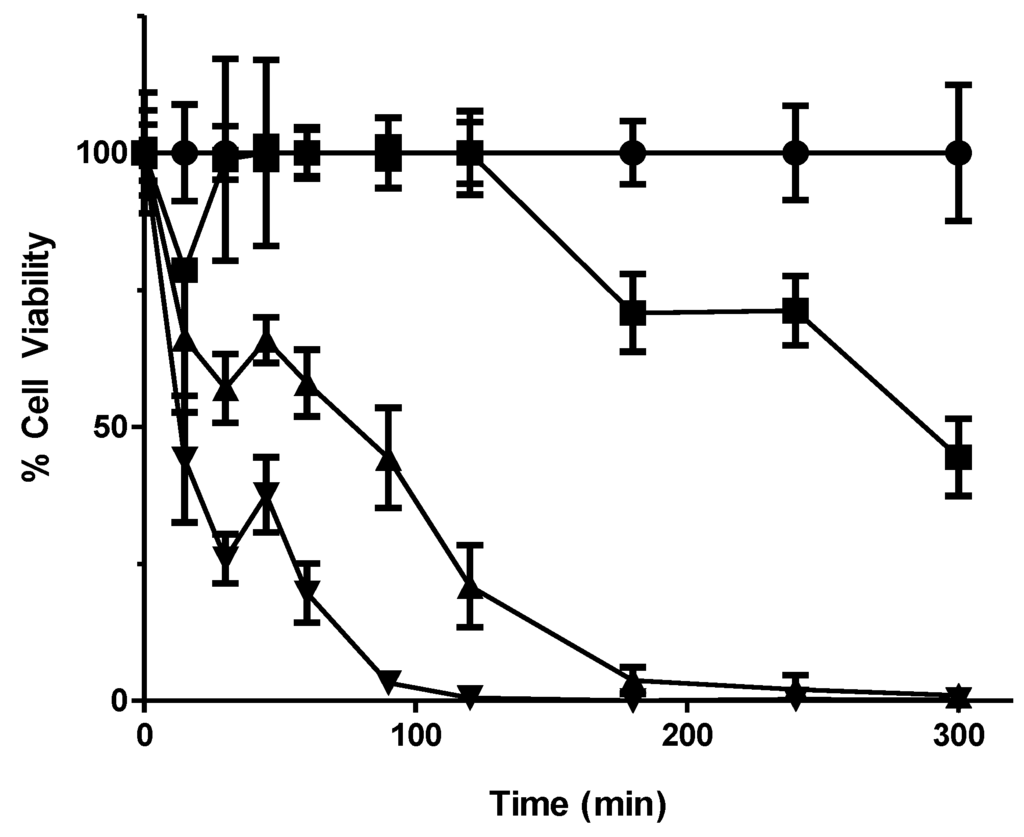
Figure 1.
Kill curve using wildtype BY4743 to determine yeast deletion mutant pool test parameters. A culture was grown to OD 1, divided into separate subcultures and exposed to 0, 4, 6 and 8 mM of 56MESS. Treated culture samples were plated onto yeast extract/peptone/dextrose (YEPD) plates over 5 h. The resultant colonies after 48 h of incubation at 30 °C were counted to determine the cell viability for each culture exposed to each 56MESS concentration. Error bars represent standard error of the mean (SEM) (n = 3) ● Solvent; ■ 4 mM 56MESS; ▲ 6 mM 56MESS; ▼ 8 mM 56MESS.
The functional group with the highest representation (31.3%) among the confirmed sensitive mutants was the vacuolar trafficking system (Figure 3), representing the primary cyclic exo- and endocytotic pathways within these cells. Other functional groups represented in the 56MESS sensitive deletion mutant collection included cell cycle genes (16.7%) and those involved in ion homeostasis (10.4%). Among the other groups, genes linked to biomolecule metabolism (8.3%), the peroxisome (6.3%) and protein degradation (4.2%) also increased sensitivity to 56MESS when deleted. No major DNA repair mechanisms were present among the confirmed sensitive gene deletions in stark contrast to what is known about the mechanism of action of cisplatin. Taking all genes involved in protein trafficking, handling and/or degradation into account, there is a high proportion (41.8% total) that contribute to 56MESS sensitivity. This potentially implicates these processes and proteins themselves as significant potential targets of 56MESS. This also indicates that DNA may not be the sole target of the anticancer compound 56MESS. Furthermore, as proteins (i.e., gene products) are themselves the active arbiters of cellular function and response, it was also deemed necessary to analyse the proteome using both 1- and 2D SDS PAGE techniques. As cisplatin is considered the leading industry standard, it was also included for direct comparison with 56MESS.
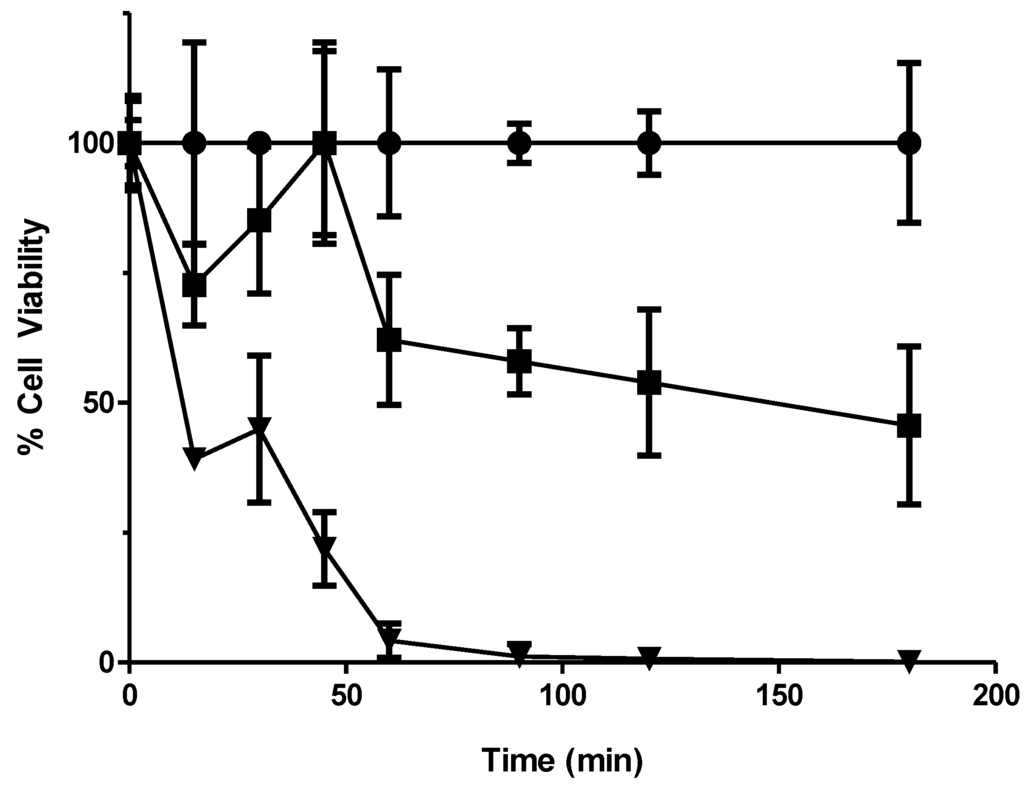
Figure 2.
Kill curve using the yeast deletion mutant pool conducted to relate sampling times to losses in cell viability. Cultures were exposed to 0, 4 and 8 mM 56MESS and aliquots taken over 3 h were plated onto YEPD. Following 48 h of incubation at 30 °C, plated colonies were counted to determine cell viability. Error bars represent SEM (n = 3). ● Solvent; ■ 4 mM 56MESS; ▼ 8 mM 56MESS.
