Dynamic New World: Refining Our View of Protein Structure, Function and Evolution
Abstract
:1. Introduction
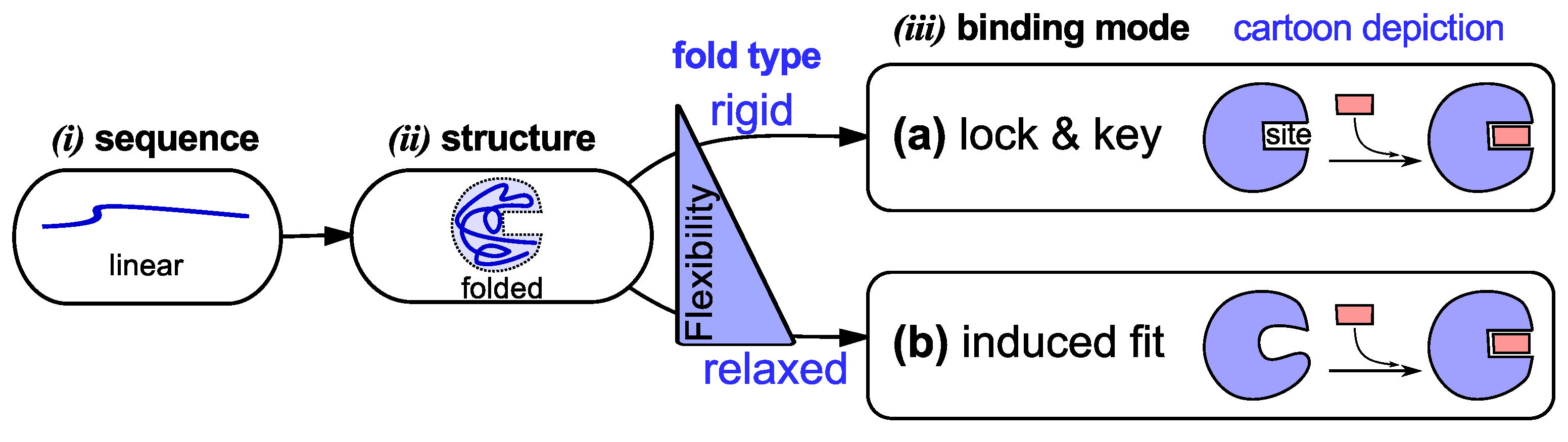
1.1. Sequence → Ordered Structure
1.2. Ordered Structure → Specific Function (Binding)
2. First “New View”: Protein Dynamism and Structure
2.1. The Increasingly Disordered View of “Perfect” Protein Structure
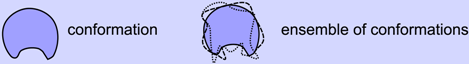
2.2. Refreshing the First Textbook Definition: Protein Structure is an Ensemble
- Structure ≠ conformation
- Structure = ensemble of conformations
- Unlike the synonymous use of the words “structure” and “conformation” in the macroscopic world (e.g., the structure of a building and its conformation/configuration are indistinguishable), at the molecular level, a structure is a collection of accessible conformations that together constitute the temperature-dependent native state structural ensemble. The native state ensembles of some proteins are more diverse than others, making some proteins more dynamic than others.
2.3. Intrinsically Disordered Regions (IDRs) Are Important to Function
2.4. Intrinsically Disordered Proteins (IDPs) Bolster the Dynamic View
2.5. Refreshing the Second Textbook Definition: Proteins versus Peptides
- Both peptides and proteins are linear chains of amino acids. What distinguishes a protein from a peptide?
- Expanded distinction: A protein is a peptide chain that folds [72] reliably into a single (or few) distinct conformation(s) or binds reliably to at least one specific cognate partner.
- The new view assigns highly dynamic, often disordered, but still functional chains—a legitimate entity in the proteome—as legitimate within the protein family.
3. Second “New View”: Protein Dynamism and Promiscuous Function
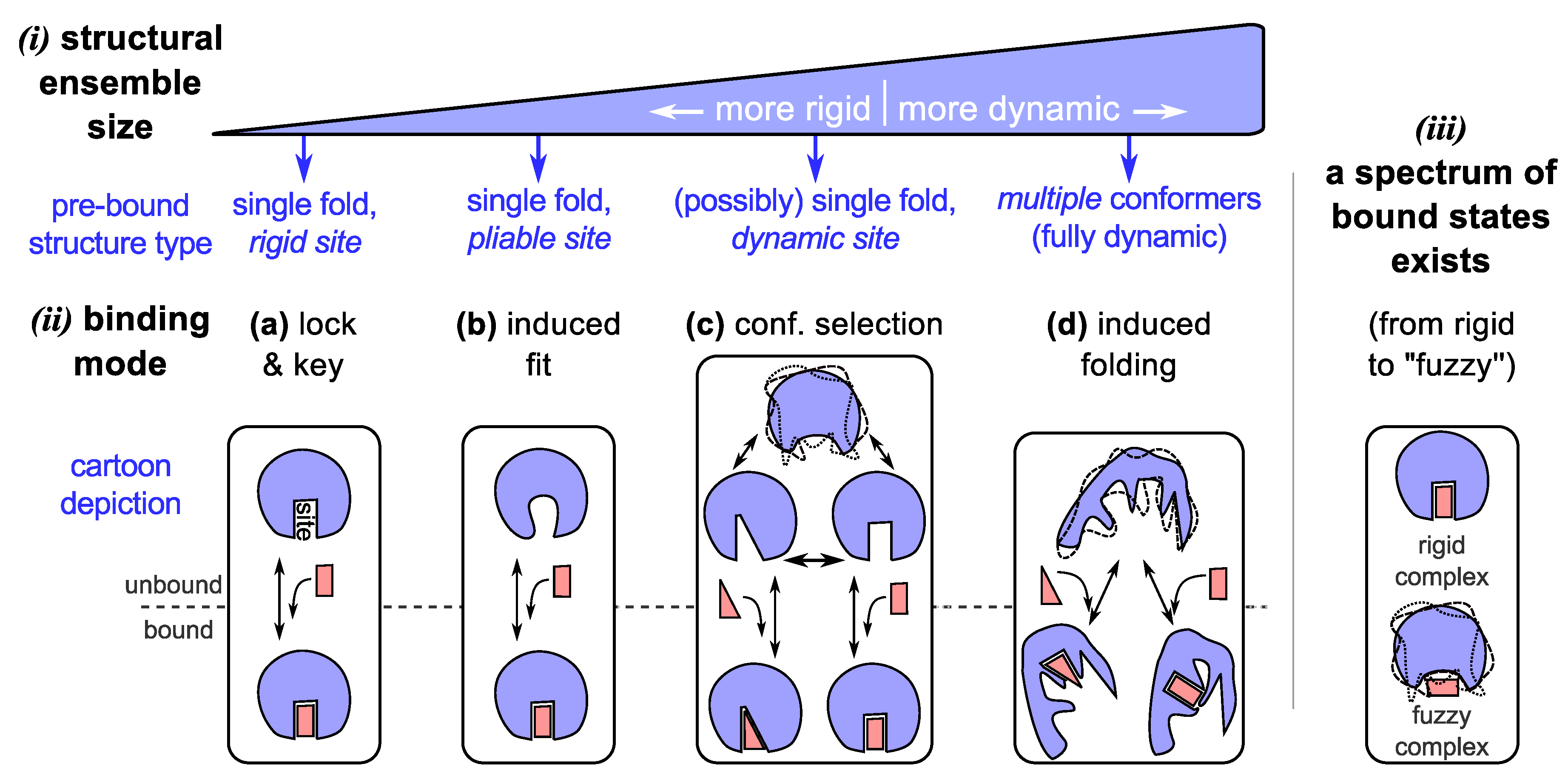
3.1. Binding Model #3: Conformational Selection
3.2. Promiscuous Binding
3.3. Binding Model #4: IDPs Often Bind by Losing Structural Diversity
3.4. Binding Modes Describe a Spectrum, as Do Bound Complexes
3.5. Sequence Determinants (and Bioinformatics) of Promiscuous Function and Dynamic Structure
4. Third “New View”: The Role of Protein Dynamism in Evolution
4.1. Dynamism and Promiscuity Hastens Evolution of New Functionality
4.2. Molecular (Sequence) Evolution from a Historical Perspective
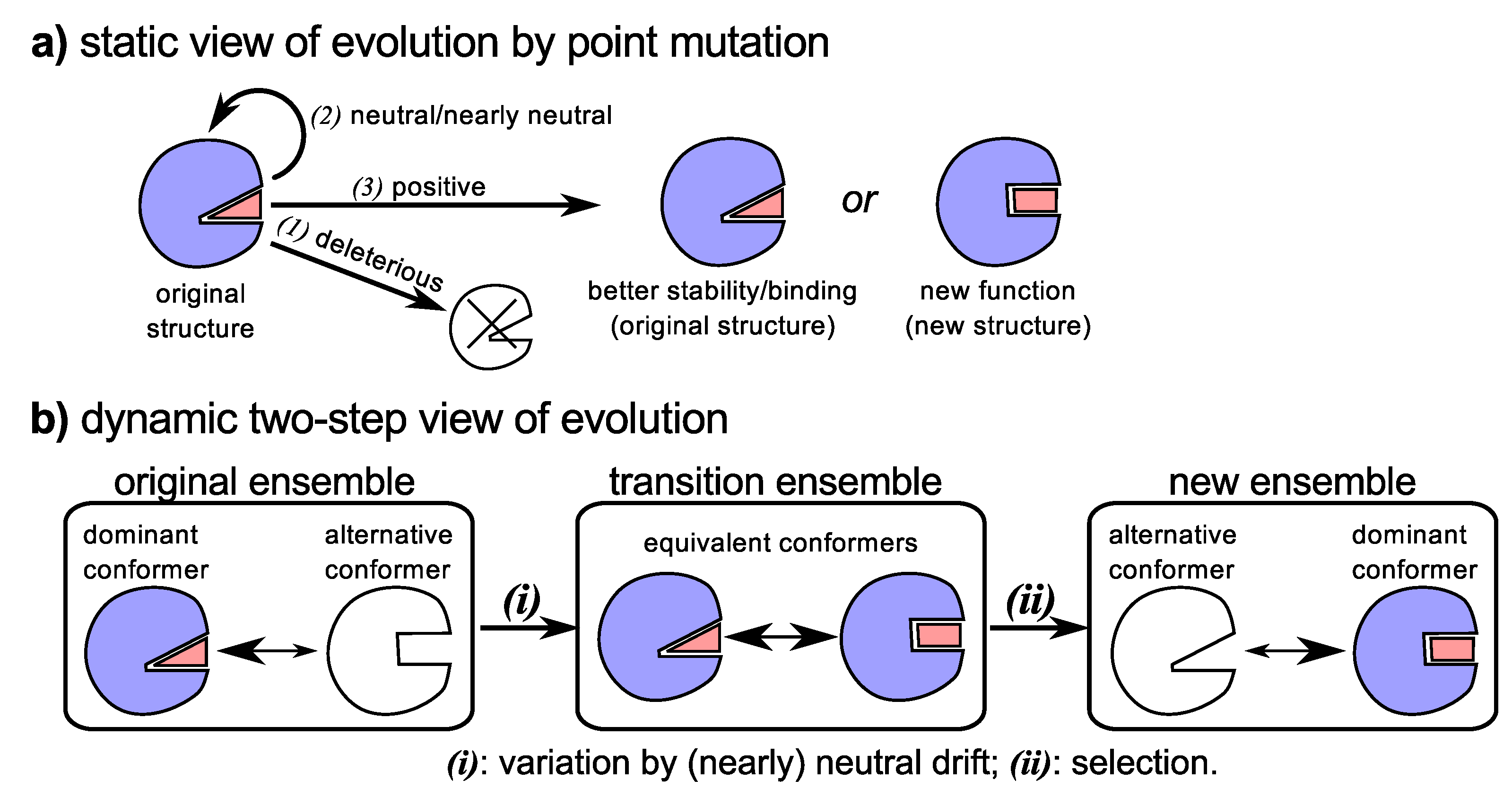
4.3. Neutral Drift Increases Evolutionary Fodder (Structural Dynamism and Functional Promiscuity)
5. Antibody Maturation: A Single System Describing Many Crucial Elements of Dynamism
5.1. Possibly the First Reference to IDPs and IDRs
5.2. Antibody Structure and Function Today
6. From Protein Evolution to Protein Origination
6.1. Differentiation
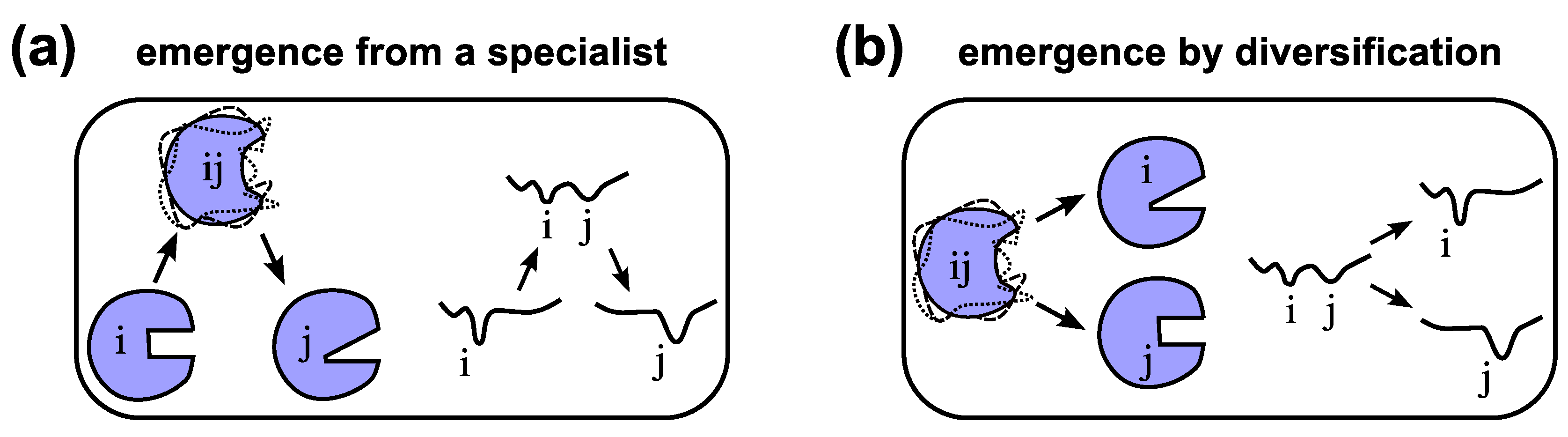
6.2. Origination

7. New Connections and Questions: Links to the Advent of Complex Organisms and Diseases
8. Final Words
Acknowledgments
Conflicts of Interest
References
- Chothia, C.; Hubbard, T.; Brenner, S.; Barns, H.; Murzin, A. Protein folds in the all-beta and all-alpha classes. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 597–627. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, International Edition, 7th ed.; WH Freeman & Co.: New York, NY, USA, 2010. [Google Scholar]
- Cooper, G.M.; Hausman, R.E. The Cell: A Molecular Approach, 6th ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2013. [Google Scholar]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradović, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002, 323, 573–584. [Google Scholar] [CrossRef]
- Babu, M.M.; van der Lee, R.; de Groot, N.S.; Gsponer, J. Intrinsically disordered proteins: Regulation and disease. Curr. Opin. Struct. Biol. 2011, 21, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Eliezer, D. Biophysics of Parkinsons Disease: Structure and Aggregation of-Synuclein. Curr. Protein Pept. Sci. 2009, 10, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Midic, U.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Protein disorder in the human diseasome: Unfoldomics of human genetic diseases. BMC Genomics 2009, 10, S12. [Google Scholar] [CrossRef] [PubMed]