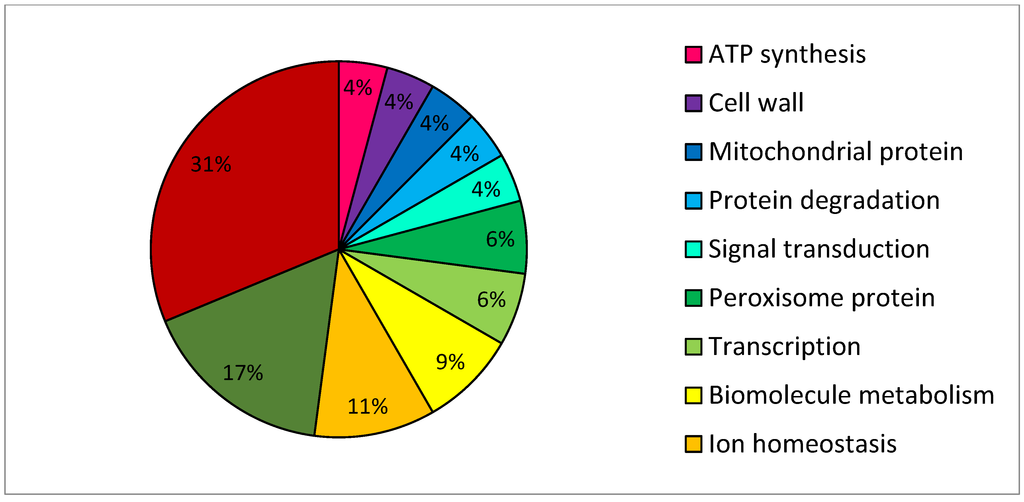
Figure 3.
Distribution of gene function amongst confirmed 56MESS sensitive deletion mutants. The top 200 sensitive mutants as identified by microarray were submitted to individual 56MESS sensitivity testing in microtitre plates. A subset of 48 mutants were confirmed for sensitivity to 56MESS.
Yeast proteins from cultures exposed to solvent (i.e., control), cisplatin, and 56MESS for 4 h were resolved using 1D SDS-PAGE (Figure 4A). Concentrations were selected based on cell viability in response to a range of 56MESS and cisplatin concentrations, and also produced the most similar decreases in viability (Figure S2). The cisplatin-exposed samples yielded a protein band pattern comparable to the control after 4 h (Figure 4A). The addition of cisplatin to growing conditions did not alter the protein complement of yeast in comparison to the control; the addition of 56MESS reduced the number of detectable proteins. When this was quantified as a ratio of fluorescent signal associated with samples treated either with cisplatin or 56MESS over that of the total protein extracted from control samples, it indicated that significantly less protein was detected in the 56MESS exposed samples (p = 0.0064). Furthermore, it was also evident that while there was less protein in the 25–100 kDa range, the samples that had been exposed to 56MESS also had a diffuse protein band located beyond the dye front. Quantification of this low molecular weight material indicated that a high proportion (40 ± 1% of total signal) of the total protein sample was present in the diffuse band, thus accounting for the differences in the density of resolved proteins despite equivalent total protein loads of each sample (i.e., as determined using a general protein detection reagent) (Figure 4B). No similar band was detected in the cisplatin-exposed sample.
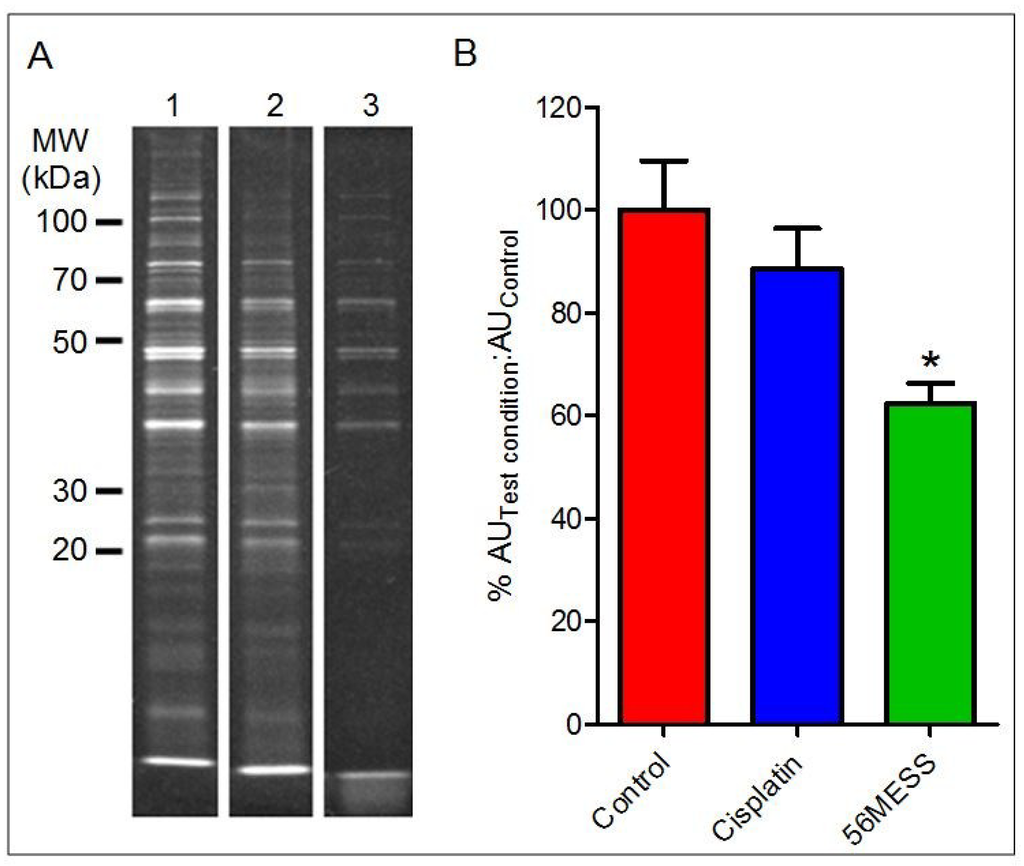
Figure 4.
1D SDS-PAGE of total protein extracts after 4 h exposure of cells to control, cisplatin or 56MESS treatments. Equal protein loads as measured by EZQ were resolved and detected with SYPRO Ruby and gel images were analysed using Multi Gauge. (A) Lane 1: Control; Lane 2: Cisplatin; Lane 3: 56MESS; (B) Ratio of the fluorescent signal of the test condition protein sample over the fluorescent signal of the control protein sample after 4 h of drug exposure.  control;
control;  1 mM cisplatin;
1 mM cisplatin;  2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p = 0.0064 (n = 3).
2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p = 0.0064 (n = 3).
 control;
control;  1 mM cisplatin;
1 mM cisplatin;  2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p = 0.0064 (n = 3).
2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p = 0.0064 (n = 3).
The presence of low molecular weight (MW) fragments suggested that a method capable of resolving protein/peptides in this MW range was required. Thus, tris-tricine electrophoresis was employed to resolve any low MW peptide material within these samples. The diffuse band below the dye front in the 56MESS 1D gel lane (Figure 5A) was also apparent in the 1 kDa range following resolution using tris-tricine gels, consistent with peptide-size protein fragments and/or the end products of protein degradation (Figure 5). Furthermore, extracts from the control and cisplatin exposed samples did not have any such prominent low molecular weight band. Quantification of this diffuse protein band indicated that this proteinaceous material was ~1.8-fold more abundant than in the corresponding solvent control sample extracts as indicated by fluorescent signal intensity (Figure 5B).
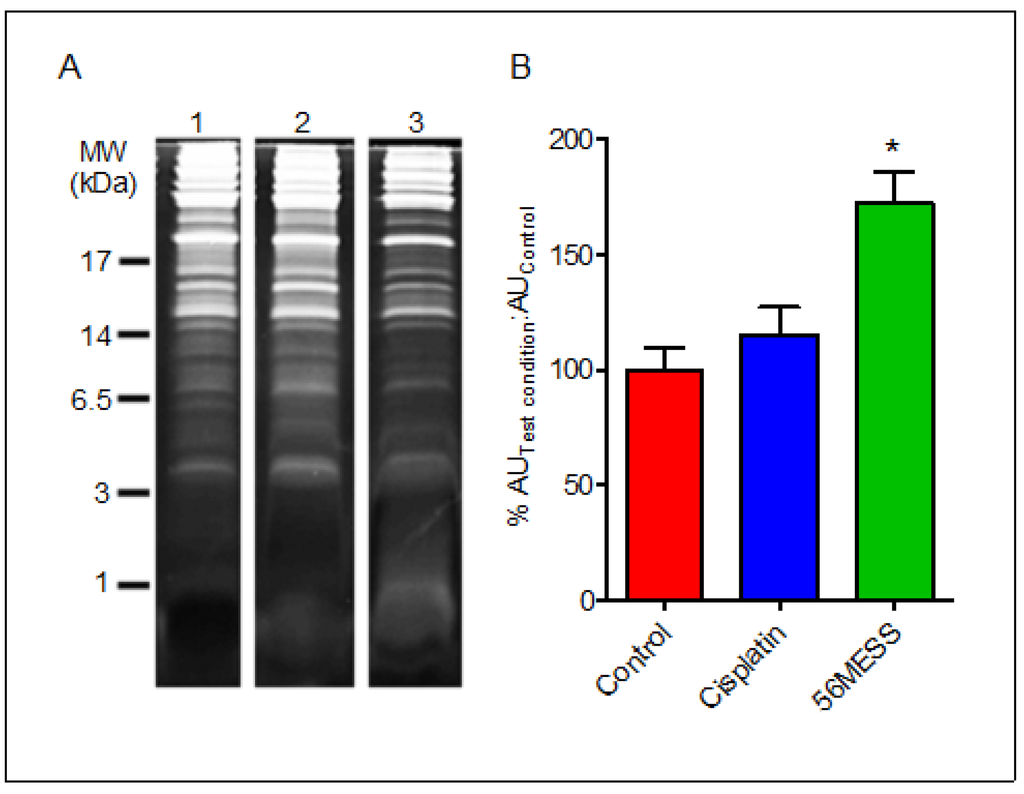
Figure 5.