- James, L.C.; Tawfik, D.S. The specificity of cross-reactivity: Promiscuous antibody binding involves specific hydrogen bonds rather than nonspecific hydrophobic stickiness. Protein Sci. 2003, 12, 2183–2193. [Google Scholar] [CrossRef] [PubMed]
- Vértessy, B.G.; Orosz, F. From “fluctuation fit” to “conformational selection”: Evolution, rediscovery, and integration of a concept. Bioessays 2011, 33, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Landsteiner, K. The Specificity of Serological Reactions; Dover Publications (reprinted 1962): Mineola, NY, USA, 1936. [Google Scholar]
- Pauling, L. A theory of the structure and process of formation of antibodies*. J. Am. Chem. Soc. 1940, 62, 2643–2657. [Google Scholar] [CrossRef]
- O’Brien, P.J.; Herschlag, D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 1999, 6, R91–R105. [Google Scholar] [CrossRef]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: An overview. Cell Res. 2009, 19, 929–949. [Google Scholar] [CrossRef] [PubMed]
- James, L.C.; Tawfik, D.S. Conformational diversity and protein evolution—A 60-year-old hypothesis revisited. Trends Biochem. Sci. 2003, 28, 361–368. [Google Scholar] [CrossRef]
- Tokuriki, N.; Tawfik, D.S. Protein dynamism and evolvability. Science 2009, 324, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Schad, E.; Tompa, P.; Hegyi, H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011, 12, R120. [Google Scholar] [CrossRef] [PubMed]
- Copley, S.D. Enzymes with extra talents: Moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 2003, 7, 265–272. [Google Scholar] [CrossRef]
- Khersonsky, O.; Roodveldt, C.; Tawfik, D.S. Enzyme promiscuity: Evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 2006, 10, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Obradovic, Z.; Smith, D.K.; Zhu, G.; Vucetic, S.; Brown, C.J.; Lawson, J.D.; Dunker, A.K. Protein flexibility and intrinsic disorder. Protein Sci. 2004, 13, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Radivojac, P.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Prediction of intrinsic disorder and its use in functional proteomics. Methods Mol. Biol. 2007, 408, 69–92. [Google Scholar] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 2011, 43, 1090–1103. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Arnold, F.H. In the light of directed evolution: Pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 9995–10000. [Google Scholar] [CrossRef]
- Linderstrøm-Lang, K.U. Lane Medical Lectures: Proteins and Enzymes; Stanford University Press: Redwood City, CA, USA, 1952; Volume 6. [Google Scholar]
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Bragg, L.; Kendrew, J.C.; Perutz, M.F. Polypeptide chain configurations in crystalline proteins. Proc. R. Soc. Lond. Ser. A Math. Phys. Sci. 1950, 203, 321–357. [Google Scholar] [CrossRef]
- Eisenberg, D. The discovery of the α-helix and β-sheet, the principal structural features of proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 11207–11210. [Google Scholar] [CrossRef] [PubMed]
- Pauling et al. [27] adhered to basic but still relatively new chemical principles such as planarity of the amide bond within each amino acid and linear hydrogen bonding rules (notions that were omitted in a previously failed attempt a year earlier by luminaries Bragg, Kendrew, and Perutz [28]).
- Kendrew, J.C.; Bodo, G.; Dintzis, H.M.; Parrish, R.G.; Wyckoff, H.; Phillips, D.C. A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature 1958, 181, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Anfinsen, C.B.; Haber, E. Studies on the reduction and re-formation of protein disulfide bonds. J. Biol. Chem. 1961, 236, 1361–1363. [Google Scholar] [PubMed]
- Fischer, E. Einfluss der configuration auf die wirkung der enzyme. Ber. Dt. Chem. Ges. 1894, 27, 2985–2993. [Google Scholar] [CrossRef]
- Behr, J.P. The Lock-and-Key Principle, The State of the Art–100 Years On; John Wiley & Sons: Chichester, UK, 2008; Volume 1. [Google Scholar]
- Koshland, D., Jr. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA 1958, 44, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)—Round IX. Proteins: Struct. Funct. Bioinform. 2011, 79, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.S.; Milburn, D.; Gerstein, M. Conformational changes associated with protein-protein interactions. Curr. Opin. Struct. Biol. 2004, 14, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Kendrew’s structure [31] predated the Protein Databank [36]; however, Watson and Kendrew eventually deposited into the PDB a refined version of the original structure in 1973; PDB ID: 1MBN.
- For example, the first decade of structure deposition in the PDB–the 1970’s–witnessed only one of 92 deposited protein chains (PDB ID 1CHG, chain A) that displayed a substantial number of residues (˃5%) with missing backbone coordinates.