(A) Tris-tricine PAGE gel of proteins extracted from solvent, cisplatin and 56MESS exposed cultures. Equal protein loads as measured by EZQ were resolved and detected with SYPRO Ruby and gel images were analysed using Multi Gauge. 1. Control; 2. Cisplatin; 3. 56MESS; (B) Ratio of the fluorescent signal of the test condition protein sample to that of the control protein sample after 4 h of drug exposure. Quantitation was limited to the 1 kDa region of the lane to focus on the fluorescence associated with the low molecular weight material in the 56MESS sample.  Control;
Control;  1 mM cisplatin;
1 mM cisplatin;  2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p < 0.05 (n = 3).
2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p < 0.05 (n = 3).
 Control;
Control;  1 mM cisplatin;
1 mM cisplatin;  2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p < 0.05 (n = 3).
2 mM 56MESS. Error bars represent SEM. ‘*’ indicates p < 0.05 (n = 3).
As substantial changes in protein abundance (e.g., increased protein degradation) were suggested by the initial proteomic assessments (Figure 4 and Figure 5), a more comprehensive examination of the yeast proteome following exposure to 56MESS was undertaken using a highly refined, quantitative top-down analytical approach [35,41,44,45,46] — 2D gel electrophoresis (2DE) — coupled with mass spectrometry for protein identification (2DE-MS) (Figure 6). The resulting gel images were analysed to determine the total number of protein spots (i.e., species) resolved and any differential changes in spot volume (i.e., amount of protein) between conditions. Extracts from cultures exposed to 56MESS had the lowest number of detectable proteins (658 ± 6, mean ± SEM), and this was significantly different (p = 0.0041) from the cisplatin-treated and control cultures (820 ± 26 and 797 ± 27, respectively; Figure 6D). This represents a potential change in expression, processing, modification or degradation of ~20% of the proteome. This is far from a minor effect. Furthermore, direct comparison of individual spots between treatments showed that the majority were of lower volume in the 56MESS samples than in the extracts from cisplatin-treated cells. Differentially abundant protein spots (Figure 6; indicated with arrows) were further characterised according to apparent molecular weight, isoelectric point, and fold-change following drug treatment (Tables S2 and S3). Select protein spots were submitted for MS analysis based on all or none changes between the control and 56MESS conditions. Several proteins that were more abundant in the 56MESS samples relative to the control were identified as heat shock proteins (Table 1). Two enolase proteins and a pyruvate kinase involved in glycolysis and gluconeogenesis, as well as a ribosomal protein involved in translational elongation were also identified (Table 1). There were no overlapping hits between the 48 confirmed 56MESS sensitive mutants (Figure 3; Table S4) and the proteins identified by 2DE-MS. However, the experimental molecular weight and isoelectric point values of differentially abundant protein spots can be compared with the reported values of the sensitive deletion mutants and some showed good correlation (marked with a “*” in Table S4).
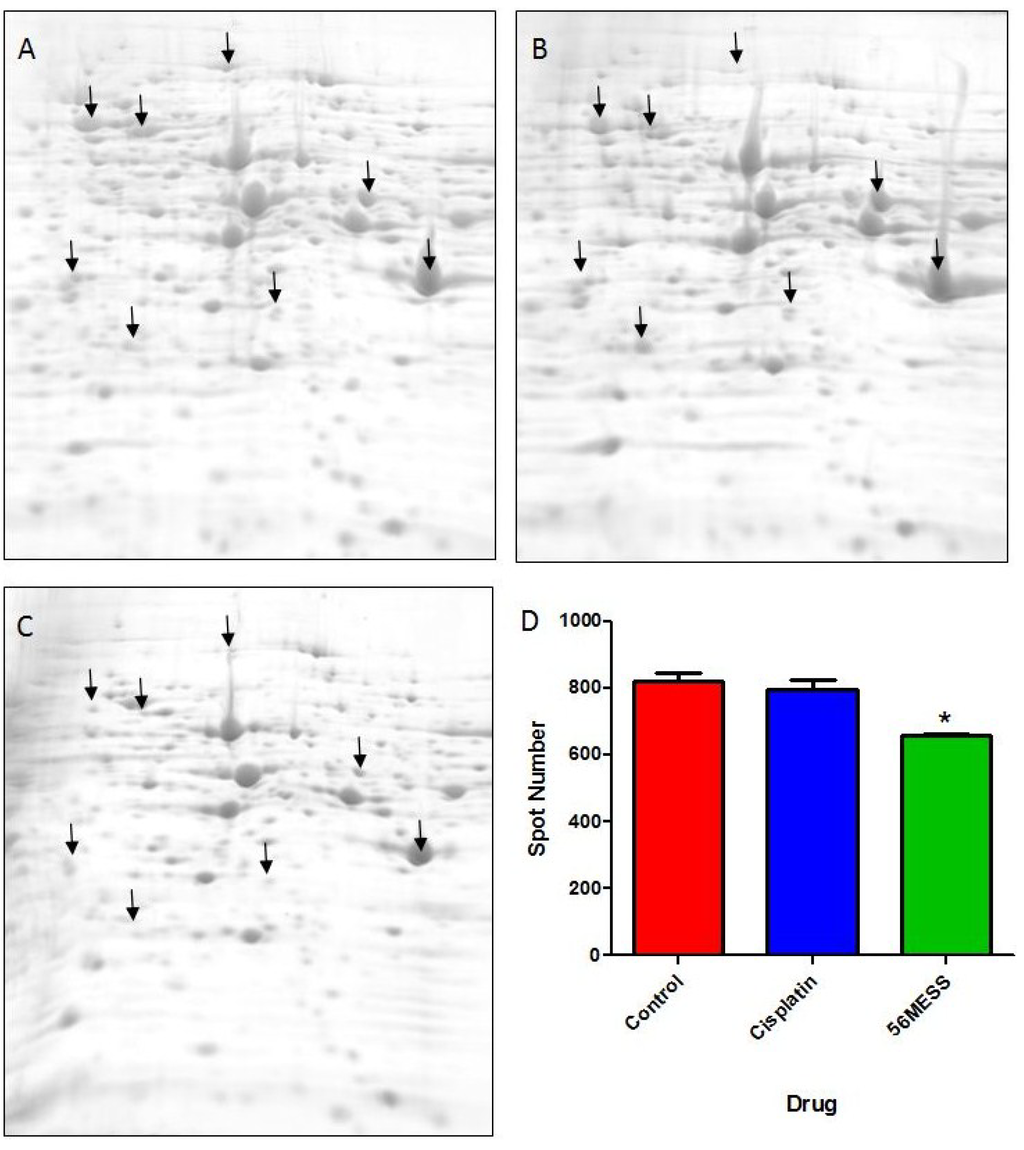
Figure 6.
Representative 2DE gel images for each of the proteomes extracted from a control yeast culture (A) and cultures exposed to cisplatin (B) or 56MESS (C), and protein spot counts (D) Error bars represent SEM. ‘*’ indicates p = 0.0041 (n = 3). Arrows indicate some representative protein spots (i.e., protein species) that are differentially abundant between experimental and control conditions.

Table 1.