- Doerr, A. Widening the protein crystallization bottleneck. Nat. Methods 2006, 3, 961. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Furletova, E.I.; Bogatyreva, N.S.; Roytberg, M.A.; Galzitskaya, O.V. Library of disordered patterns in 3D protein structures. PLoS Comput. Biol. 2010, 6, e1000958. [Google Scholar] [CrossRef] [PubMed]
- Amadei, A.; Linssen, A.B.; Berendsen, H.J. Essential dynamics of proteins. Proteins 1993, 17, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Atilgan, A.R.; Durell, S.R.; Jernigan, R.L.; Demirel, M.C.; Keskin, O.; Bahar, I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001, 80, 505–515. [Google Scholar] [CrossRef]
- Le Gall, T.; Romero, P.R.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in the Protein Data Bank. J. Biomol. Struct. Dyn. 2007, 24, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.; Olson, A.; Schutt, C.; Winkler, F.; Bricogne, G. Tomato bushy stunt virus at 2.9 Å resolution. Nature 1978, 276, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Breinl, F.; Haurowitz, F. Chemische untersuchung des präzipitates aus Hämoglobin und anti-Hämoglobin-serum und bemerkungen über die natur der antikörper. Hoppe-Seyler’s Z. Physiol. Chem. 1930, 192, 45–57. [Google Scholar] [CrossRef]
- Alexander, J. Some intracellular aspects of life and disease. Protoplasma 1932, 14, 296–306. [Google Scholar] [CrossRef]
- Mudd, S. A hypothetical mechanism of antibody formation. J. Immunol. 1932, 23, 423–427. [Google Scholar]
- Wüthrich, K.; Wagner, G. Internal motion in globular proteins. Trends Biochem. Sci. 1978, 3, 227–230. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Best, R.B.; Depristo, M.A.; Dobson, C.M.; Vendruscolo, M. Simultaneous determination of protein structure and dynamics. Nature 2005, 433, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Lange, O.F.; Lakomek, N.A.; Farès, C.; Schröder, G.F.; Walter, K.F.A.; Becker, S.; Meiler, J.; Grubmüller, H.; Griesinger, C.; de Groot, B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 2008, 320, 1471–1475. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Vucetic, S.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Flavors of protein disorder. Proteins 2003, 52, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: Which way to go? Cell. Mol. Life Sci. 2003, 60, 1852–1871. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005, 15, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Kissinger, C.R.; Villafranca, J.E.; Dunker, A.K. Identifying disordered regions in proteins from amino acid sequences. In Proceedings of the IEEE International Conference on Neural Networks, Houston, TX, USA, 9–12 June 1997; Volume 1, pp. 90–95.
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder and functional proteomics. Biophys. J. 2007, 92, 1439–1456. [Google Scholar] [CrossRef] [PubMed]
- Dosztnyi, Z.; Mszros, B.; Simon, I. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief. Bioinform. 2010, 11, 225–243. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005, 579, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Hazy, E.; Tompa, P. Limitations of induced folding in molecular recognition by intrinsically disordered proteins. ChemPhysChem 2009, 10, 1415–1419. [Google Scholar] [CrossRef] [PubMed]
- Orosz, F.; Ovádi, J. Proteins without 3D structure: Definition, detection and beyond. Bioinformatics 2011, 27, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.; Babu, M.; Barbar, E.; Blackledge, M.; Bondos, S.; Dosztányi, Z.; Dyson, H.; Forman-Kay, J.; Fuxreiter, M.; Gsponer, J.; et al. Whats in a name? Why these proteins are intrinsically disordered? Intrinsically Disord. Proteins 2013, 1, e24157. [Google Scholar] [CrossRef]
- Tompa, P. Unstructural biology coming of age. Curr. Opin. Struct. Biol. 2011, 21, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Farrell, S. Biochemistry; Thomson Brooks/Cole: Belmont, CA, USA, 2006. [Google Scholar]
- Klug, A.; Rhodes, D. Zinc fingers: A novel protein motif for nucleic acid recognition. Trends Biochem. Sci. 1987, 12, 464–469. [Google Scholar] [CrossRef]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Liu, Y.; Liao, J.; Zhu, B.; Wang, E.; Ding, J. Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination. Biochem. J. 2006, 394, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, A.; An, S.; Rosen, A.; Martinis, S.; Musier-Forsyth, K. Fidelity Mechanisms of the Aminoacyl-tRNA Synthetases. In Protein Engineering; Springer: Berlin, Germany, 2009; pp. 155–203. [Google Scholar]
- Morrison, J.L.; Breitling, R.; Higham, D.J.; Gilbert, D.R. A lock-and-key model for protein-protein interactions. Bioinformatics 2006, 22, 2012–2019. [Google Scholar] [CrossRef] [PubMed]
- Lagunas, R.; Sols, A.; DelaFuente, G. Induced fit in yeast hexokinase. Eur. J. Biochem. 1970, 16, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Kuser, P.; Cupri, F.; Bleicher, L.; Polikarpov, I. Crystal structure of yeast hexokinase PI in complex with glucose: A classical “induced fit” example revised. Proteins: Struct. Funct. Bioinform. 2008, 72, 731–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, G.E.; Müller, C.W.; Diederichs, K. Induced-fit movements in adenylate kinases. J. Mol. Biol. 1990, 213, 627–630. [Google Scholar] [CrossRef]
- Peters, J.H.; de Groot, B.L. Ubiquitin dynamics in complexes reveal molecular recognition mechanisms beyond induced fit and conformational selection. PLoS Comput. Biol. 2012, 8, e1002704. [Google Scholar] [CrossRef] [PubMed]