Differentially expressed proteins submitted for MS analysis. All identified proteins were S. cerevisiae in origin.
| Mascot ID | Gene | Score | Theoretical | Observed | Coverage | Peptides | E-value | ||
|---|---|---|---|---|---|---|---|---|---|
| P10591 | SSA1/member of the heat shock family | 1551 | 69.6 | 5.0 | 72.2 | 4.4 | 42% | K.ATAGDTHLGGEDFDNR.L | 9.50E-06 |
| K.SQVDEIVLVGGSTR.I | 6.20E-05 | ||||||||
| K.LVTDYFNGKEPNR.S + Deamidated (NQ) | 3.80E-04 | ||||||||
| K.NQAAMNPSNTVFDAK.R + Oxidation (M) | 1.70E-03 | ||||||||
| R.SINPDEAVAYGAAVQAAILTGDESSK.T | 2.00E-03 | ||||||||
| K.DAGTIAGLNVLR.I | 2.30E-03 | ||||||||
| K.ATAGDTHLGGEDFDNR.L | 1.00E-02 | ||||||||
| K.ELQDIANPIMSK.L + Oxidation (M) | 1.00E-02 | ||||||||
| R.IINEPTAAAIAYGLDK.K | 4.90E-02 | ||||||||
| P00925 | ENO2/Enolase II, phosphopyruvate hydratase | 997 | 47 | 5.6 | 31.4 | 5.1 | 37% | K.TAGIQIVADDLTVTNPAR.I | 1.10E-05 |
| K.DGKYDLDFKNPESDK.S | 2.80E-05 | ||||||||
| K.AVDDFLLSLDGTANK.S | 4.40E-05 | ||||||||
| R.SGETEDTFIADLVVG.L | 5.80E-05 | ||||||||
| K.LGANAILGVSMAAAR.A + Oxidation (M) | 8.40E-04 | ||||||||
| K.IGLDCASSEFFK.D + Propionamide (C) | 2.30E-03 | ||||||||
| K.VNQIGTLSESIK.A | 3.30E-02 | ||||||||
| P00560 | PGK1/3-phosphoglycerate kinase | 494 | 44.7 | 7.1 | 19.7 | 5.8 | 23% | K.ASAPGSVILLENLR.Y | 1.70E-02 |
| K.VLENTEIGDSIFDK.A | 1.30E-05 | ||||||||
| E7NIQ9 | ENO1/Enolase I, phosphopyruvate hydratase | 309 | 34 | 5.7 | 17.6 | 6.7 | 16% | R.GNPTVEVELTTEK.G | 1.60E-02 |
| K.AVDDFLLSLDGTANK.S | 2.60E-04 | ||||||||
| P10592 | SSA2/ stress induced heat shock protein | 4809 | 69.4 | 5.0 | 72.2 | 4.8 | 67% | K.KAEETIAWLDSNTTATKEEFDDQLK.E | 1.50E-08 |
| K.NTISEAGDKLEQADKDAVTK.K + Deamidated (NQ) | 5.90E-08 | ||||||||
| R.SINPDEAVAYGAAVQAAILTGDESSK.T | 2.40E-07 | ||||||||
| K.LDKSQVDEIVLVGGSTR.I | 3.40E-07 | ||||||||
| K.ATAGDTHLGGEDFDNR.L | 1.80E-06 | ||||||||
| K.AVGIDLGTTYSCVAHFSNDR.V + Propionamide (C) | 3.20E-06 | ||||||||
| K.NQAAMNPANTVFDAKR.L + Oxidation (M) | 4.90E-06 | ||||||||
| K.TQDLLLLDVAPLSLGIETAGGVMTK.L + Oxidation (M) | 7.70E-06 | ||||||||
| R.IINEPTAAAIAYGLDKK.G | 7.80E-06 | ||||||||
| K.LVTDYFNGKEPNR.S + Deamidated (NQ) | 3.40E-05 | ||||||||
| K.KSEVFSTYADNQPGVLIQVFEGER.A | 8.90E-05 | ||||||||
| K.ATAGDTHLGGEDFDNR.L | 1.10E-04 | ||||||||
| K.MKETAESYLGAK.V + Oxidation (M) | 2.40E-04 | ||||||||
| K.SQVDEIVLVGGSTR.I | 3.00E-04 | ||||||||
| K.NFTPEQISSMVLGK.M + Oxidation (M) | 3.60E-04 | ||||||||
| K.KAEETIAWLDSNTTATKEEFDDQLK.E | 1.50E-08 | ||||||||
| P32589 | SSE1/ATPase involved in the Hsp90 complex | 1986 | 77.3 | 5.2 | 82.3 | 4.9 | 36% | F.GLDLGNNNSVLAVAR.N | 6.40E-07 |
| K.KDDLTIVAHTFGLDAK.K | 1.00E-06 | ||||||||
| K.DDLTIVAHTFGLDAK.K | 6.90E-06 | ||||||||
| K.PLSTTLNQDEAIAK.G | 1.00E-05 | ||||||||
| F.GLDLGNNNSVLAVAR.N + Deamidated (NQ) | 2.70E-05 | ||||||||
| K.HVFSATQLAAMFIDK.V + Oxidation (M) | 1.70E-04 | ||||||||
| R.EELEELVKPLLER.V | 2.70E-04 | ||||||||
| R.GIDIVVNEVSNR.S | 4.90E-04 | ||||||||
| K.QVEDEDHMEVFPAGSSFPSTK.L + Oxidation (M) | 5.20E-04 | ||||||||
| R.IVNDVTAAGVSYGIFK.T | 7.50E-04 | ||||||||
| R.KNTLEEYIYTLR.G | 8.00E-04 | ||||||||
| K.QSISEAFGKPLSTTLNQDEAIAK.G | 1.30E-03 | ||||||||
| R.GKLEEEYAPFASDAEK.T | 2.40E-03 | ||||||||
| K.YEELASLGNIIR.G | 2.60E-03 | ||||||||
| R.IIGLDYHHPDFEQESK.H | 3.60E-03 | ||||||||
| K.GAAFICAIHSPTLR.V + Propionamide (C) | 3.90E-03 | ||||||||
| K.LNELIEKENEMLAQDK.L + Oxidation (M) | 2.40E-02 | ||||||||
| E7KFS3 | HSP60/Chaperonin mediates protein refolding after stress | 2189 | 60.7 | 5.2 | 61.2 | 4.8 | 52% | K.TNEAAGDGTTSATVLGR.A | 5.30E-10 |
| K.QIIENAGEEGSVIIGK.L | 3.90E-08 | ||||||||
| K.EITTSEEIAQVATISANGDSHVGK.L + Deamidated (NQ) | 1.20E-06 | ||||||||
| K.GVETLAEAVAATLGPK.G | 2.80E-06 | ||||||||
| K.SEYTDMLATGIIDPFK.V + Oxidation (M) | 1.50E-05 | ||||||||
| R.TLEDELEVTEGMR.F + Oxidation (M) | 1.00E-04 | ||||||||
| K.VEFEKPLLLLSEK.K | 1.20E-04 | ||||||||
| K.DRYDDALNATR.A | 1.60E-04 | ||||||||
| R.VGGASEVEVGEK.K | 5.00E-04 | ||||||||
| K.GSIDITTTNSYEK.E + Deamidated (NQ) | 2.10E-03 | ||||||||
| R.VLDEVVVDNFDQK.L | 2.00E-02 | ||||||||
| P05317 | RPP0/Conserved ribosomal protein involved in translational elongation | 582 | 33.7 | 4.8 | 24.4 | 4.7 | 22% | K.SLFVVGVDNVSSQQMHEVR.K | 3.70E-09 |
| K.TSFFQALGVPTK.I | 8.50E-04 | ||||||||
| K.GNVGFVFTNEPLTEIK.N | 3.30E-03 | ||||||||
| R.GTIEIVSDVK.V | 4.20E-02 | ||||||||
| P00549 | CDC19/Pyruvate kinase | 3074 | 54.5 | 7.6 | 57.0 | 7.2 | 74% | F.VFEKEPVSDWTDDVEAR.I | 8.50E-04 |
| K.ACDDKIMYVDYK.N + Oxidation (M); Propionamide (C) | 2.90E-02 | ||||||||
| K.AIIVLSTSGTTPR.L | 4.30E-04 | ||||||||
| K.EPVSDWTDDVEAR.I | 3.60E-04 | ||||||||
| K.GVNLPGTDVDLPALSEK.D | 1.20E-02 | ||||||||
| K.IENQQGVNNFDEILK.V | 3.00E-03 | ||||||||
| K.KGDTYVSIQGFK.A | 3.60E-07 | ||||||||
| K.NGVHMVFASFIR.T + Deamidated (NQ); Oxidation (M) | 4.00E-02 | ||||||||
| K.PTSTTETVAASAVAAVFEQK.A | 4.10E-04 | ||||||||
| K.SEELYPGRPLAIALDTK.G | 2.60E-02 | ||||||||
| K.SNLAGKPVICATQMLESMTYNPR.P + 2 Oxidation (M); Propionamide (C) | 2.10E-05 | ||||||||
| K.TNNPETLVALR.K | 2.10E-03 | ||||||||
| R.AEVSDVGNAILDGADCVMLSGETAK.G + Oxidation (M); Propionamide (C) | 3.20E-04 | ||||||||
| R.EVLGEQGKDVK.I | 4.50E-02 | ||||||||
| R.KSEELYPGRPLAIALDTK.G | 2.70E-04 | ||||||||
| R.LTSLNVVAGSDLR.R | 4.40E-04 | ||||||||
| R.NCTPKPTSTTETVAASAVAAVFEQK.A + Propionamide (C) | 5.50E-03 | ||||||||
4. Discussion
Proteome analysis of yeast challenged with 56MESS identified a protein-based response that was demonstrably different from the cellular response to the current clinical treatment standard, cisplatin. Total numbers of protein species as well as overall protein abundance were reduced, suggesting a role for 56MESS in triggering protein degradation or affecting protein translation. Of the differentially abundant protein species, most were primarily implicated in the known generalised stress response.