- Keskin, O. Binding induced conformational changes of proteins correlate with their intrinsic fluctuations: A case study of antibodies. BMC Struct. Biol. 2007, 7, e31. [Google Scholar] [CrossRef] [PubMed]
- Bienkiewicz, E.A.; Adkins, J.N.; Lumb, K.J. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1). Biochemistry 2002, 41, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Verkhivker, G.M.; Bouzida, D.; Gehlhaar, D.K.; Rejto, P.A.; Freer, S.T.; Rose, P.W. Simulating disorder–order transitions in molecular recognition of unstructured proteins: Where folding meets binding. Proc. Natl. Acad. Sci. USA 2003, 100, 5148–5153. [Google Scholar] [PubMed]
- Lacy, E.R.; Filippov, I.; Lewis, W.S.; Otieno, S.; Xiao, L.; Weiss, S.; Hengst, L.; Kriwacki, R.W. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol. 2004, 11, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Mok, K.H.; Muhandiram, R.; Park, K.H.; Suk, J.E.; Kim, D.H.; Chang, J.; Sung, Y.C.; Choi, K.Y.; Han, K.H. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J. Biol. Chem. 2000, 275, 29426–29432. [Google Scholar] [CrossRef] [PubMed]
- Aguado-Llera, D.; Hamidi, T.; Domnech, R.; Pantoja-Uceda, D.; Gironella, M.; Santoro, J.; Velzquez-Campoy, A.; Neira, J.L.; Iovanna, J.L. Deciphering the Binding between Nupr1 and MSL1 and Their DNA-Repairing Activity. PLoS One 2013, 8, e78101. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M.; Tompa, P. Fuzzy complexes: A more stochastic view of protein function. Adv. Exp. Med. Biol. 2012, 725, 1–14. [Google Scholar] [PubMed]
- Tompa, P.; Fuxreiter, M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008, 33, 2–8. [Google Scholar] [CrossRef] [PubMed]
- James, L.C.; Roversi, P.; Tawfik, D.S. Antibody multispecificity mediated by conformational diversity. Science 2003, 299, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- Straub, F.B.; Szabolcsi, G. Molecular Biology: Problems and Perspectives; Chapter Remarks on the Dynamic Aspects of Enzyme Structure (in Russian): Nauka, Moscow, 1964; pp. 182–187. [Google Scholar]
- Straub, F. SH groups and SS bridges in the structure of enzymes. In Proceedings of the 7th International Congress of Biochemistry, Tokyo, Japan, 19–25 August 1967; pp. 41–50.
- Chang, C.E.A.; McLaughlin, W.A.; Baron, R.; Wang, W.; McCammon, J.A. Entropic contributions and the influence of the hydrophobic environment in promiscuous protein-protein association. Proc. Natl. Acad. Sci. USA 2008, 105, 7456–7461. [Google Scholar] [CrossRef] [PubMed]
- Khersonsky, O.; Tawfik, D.S. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010, 79, 471–505. [Google Scholar] [PubMed]
- Shoemaker, B.A.; Portman, J.J.; Wolynes, P.G. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl. Acad. Sci. USA 2000, 97, 8868–8873. [Google Scholar] [CrossRef] [PubMed]
- Sugase, K.; Dyson, H.J.; Wright, P.E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 2007, 447, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M.; Simon, I.; Friedrich, P.; Tompa, P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 2004, 338, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Receveur-Brchot, V.; Bourhis, J.M.; Uversky, V.N.; Canard, B.; Longhi, S. Assessing protein disorder and induced folding. Proteins 2006, 62, 24–45. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Z. Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: A critical assessment of the “fly-casting” mechanism. J. Mol. Biol. 2009, 393, 1143–1159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X. From induced fit to conformational selection: A continuum of binding mechanism controlled by the timescale of conformational transitions. Biophys. J. 2010, 98, L15–L17. [Google Scholar] [CrossRef] [PubMed]
- Csermely, P.; Palotai, R.; Nussinov, R. Induced fit, conformational selection and independent dynamic segments: An extended view of binding events. Trends Biochem. Sci. 2010, 35, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Fermani, S.; Trivelli, X.; Sparla, F.; Thumiger, A.; Calvaresi, M.; Marri, L.; Falini, G.; Zerbetto, F.; Trost, P. Conformational selection and folding-upon-binding of intrinsically disordered protein CP12 regulate photosynthetic enzymes assembly. J. Biol. Chem. 2012, 287, 21372–21383. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chu, X.; Longhi, S.; Roche, P.; Han, W.; Wang, E.; Wang, J. Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc. Natl. Acad. Sci. USA 2013, 110, E3743–E3752. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M. Fuzziness: Linking regulation to protein dynamics. Mol. Biosyst. 2012, 8, 168–177. [Google Scholar] [PubMed]
- Bhattacharyya, R.P.; Remnyi, A.; Good, M.C.; Bashor, C.J.; Falick, A.M.; Lim, W.A. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 2006, 311, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Nobeli, I.; Favia, A.D.; Thornton, J.M. Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 2009, 27, 157–167. [Google Scholar] [CrossRef]
- From Uversky et al.’s findings, a protein with high net charge magnitude (R) could remain folded if it were countered by high hydrophobicity (H; on a Kyte-Doolittle scale normalized to range between 0 and 1[64]); accordingly, the remarkable boundary line on the H-R landscape that distinguishes folding vs. disordered proteins was reported [64] to be: (R) = 2.785(H) − 1.151.
- Yamada, J.; Phillips, J.L.; Patel, S.; Goldfien, G.; Calestagne-Morelli, A.; Huang, H.; Reza, R.; Acheson, J.; Krishnan, V.V.; Newsam, S.; et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 2010, 9, 2205–2224. [Google Scholar] [CrossRef] [PubMed]
- Bridgham, J.T.; Carroll, S.M.; Thornton, J.W. Evolution of hormone-receptor complexity by molecular exploitation. Science 2006, 312, 97–101. [Google Scholar] [PubMed]
- Adami, C. Reducible complexity. Science 2006, 312, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Galvão, T.C.; de Lorenzo, V. Transcriptional regulators a la carte: Engineering new effector specificities in bacterial regulatory proteins. Curr. Opin. Biotechnol. 2006, 17, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. Evol. Genes Proteins 1965, 97, 97–166. [Google Scholar]
- Zuckerkandl, E.; Pauling, L.B. Molecular Disease, Evolution, and Genetic Heterogeneity; Academic Press: New York, NY, USA, 1962; pp. 189–225. [Google Scholar]
- Marigoliash, E. Primary structure and evolution of cytochrome C. Proc. Natl. Acad. Sci. USA 1963, 50, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Molecules as documents of evolutionary history. J. Theor. Biol. 1965, 8, 357–366. [Google Scholar] [CrossRef]
- Bromham, L.; Penny, D. The modern molecular clock. Nat. Rev. Genet. 2003, 4, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. The neutral theory of molecular evolution and the world view of the neutralists. Genome 1989, 31, 24–31. [Google Scholar] [CrossRef]
- Kimura, M. Evolutionary rate at the molecular level. Nature 1968, 217, 624–626. [Google Scholar] [CrossRef] [PubMed]
- King, J.L.; Jukes, T.H. Non-Darwinian evolution. Science 1969, 164, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. Model of effectively neutral mutations in which selective constraint is incorporated. Proc. Natl. Acad. Sci. USA 1979, 76, 3440–3444. [Google Scholar] [CrossRef]
- Ohta, T. Slightly deleterious mutant substitutions in evolution. Nature 1973, 246, 96–98. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Ota, T. On some principles governing molecular evolution. Proc. Natl. Acad. Sci. USA 1974, 71, 2848–2852. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. Possibility of extensive neutral evolution under stabilizing selection with special reference to nonrandom usage of synonymous codons. Proc. Natl. Acad. Sci. USA 1981, 78, 5773–5777. [Google Scholar] [CrossRef]
- Ohta, T.; Gillespie, J.H. Development of Neutral and Nearly Neutral Theories. Theor. Popul. Biol. 1996, 49, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Raval, A.; Wilke, C.O. Thermodynamics of neutral protein evolution. Genetics 2007, 175, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.H. The molecular clock may be an episodic clock. Proc. Natl. Acad. Sci. USA 1984, 81, 8009–8013. [Google Scholar] [CrossRef]
- Gillespie, J.H. The status of the neutral theory: The neutral theory of molecular evolution. Science 1984, 224, 732–733. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.H. Natural selection and the molecular clock. Mol. Biol. Evol. 1986, 3, 138–155. [Google Scholar] [PubMed]
- Nilsson, J.; Grahn, M.; Wright, A.P.H. Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 2011, 12, R65. [Google Scholar] [CrossRef]
- Blum, D. Scientists in Open War over “Neutral Theory” of genetics. Sacramento Bee 1992, A1. [Google Scholar]
- Mindell, D.; Sites, J.; Graur, D. Mode of allozyme evolution: Increased genetic distance associated with speciation events. J. Evol. Biol. 1990, 3, 125–131. [Google Scholar] [CrossRef]
- Mindell, D.P.; Sites, J.W., Jr.; Graur, D. Assessing The Relationship Between Speciation And Evolutionary Change. Cladistics 1990, 6, 393–398. [Google Scholar] [CrossRef]
- Barraclough, T.G.; Savolainen, V. Evolutionary rates and species diversity in flowering plants. Evolution 2001, 55, 677–683. [Google Scholar] [CrossRef]
- Lanfear, R.; Ho, S.Y.W.; Love, D.; Bromham, L. Mutation rate is linked to diversification in birds. Proc. Natl. Acad. Sci. USA 2010, 107, 20423–20428. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Bromham, L.; Ho, S.Y.W. Reply to Englund: Molecular evolution and diversification—Counting species is better than counting nodes when the phylogeny is unknown. Proc. Natl. Acad. Sci. USA 2011, 108, E85–E86. [Google Scholar] [CrossRef]
- Eo, S.H.; DeWoody, J.A. Evolutionary rates of mitochondrial genomes correspond to diversification rates and to contemporary species richness in birds and reptiles. Proc. Biol. Sci. 2010, 277, 3587–3592. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Gaidukov, L.; Khersonsky, O.; Gould, S.M.; Roodveldt, C.; Tawfik, D.S. The “evolvability” of promiscuous protein functions. Nat. Genet. 2004, 37, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Bershtein, S.; Goldin, K.; Tawfik, D.S. Intense neutral drifts yield robust and evolvable consensus proteins. J. Mol. Biol. 2008, 379, 1029–1044. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Romero, P.A.; Lu, Z.; Arnold, F.H. Neutral genetic drift can alter promiscuous protein functions, potentially aiding functional evolution. Biol. Direct 2007, 2, e17. [Google Scholar] [CrossRef]