4.1. Yeast as a Tool for Systems Biology Analyses of Drug Actions
As a simple and readily available eukaryote, S. cerevisiae is used to investigate fundamental processes of wider biological relevance [53]. High rates of homologous recombination and a completely sequenced genome facilitate high levels of experimental control over genetic content. This is particularly relevant as 31% of yeast proteins have a human orthologue and 50% of human disease genes have yeast orthologues [54]. Yeast is thus a valuable model to initially identify the potential targets and/or mechanisms of action of novel anticancer agents.
As cancer poses a continuing threat to the population, innovative chemotherapeutic approaches must be sought [1,55]. The novel anticancer compound, 56MESS, has a potent activity in both cisplatin-susceptible and -resistant cell lines. However, as unknown mechanisms of action are often limiting factors in the progression of compounds to approved treatments, this compound requires further characterisation.
4.2. The Yeast Response to 56MESS
An initial broader spectrum, high throughput microarray approach was useful as a preliminary investigation into the activity of 56MESS. The deletion library was a valuable resource for the concurrent interrogation of the entire deletion mutant complement of the yeast genome. Each of these gene deletion mutants is identified by a molecular barcode, enabling isolation and identification of strains (and thus genes) potentially involved in the response to different physiological challenges [26]. Screening the yeast deletion mutant library, which represented 96% of the ORFs, reduced the field of investigation from approximately 6000 genes in the complete genome [56,57] to 133 mutants (Figure S1) and facilitated the individual examination of mutant sensitivities. This resulted in 48 confirmed sensitive deletion mutants (Figure 3; Table S4).
Deletion mutant studies of cisplatin primarily have identified genes involved in different repair pathways (i.e., nucleotide excision, recombinational, post replication, DNA inter-strand cross-link, and oxidative damage); these gene deletions sensitize yeast to cisplatin, suggesting DNA as an important target and its repair as vital for cell survival [30]. None of these genes or processes were implicated by the 56MESS sensitive deletion mutants. The overlap between genes affecting cisplatin sensitivity and those influencing 56MESS sensitivity was marginal at 4%. Only two genes were common in the 56MESS and cisplatin sensitive deletion mutants, IMP2` and HAL5 [30]. IMP2` is a transcriptional activator involved in protection against oxidative DNA damage as well as ion homeostasis [58]. HAL5 is a protein kinase involved in transport regulation, which causes sensitivity to acidic pH and cations when deleted [59]. Both genes have regulatory functions and could represent a general cell signalling response to the cationic charge of both drugs rather than to the cytotoxic effects of either. Thus, identification of common genes related to the cellular responses to cisplatin and 56MESS does not preclude different mechanisms of action for these compounds.
Here, deletion of various vacuolar protein sorting (VPS) genes resulted in increased sensitivity to 56MESS (Figure 3, Table S1). Traffic from the Golgi to the vacuole relies on products of VPS genes [60], and these are also required for broad resistance to oxidative stress [61]. VAM7, VTC1, VPS30 and VPS41 genes (Figure 3; Table S4) code for subunits of vacuolar transporter chaperones, receptors, tethering complexes and a protein involved in intra-vacuolar processing [62,63,64,65,66,67]. Other related gene deletions also implicated wider protein sorting and trafficking pathways in 56MESS sensitivity including membrane fusion, ubiquitin dependent protein sorting and Golgi protein recycling [68,69]. In direct contrast to 56MESS, the deletion of vesicle-mediated transport/trafficking related genes increased resistance to cisplatin when deleted [70]. Cumulatively, these results suggest that intracellular protein trafficking mechanisms are particularly critical for resistance to 56MESS such that inhibition of these functions results in sensitivity to this agent (Figure 3; Table S4). The prevalence of protein handling genes as TAG3 microarray hits suggested that analysis of the yeast proteome following 56MESS challenge would be useful.
Exposure to 56MESS decreased the number of detectable protein species (Figure 4, Figure 5 and Figure 6). The exact mechanism of this broad protein depletion is unclear. However, as the overall abundance of proteins was affected, it is possible that 56MESS is interacting with cellular components other than, or in addition to, DNA [17,71]. Considering the low molecular weight protein fragments detected (Figure 4), it is plausible that 56MESS exposure induced some type of protein degradation and/or transcriptional stall. Thus, protein trafficking, degradation, or transcriptional blockade appear to be potentially critical mechanisms in the response to 56MESS; whether this is in response to the compound specifically or damage caused to cellular components by 56MESS, remains to be determined. Thus, rather than DNA damage, more direct effects of 56MESS on the proteome may be the critical factor in the loss of cell viability.
The identification of different heat shock proteins, that both increase and decrease in abundance, neither confirms nor eliminates protein as a target of 56MESS, but are indicative of cellular stress in response to the compound (Table 1). Furthermore, increased amounts of ENO and PGK proteins that are associated with glycolysis and gluconeogenesis [72], implicate glucose metabolism in the anticancer effects of 56MESS. Mammalian tumours demonstrate increased rates of glycolysis for ATP production in a hypoxic environment [73,74]. By altering the abundance of glycolytic proteins, 56MESS may affect the ability of tumour cells to generate energy thus limiting further growth. Decreased amounts of the translational elongation protein, RRP0p, are, however, consistent with the working hypothesis that the inhibition of transcription has a role in the yeast response to 56MESS. There is little to no correlation between the proteins identified by 2DE-MS and those identified using the deletion mutant library. However, some experimentally determined molecular weight and pI values of differentially abundant proteins (Tables S2 and S3) are comparable to potential gene products (i.e., proteins) identified using the deletion mutant library (Table S4). Thus, were somewhat less stringent criteria used for 2DE ‘spot’ selection (i.e., for subsequent MS analyses), genes/proteins common to both the genomic and proteomic datasets may have been identified. However, using the low stringency of microarray analyses (i.e., identifying all changes ≥1.5-fold) would require the MS analysis of 76% of the detectable proteome. Obviously this would not have been an efficient means to define mechanisms of action. The routine initial analysis used here indicated involvement of proteins including enolase, phosphopyruvate hydratases, and heat shock proteins that should be further assessed in larger scale future analyses.
The gene products identified here using genomic and proteomic approaches can also be compared to transcriptomic data from yeast exposed to 56MESS [21]. There are no common genes/proteins between that previous study and the results here; however, Yeast GO-Sim Mapping indicated a number of overlapping cellular processes. Ion transport represented 8.3% of the genes identified in this study compared to 15.4% in the transcriptomic data; this was likely implicated as cells attempt to sequester 56MESS in organelles for degradation to counter its toxicity. This is mirrored by cellular ion homeostasis: 8.3 and 12.1%, respectively. The gene ontology term, ‘response to chemical’ represented by 16.7% (genomic) and 7.7% (transcriptomic) of genes in each study, is also common, as is lipid metabolic process (8.3 and 5.5%, respectively) and organelle fission (4.2 and 3.3%, respectively). Nucleobase small molecule metabolism (6.3% / 3.3%) and transcription from RNA polymerase II (12.5% / 2.2%) were also mapped to both transcriptomic and proteomic datasets. Transmembrane transport, however, was found in each of the transcriptomic, genomic and proteomic datasets at 9.9%, 6.3% and 33.3%, respectively. Carbohydrate metabolic process and mitochondrial organisation were also common to all three datasets although this was represented by only a single gene in the genomic and transcriptomic lists, respectively. Many of these common processes could be related to gathering resources in response to chemical insult. The processes common to both genomic and proteomic datasets included protein folding, protein targeting, vacuole organisation and organelle fusion. Many of the differences between the transcriptomic study [21] and the results here could arise from the difference between challenging a fully functioning cell and deleting a gene entirely before challenge. That there were very few common genes between the genomic and transcriptomic studies is likely due to the dissimilar targets of these techniques; while microarrays assess changes in mRNA expression triggered by an external stimulus, gene deletion removes a protein and alters one or more pathways (i.e., likely broader responses in terms of a system already in stress). The changes in cell viability and protein abundance as measured by proteomics may thus be a more rational representation of the yeast response to 56MESS as they correspond directly to a struggle to maintain cellular functions using an unaltered genome. The effect of post-translational modifications must also not be ignored. As transcriptomic techniques can only detect relative changes in gene expression, gene products that are transcribed but remain inactive pending post-translational modification would also never be detected by these methods. This suggests that the real strength of genomic, transcriptomic and proteomic methods is their use collectively, in understanding changes across the system in order to focus in on and identify those specifically critical to the physiological challenge in question. It may also be that specific mechanisms tend to be obscured by the use of genomic or transcriptomic analyses as these assess molecules that tend to (i) be distinct from active cellular functions; (ii) not to correlate directly with protein levels or active species (e.g., post-translationally modified); and (iii) undergo transient changes in expression as systems seek a new equilibrium in response to physiological challenges.
While proteomic studies to characterise chemotherapeutic triggered changes in protein abundance primarily focus on mammalian cell lines [75], the interactions of individual proteins with cisplatin adducted DNA in yeast have been highlighted [23,76,77,78,79]. The proteins are usually critical for DNA repair and have a high affinity for cisplatin adducted DNA [76,80,81]. Some cisplatin sensitivity is mediated by Ctr1p, a high affinity copper transporter [23]; loss of this protein results in increased yeast resistance to cisplatin, possibly as a result of reduced influx and accumulation of cisplatin [23]. Proteasome inhibition also increases cisplatin resistance [82]. Screening for cisplatin-sensitive mutants using a non-essential haploid mutant library also implicated the ubiquitin-proteasome pathway [83]. While this suggested a link between DNA repair and degradation processes in the yeast response to cisplatin, an effect on the resolved proteome was not apparent (Figure 6).
Overall, in terms of mechanisms of action, the results concerning 56MESS are informative in comparison to cisplatin, but also relative to other intercalators developed as anticancer therapeutics. Intercalator-protein interactions have been described before, and include effects on double strand breaks, inhibition of ribosomal RNA formation, metabolic processes of proteins and nucleic acids, cytoskeleton organisation, and the ubiquitin proteasome pathway [84,85,86,87,88]. Thus, intercalator–protein interactions are likely, usually with a specific or subsequent association with DNA. The effects of 56MESS identified here, suggest a role for such alternate intercalators in the lifecycle of proteins, perhaps at or near synthesis. Thus, the current results support the idea that intercalators interact directly with proteins, and that 56MESS affects proteins in a way that is different to both cisplatin and other intercalators. It is also possible that some intercalators specifically affect transcription of particular gene products; binding specificity can be via interaction with guanine bases in the minor groove or targeting of GC-rich regions [89,90,91]. However, targeting a particular sequence of DNA is far removed from targeting an actual gene sequence. Longer sequences are required to focus the interaction of intercalators to a specific site [92,93]. While intercalation is not yet selective for specific sites on DNA and thus for specific genes (and, subsequently, gene products), there is potential for this specificity to be developed. 56MESS may thus function in a similar albeit less selective manner, affecting a much wider range of genes and subsequent gene products.
5. Conclusions
The yeast model has proven informative in terms of exploring potential mechanisms of drug action using an integrated genomic and chemical proteomic approach, and in assessing the similarities and differences in such ‘Omic’ datasets. The differences in mechanism of action of 56MESS relative to cisplatin are thus now characterized according to multiple criteria; rather than DNA repair, gene deletions in protein trafficking pathways induced sensitivity to 56MESS. Higher drug concentrations were used to induce this sensitivity than comparable studies; however, the yeast cell wall and potentially poor metal uptake are possible causes for this difference. A top-down proteomic analysis yielded evidence of protein degradation or interruption of synthesis. Thus, the mechanisms underlying the superior cytotoxicity of 56MESS are very different from those of the widely used clinical standard, cisplatin. This suggests that use of 56MESS in chemotherapy could potentially counter cisplatin resistance in many cancers. The wider implication of these integrated molecular assessments is that 56MESS may provide an avenue for more effective treatment, and for further development of targeted therapeutics. Although additional analyses using mammalian cells are clearly required, the yeast work has now substantially focused this research on alternative targets for next generation anticancer agents. In this regard, the results also emphasize the importance of validating findings at the protein level, indicating that top-down proteomics should constitute a more routine and expected role in studies utilising high-throughput genomic and transcriptomic analyses.
Supplementary Materials
Supplementary File 1Abbreviations
| 56MESS | [(5,6-dimethyl-1,10-phenanthroline)(1S,2S-diaminocyclohexane)platinum(II)]2+ |
| AFD | automated frozen disruption |
| Camp | cyclic adenosine monophosphate |
| CHAPS | 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate |
| DTT | dithiothreitol |
| IEF | isoelectric focusing |
| IPG | immobilised pH gradient |
| MW | molecular weight |
| PM | perfect match |
| RT | room temperature |
| SDS PAGE | sodium dodecyl sulphate polyacrylamide gel electrophoresis |
| SGD | Saccharomyces Genome Database |
| SNARE | Soluble N-ethylmaleimide sensitive fusion protein Attachment Protein Receptor |
| SRXRF | synchrotron radiation X-ray fluorescence |
| YPD | Yeast Proteome Database |
Acknowledgements
All authors contributed to the writing of the manuscript, and all gave final approval to the submitted version. The authors gratefully acknowledge an Australian International Science Linkage grant (Department of Innovation, Industry, Science and Research) and an Australian Postgraduate Award to EPW which both supported initial aspects of this work. The authors note critical support from the University of Western Sydney, the UWS Molecular Medicine Research Group, and the School of Medicine.