- Morange, M. History of Molecular Biology; Harvard University Press: Cambridge, MA, USA, 1998; Cobb, M., Translator. [Google Scholar]
- Clark, L.A.; Ganesan, S.; Papp, S.; van Vlijmen, H.W.T. Trends in antibody sequence changes during the somatic hypermutation process. J. Immunol. 2006, 177, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Teng, G.; Papavasiliou, F.N. Immunoglobulin somatic hypermutation. Annu. Rev. Genet. 2007, 41, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Oakman, E.L.; Thorpe, I.F.; Shi, X.; Abbyad, P.; Brooks, C.L., III; Boxer, S.G.; Romesberg, F.E. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. USA 2006, 103, 13722–13727. [Google Scholar] [CrossRef]
- Thorpe, I.F.; Brooks, C.L., III. Molecular evolution of affinity and flexibility in the immune system. Proc. Natl. Acad. Sci. USA 2007, 104, 8821–8826. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976, 30, 409–425. [Google Scholar] [PubMed]
- Yčas, M. On earlier states of the biochemical system. J. Theor. Biol. 1974, 44, 145–160. [Google Scholar] [CrossRef]
- Jensen, R.A.; Gu, W. Evolutionary recruitment of biochemically specialized subdivisions of Family I within the protein superfamily of aminotransferases. J. Bacteriol. 1996, 178, 2161–2171. [Google Scholar] [PubMed]
- Glasner, M.E.; Gerlt, J.A.; Babbitt, P.C. Evolution of enzyme superfamilies. Curr. Opin. Chem. Biol. 2006, 10, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Glasner, M.E.; Gerlt, J.A.; Babbitt, P.C. Mechanisms of protein evolution and their application to protein engineering. In Advances in Enzymology and Related Areas of Molecular Biology; Volume 75, Toone, E.J., Ed.; Wiley: Hoboken, NJ, USA, 2007; pp. 193–239. [Google Scholar]
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef] [PubMed]
- Matthews, C.N.; Moser, R.E. Peptide synthesis from hydrogen cyanide and water. Nature 1967, 215, 1230–1234. [Google Scholar] [CrossRef] [PubMed]
- Anders, E. Pre-biotic organic matter from comets and asteroids. Nature 1989, 342, 255–257. [Google Scholar] [CrossRef] [PubMed]
- Zahnle, K.; Grinspoon, D. Comet dust as a source of amino acids at the Cretaceous/Tertiary boundary. Nature 1990, 348, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Glavin, D.; Dworkin, J.; Sandford, S. Detection of cometary amines in samples returned by Stardust. Meteorit. Planet. Sci. 2008, 43, 399–413. [Google Scholar] [CrossRef]
- Wachtershauser, G. On the chemistry and evolution of the pioneer organism. Chem. Biodivers. 2007, 4, 584–602. [Google Scholar] [CrossRef] [PubMed]
- Plankensteiner, K.; Reiner, H.; Rode, B.M. Catalytically increased prebiotic peptide formation: Ditryptophan, dilysine, and diserine. Orig. Life Evol. Biosph. 2005, 35, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Leman, L.; Orgel, L.; Ghadiri, M.R. Carbonyl sulfide-mediated prebiotic formation of peptides. Science 2004, 306, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Abkevich, V.I.; Gutin, A.M.; Shakhnovich, E.I. How the first biopolymers could have evolved. Proc. Natl. Acad. Sci. USA 1996, 93, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Mannige, R.V. Origin of the protein fold repertoire from oily pluripotent peptides. Proteomes 2014. submitted for publication. [Google Scholar]
- Mannige, R.V. Two regimes of protein evolution and their unique dependencies on sequence composition. Phys. Rev. E 2013, 87, e062714. [Google Scholar] [CrossRef]
- Mannige, R.V.; Brooks, C.L., III; Shakhnovich, E.I. A universal trend among proteomes indicates an oily last common ancestor. PLoS Comput. Biol. 2012, 8, e1002839. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X. Intrinsic disorder: Signaling via highly specific but short-lived association. Trends Biochem. Sci. 2012, 37, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Nakamura, H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006, 580, 2041–2045. [Google Scholar] [CrossRef] [PubMed]
- Manna, B.; Bhattacharya, T.; Kahali, B.; Ghosh, T.C. Evolutionary constraints on hub and non-hub proteins in human protein interaction network: Insight from protein connectivity and intrinsic disorder. Gene 2009, 434, 50–55. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mannige, R.V. Dynamic New World: Refining Our View of Protein Structure, Function and Evolution. Proteomes 2014, 2, 128-153. https://doi.org/10.3390/proteomes2010128
Mannige RV. Dynamic New World: Refining Our View of Protein Structure, Function and Evolution. Proteomes. 2014; 2(1):128-153. https://doi.org/10.3390/proteomes2010128
Chicago/Turabian StyleMannige, Ranjan V. 2014. "Dynamic New World: Refining Our View of Protein Structure, Function and Evolution" Proteomes 2, no. 1: 128-153. https://doi.org/10.3390/proteomes2010128
APA StyleMannige, R. V. (2014). Dynamic New World: Refining Our View of Protein Structure, Function and Evolution. Proteomes, 2(1), 128-153. https://doi.org/10.3390/proteomes2010128