Author Contributions
J.R.A.W., V.J.H. and J.R.C. conceived and designed the experiments. E.P.W. performed the experiments; E.P.W. and M.P.P. analysed the data; M.P.P. contributed analysis tools; E.P.W., J.R.A.W., V.J.H., M.P.P., and J.R.C. wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kostova, I. Platinum complexes as anticancer agents. Recent Pat. Anticancer Drug Discov. 2006, 1, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Boulikas, T.; Vougiouka, M. Cisplatin and platinum drugs at the molecular level. Oncol. Rep. 2003, 10, 1663–1682. [Google Scholar] [PubMed]
- Reedijk, J. New clues for platinum antitumor chemistry: Kinetically controlled metal binding to DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 3611–3616. [Google Scholar] [CrossRef] [PubMed]
- Reedijk, J. Improved understanding in platinum antitumour chemistry. Chem. Commun. 1996, 801–806. [Google Scholar] [CrossRef]
- Brabec, V.; Kasparkova, J. Molecular aspects of resistance to antitumor platinum drugs. Drug Resist. Updates 2002, 5, 147–161. [Google Scholar] [CrossRef]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.; Giandomenico, C.M. Current status of platinum-based antitumor drugs. Chem. Rev. 1999, 99, 2451–2466. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Williams, G.; Pazdur, R. End points and United States food and drug administration approval of oncology drugs. J. Clin. Oncol. 2003, 21, 1404–1411. [Google Scholar] [CrossRef] [PubMed]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautum, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef]
- Butcher, E.C.; Berg, E.L.; Kunkel, E.J. Systems biology in drug discovery. Nat. Biotech. 2004, 22, 1253–1259. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 2011, 80, 273–299. [Google Scholar] [CrossRef] [PubMed]
- Wheate, N.J.; Brodie, C.R.; Collins, J.G.; Kemp, S.; Aldrich-Wright, J.R. DNA intercalators in cancer therapy: Organic and inorganic drugs and their spectroscopic tools of analysis. Mini-Rev. Med. Chem. 2007, 7, 627–648. [Google Scholar] [CrossRef] [PubMed]
- Wan, K.X.; Shibue, T.; Gross, M.L. Non-covalent complexes between DNA-binding drugs and double-stranded oligodeoxynucleotides: A study by ESI ion-trap mass spectrometry. J. Am. Chem. Soc. 2000, 122, 300–307. [Google Scholar] [CrossRef]
- Cusumano, M.; Di Pietro, M.L.; Giannetto, A.; Vainiglia, P.A. The intercalation to DNA of bipyridyl complexes of platinum(II) with thioureas. J. Inorg. Biochem. 2005, 99, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Tse, W.C.; Boger, D.L. Sequence-selective DNA recognition: Natural products and nature’s lessons. Chem. Biol. 2004, 11, 1607–1617. [Google Scholar] [CrossRef] [PubMed]
- Wheate, N.J.; Taleb, R.I.; Krause-Heuer, A.M.; Cook, R.L.; Wang, S.; Higgins, V.J.; Aldrich-Wright, J.R. Novel platinum(II)-based anticancer complexes and molecular hosts as their drug delivery vehicles. Dalton Trans. 2007, 43, 5055–5064. [Google Scholar] [CrossRef] [PubMed]
- Garbutcheon-Singh, K.B.; Leverett, P.; Myers, S.; Aldrich-Wright, J.R. Cytotoxic platinum(II) intercalators that incorporate 1R,2R-diaminocyclopentane. Dalton Trans. 2013, 42, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Fisher, D.M.; Bednarski, P.J.; Grünert, R.; Turner, P.; Fenton, R.R.; Aldrich-Wright, J.R. Chiral platinum(II) metallointercalators with potent in vitro cytotoxic activity. ChemMedChem 2007, 2, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.J.; Carrall, J.A.; Lai, B.; Aldrich-Wright, J.R.; Ralph, S.F.; Dillon, C.T. Does cytotoxicity of metallointercalators correlate with cellular uptake or DNA affinity? Dalton Trans. 2012, 41, 9417–9426. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Higgins, V.; Aldrich-Wright, J.; Wu, M. Identification of the molecular mechanisms underlying the cytotoxic action of a potent platinum metallointercalator. J. Chem. Biol. 2012, 5, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Okuda, T.; Holzer, A.; Howell, S.B. The copper transporter CTR1 regulates cisplatin uptake in Saccharomyces cerevisiae. Mol. Pharmacol. 2002, 62, 1154–1159. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Lee, J.; Thiele, D.J.; Herskowitz, I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc. Natl. Acad. Sci. USA 2002, 99, 14298–14302. [Google Scholar] [CrossRef] [PubMed]
- Sadler, W.R.; Trudinger, P.A. The inhibition of microorganisms by heavy metals. Mineral. Deposita 1967, 2, 158–168. [Google Scholar] [CrossRef]
- Bleackley, M.; MacGillivray, R.A. Transition metal homeostasis: From yeast to human disease. Biometals 2011, 24, 785–809. [Google Scholar] [CrossRef] [PubMed]
- Delneri, D. The use of yeast mutant collections in genome profiling and large-scale functional analysis. Curr. Genomics 2004, 5, 59–65. [Google Scholar] [CrossRef]
- Parsons, A.B.; Geyer, R.; Hughes, T.R.; Boone, C. Yeast genomics and proteomics in drug discovery and target validation. Prog. Cell Cycle Res. 2003, 5, 159–166. [Google Scholar] [PubMed]
- Kurtz, J.E.; Dufour, P.; Bergerat, J.P.; Exinger, F. Saccharomyces cerevisiae as a genetic model in anticancer therapy. Curr. Pharmocogenetics 2005, 3, 1–7. [Google Scholar] [CrossRef]
- European Saccharomyces cerevisiae Archive for Functional Analysis. Available online: http://www.uni-frankfurt.de/fb15/mikro/euroscarf/index.html (accessed on 20 December 2008).
- Wu, H.I.; Brown, J.A.; Dorie, M.J.; Lazzeroni, L.; Brown, J.M. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 2004, 64, 3940–3948. [Google Scholar] [CrossRef] [PubMed]
- Higgins, V.J.; Alic, N.; Thorpe, G.W.; Breitenbach, M.; Larsson, V.; Dawes, I.W. Phenotypic analysis of gene deletant strains for sensitivity to oxidative stress. Yeast 2002, 19, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar]
- Burke, D.; Dawson, D.; Stearns, T. Methods in Yeast Genetics, 2000 ed.; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2000; pp. 109–114. [Google Scholar]
- Shoemaker, D.D.; Lashkari, D.A.; Morris, D.; Mittmann, M.; Davis, R.W. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nature Genet. 1996, 14, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Butt, R.H.; Coorssen, J.R. Pre-extraction sample handling by automated frozen disruption significantly improves subsequent proteomic analyses. J. Proteome Res. 2006, 5, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Butt, R.H.; Pfeifer, T.A.; Delaney, A.; Grigliatti, T.A.; Tetzlaff, W.G.; Coorssen, J.R. Enabling coupled quantitative genomics and proteomics analyses from rat spinal cord samples. Mol. Cell. Proteomics 2007, 6, 1574–1588. [Google Scholar] [CrossRef] [PubMed]
- Churchward, M.; Butt, R.H.; Lang, J.; Hsu, K.; Coorssen, J. Enhanced detergent extraction for analysis of membrane proteomes by two-dimensional gel electrophoresis. Proteome Sci. 2005, 3, e5. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Cannon-Carlson, S.; Tang, J. Modification of the laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis procedure to eliminate artifacts on reducing and nonreducing gels. Anal. Biochem. 1997, 246, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Coorssen, J.R.; Blank, P.S.; Albertorio, F.; Bezrukov, L.; Kolosova, I.; Backlund, P.S.; Zimmerberg, J. Quantitative femto- to attomole immunodetection of regulated secretory vesicle proteins critical to exocytosis. Anal. Biochem. 2002, 307, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Gauci, V.J.; Padula, M.P.; Coorssen, J.R. Coomassie Blue staining for high sensitivity gel-based proteomics. J. Proteomics 2013, 90, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Judd, R. SDS-Polyacrylamide Gel Electrophoresis of Peptides. In The Protein Protocols Handbook, 2nd ed.; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2002; Volume 2, pp. 73–79. [Google Scholar]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Butt, R.H.; Coorssen, J.R. Coomassie Blue as a near-IR fluorescent stain: A systematic comparison with Sypro Ruby for in-gel protein detection. Mol. Cell. Proteomics 2013, 12, 3834–3850. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.P.; Partridge, M.A.; Padula, M.P.; Gauci, V.J.; Malladi, C.S.; Coorssen, J.R. Top-down proteomics: Enhancing 2D gel-electrophoresis from tissue processing to high sensitivity protein detection. Proteomics 2014, 14, 872–889. [Google Scholar] [CrossRef] [PubMed]
- Butt, R.H.; Coorssen, J.R. Postfractionation for enhanced proteomic analyses: Routine electrophoretic methods increase the resolution of standard 2D-PAGE. J. Proteome Res. 2005, 4, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Butt, R.H.; Lee, M.W.Y.; Pirshahid, S.A.; Backlund, P.S.; Wood, S.; Coorssen, J.R. An initial proteomic analysis of human preterm labor: Placental membranes. J. Proteome Res. 2006, 5, 3161–3172. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.R.; Churchward, M.A.; Butt, R.H.; Coorssen, J.R. Assessing detection methods for gel-based proteomic analyses. J. Proteome Res. 2007, 6, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.P.; Prasad, K.A.G.; Padula, M.P.; Coorssen, J.R. Deep imaging: How much of the proteome does current top-down technology already resolve? PLoS One 2014, 9, e86058. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef] [PubMed]
- Hodges, P.E.; McKee, A.H.Z.; Davis, B.P.; Payne, W.E.; Garrels, J.I. The Yeast Proteome Database (YPD): A model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 1999, 27, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef]
- Menacho-Márquez, M.; Murguía, J. Yeast on drugs: Saccharomyces cerevisiae as a tool for anticancer drug research. Clin. Transl. Oncol. 2007, 9, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Foury, F. Human genetic diseases: A cross-talk between man and yeast. Gene 1997, 195, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, J. Current issues in cancer: Cancer chemotherapy: Identifying novel anticancer drugs. Br. Med. J. 1994, 308, 1288–1290. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [PubMed]
- Pierce, S.E.; Fung, E.L.; Jaramillo, D.F.; Chu, A.M.; Davis, R.W.; Nislow, C.; Giaever, G. A unique and universal molecular barcode array. Nat. Meth. 2006, 3, 601–603. [Google Scholar] [CrossRef]
- Masson, J.; Ramotar, D. The Saccharomyces cerevisiae IMP2 gene encodes a transcriptional activator that mediates protection against DNA damage caused by bleomycin and other oxidants. Mol. Cell. Biol. 1996, 16, 2091–2100. [Google Scholar] [PubMed]
- Mulet, J.M.; Leube, M.P.; Kron, S.J.; Rios, G.; Fink, G.R.; Serrano, R. A novel mechanism of ion homeostasis and salt tolerance in yeast: The Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol. Cell. Biol. 1999, 19, 3328–3337. [Google Scholar] [PubMed]
- Wendland, B.; Emr, S.D.; Riezman, H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr. Opin. Cell Biol. 1998, 10, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, G.W.; Fong, C.S.; Alic, N.; Higgins, V.J.; Dawes, I.W. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: Oxidative-stress-response genes. Proc. Natl. Acad. Sci. USA 2004, 101, 6564–6569. [Google Scholar] [CrossRef] [PubMed]
- Ungermann, C.; Wickner, W. Vam7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 1998, 17, 3269–3276. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Hirata, A.; Ohsumi, Y.; Wada, Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 11344–11349. [Google Scholar] [CrossRef] [PubMed]
- Uttenweiler, A.; Schwarz, H.; Neumann, H.; Mayer, A. The Vacuolar Transporter Chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell 2007, 18, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Seaman, M.N.J.; Marcusson, E.G.; Cereghino, J.L.; Emr, S.D. Endosome to golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 1997, 137, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Kruse, K.B.; Brodsky, J.L.; McCracken, A.A. Characterization of an ERAD Gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: One for soluble Z variant of human a-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol. Biol. Cell 2006, 17, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Teter, S.A.; Eggerton, K.P.; Scott, S.V.; Kim, J.; Fischer, A.M.; Klionsky, D.J. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2001, 276, 2083–2087. [Google Scholar] [CrossRef] [PubMed]
- Bugnicourt, A.; Froissard, M.; Sereti, K.; Ulrich, H.D.; Haguenauer-Tsapis, R.; Galan, J.-M. Antagonistic roles of ESCRT and Vps Class C/HOPS complexes in the recycling of yeast membrane proteins. Mol. Biol. Cell 2004, 15, 4203–4214. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau, P.S.; Winistorfer, S.C.; Kearney, W.R.; Robertson, A.D.; Piper, R.C. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 2003, 163, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Eddy, M.; Vujcic, M.; Kowalski, D. Genome-wide screen identifies genes whose inactivation confer resistance to cisplatin in Saccharomyces cerevisiae. Cancer Res. 2005, 65, 5890–5897. [Google Scholar] [CrossRef] [PubMed]
- Brana, M.F.; Cacho, M.; Gradillas, A.; de Pascual-Teresa, B.; Ramos, A. Intercalators as anticancer drugs. Curr. Pharm. Drugs 2001, 7, 1745–1780. [Google Scholar] [CrossRef]
- Gasch, A.P. The environmental stress response: A common yeast response to diverse environmental stresses. In Yeast Stress Responses; Hohmann, S., Mager, W., Eds.; Springer: Berlin Heidelberg, Germany, 2003; Volume 1, pp. 11–70. [Google Scholar]
- Pelicano, H.; Martin, D.S.; Xu, R.H.; Huang, P. Glycolysis inhibition for anticancer treatment. Oncogene 2006, 25, 4633–4646. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.J. Glucose metabolism and cancer. Curr. Opin. Cell Biol. 2006, 18, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Möller, A.; Soldan, M.; Völker, U.; Maser, E. Two-dimensional gel electrophoresis: A powerful method to elucidate cellular responses to toxic compounds. Toxicology 2001, 160, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Kellett, P.J.; Lippard, S.J. Ixr1, a yeast protein that binds to platinated DNA and confers sensitivity to cisplatin. Science 1993, 261, 603–605. [Google Scholar] [CrossRef] [PubMed]
- McA'Nulty, M.M.; Whitehead, J.P.; Lippard, S.J. Binding of Ixr1, a yeast HMG-domain protein, to cisplatin-DNA adducts in vitro and in vivo. Biochemistry 1996, 35, 6089–6099. [Google Scholar] [CrossRef] [PubMed]
- Furuchi, T.; Ishikawa, H.; Miura, N.; Ishizuka, M.; Kajiya, K.; Kuge, S.; Naganuma, A. Two nuclear proteins, Cin5 and Ydr259c, confer resistance to cisplatin in Saccharomyces cerevisiae. Mol. Pharmacol. 2001, 59, 470–474. [Google Scholar] [PubMed]
- Hastie, C.J.; Vazquez-Martin, C.; Philp, A.; Stark, M.J.R.; Cohen, P.T.W. The Saccharomyces cerevisiae orthologue of the human protein phosphatase 4 core regulatory subunit R2 confers resistance to the anticancer drug cisplatin. FEBS J. 2006, 273, 3322–3334. [Google Scholar] [CrossRef] [PubMed]
- Kartalou, M.; Essigmann, J.M. Mechanisms of resistance to cisplatin. Mutat. Res.-Fund. Mol. Mech. Mutagen. 2001, 478, 23–43. [Google Scholar] [CrossRef]
- McA'Nulty, M.M.; Lippard, S.J. The HMG-domain protein Ixr1 blocks excision repair of cisplatin-DNA adducts in yeast. Mutat. Res.-DNA Repair 1996, 362, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Cunha, D.; Cunha, R.; Côrte-Real, M.; Chaves, S.R. Cisplatin-induced cell death in Saccharomyces cerevisiae is programmed and rescued by proteasome inhibition. DNA Repair 2013, 12, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Gatti, L.; Hoe, K.L.; Hayles, J.; Righetti, S.C.; Carenini, N.; Dal Bo, L.; Kim, D.U.; Park, H.O.; Perego, P. Ubiquitin-proteasome genes as targets for modulation of cisplatin sensitivity in fission yeast. BMC Genomics 2011, 12, e44. [Google Scholar] [CrossRef] [PubMed]
- Straetling, W.H.; Seidel, I. Relaxation of chromatin structure by ethidium bromide binding: Determined by viscometry and histone dissociation studies. Biochemistry 1976, 15, 4803–4809. [Google Scholar] [CrossRef] [PubMed]
- Zwelling, L.A.; Michaels, S.; Erickson, L.C.; Ungerleider, R.S.; Nichols, M.; Kohn, K.W. Protein-associated deoxyribonucleic acid strand breaks in L1210 cells treated with the deoxyribonucleic acid intercalating agents 4'-(9-acridinylamino)methanesulfon-m-anisidide and adriamycin. Biochemistry 1981, 20, 6553–6563. [Google Scholar] [CrossRef] [PubMed]
- Gaines, G.; Attardi, G. Intercalating drugs and low temperatures inhibit synthesis and processing of ribosomal RNA in isolated human mitochondria. J. Mol. Biol. 1984, 172, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Tyleckova, J.; Hrabakova, R.; Mairychova, K.; Halada, P.; Radova, L.; Dzubak, P.; Hajduch, M.; Gadher, S.; Kovarova, H. Cancer cell response to anthracyclines effects: Mysteries of the hidden proteins associated with these drugs. Int. J. Mol. Sci. 2012, 13, 15536–15564. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.J.; Fenselau, C. Investigation of doxorubicin resistance in MCF-7 breast cancer cells using shot-gun comparative proteomics with Proteolytic 18O labeling. J. Proteome Res. 2004, 3, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Neto, B.; Lapis, A. Recent developments in the chemistry of deoxyribonucleic acid (DNA) intercalators: Principles, design, synthesis, applications and trends. Molecules 2009, 14, 1725–1746. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R.; Denny, W.A. Genotoxicity of non-covalent interactions: DNA intercalators. Mutat. Res.-Fund. Mol. Mech. Mutagen. 2007, 623, 14–23. [Google Scholar] [CrossRef]
- Takenaka, S.; Nishira, S.; Tahara, K.; Kondo, H.; Takagi, M. Synthesis and characterization of novel tris-intercalators having potentially two different DNA binding modes. Supramol. Chem. 1993, 2, 41–46. [Google Scholar] [CrossRef]
- Guelev, V.M.; Harting, M.T.; Lokey, R.S.; Iverson, B.L. Altered sequence specificity identified from a library of DNA-binding small molecules. Chem. Biol. 2000, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Vanderwall, D.E.; Turner, C.J.; Hoehn, S.; Chen, J.; Kozarich, J.W.; Stubbe, J. Solution structure of the hydroperoxide of Co(III) phleomycin complexed with d(CCAGGCCTGG)2: Evidence for binding by partial intercalation. Nucleic Acids Res. 2002, 30, 4881–4891. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).