Cell-Type-Specific Heat-Induced Changes in the Proteomes of Pollen Mother Cells and Microspores Provide New Insights into Tomato Pollen Production Under Elevated Temperature
Abstract
1. Introduction
2. Materials and Methods
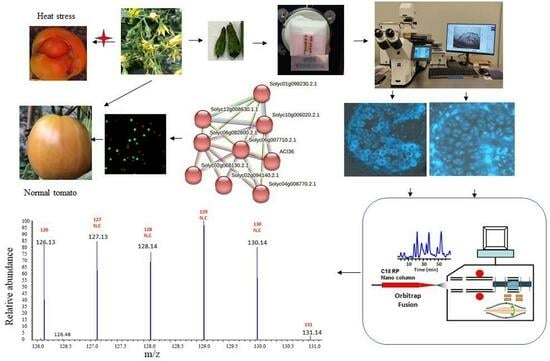
2.1. Plant Materials and Heat Treatments
2.2. Determination of Flower Bud Growth Process and Correction Between Flower Buds and Pollen Developmental Stages
2.3. Collection of Cell-Type-Specific Pollen Cell Samples Using Laser Capture Microdissection
2.4. Quantitative Proteomics Analysis
2.5. Identification of Differentially Abundant Proteins (DAPs)
2.6. Functional Analysis of Differentially Abundant Proteins (DAPs)
3. Results and Analysis
3.1. Identification of Cell-Type-Specific Proteomes in PMCs and Microspores
3.2. The Heat-Induced Proteomics Changes in PMCs
3.3. The Heat-Induced Proteomics Changes in Microspores
3.4. The Cell- and Varietal-Specific HS-Induced Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declaration
References
- FAO. Crops and Livestock Products. Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/ (accessed on 15 February 2025).
- Straits Research. Tomato Market Size, Share & Trends Analysis Report by Product (Sauces, Paste, Canned Tomatoes, Ketchup, Juice, Others) and by Region (North America, Europe, APAC, Middle East and Africa, LATAM) Forecasts, 2024–2032. Available online: https://straitsresearch.com/report/tomato-market (accessed on 15 February 2025).
- AgMRC. Tomato Profile. Agricultural Marketing Resource Center (AgMRC). Available online: https://www.agmrc.org/commodities-products/vegetables/tomatoes (accessed on 15 February 2025).
- Chaturvedi, P.; Wiese, A.J.; Ghatak, A.; Záveská Drábková, L.; Weckwerth, W.; Honys, D. Heat stress response mechanisms in pollen development. New Phytol. 2021, 231, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Peet, M.M.; Willits, D.H.; Gardner, R. Response of ovule development and post-pollen production processes in male-sterile tomatoes to chronic, sub-acute high temperature stress. J. Exp. Bot. 1997, 48, 101–111. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Iwahori, S. High-temperature injuries in tomato. IV. Development of normal flower buds and morphological abnormalities of flower buds treated with high temperatures. J. Jpn. Soc. Hort. Sci. 1966, 35, 63–71. [Google Scholar]
- Alsamir, M.; Mahmood, T.; Trethowan, R.; Ahmad, N. An overview of heat stress in tomato (Solanum lycopersicum L.). Saudi J. Biol. Sci. 2021, 28, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Peet, M.M.; Sato, S.; Gardner, R.G. Comparing heat stress effects on male-fertile and male-sterile tomatoes. Plant Cell Environ. 1998, 21, 225–231. [Google Scholar] [CrossRef]
- Graci, S.; Barone, A. Tomato plant response to heat stress: A focus on candidate genes for yield-related traits. Front. Plant Sci. 2024, 14, 1245661. [Google Scholar] [CrossRef]
- Khan, Q.; Wang, Y.; Xia, G.; Yang, H.; Luo, Z.; Zhang, Y. Deleterious effects of heat stress on the tomato, its innate responses, and potential preventive strategies in the realm of emerging technologies. Metabolites 2024, 14, 283. [Google Scholar] [CrossRef]
- Pressman, E.; Peet, M.M.; Pharr, D.M. The effect of heat stress on pollen characteristics is associated with changes in carbohydrate concentration in tomato anthers. Ann. Bot. 2002, 90, 631–636. [Google Scholar] [CrossRef]
- De Storme, N.; Geelen, D. The impact of environmental stress on male reproductive development in plants: Biological processes and molecular mechanisms. Plant Cell Environ. 2014, 37, 1–18. [Google Scholar] [CrossRef]
- Paupière, M.J.; van Haperen, P.; Rieu, I.; Visser, R.G.F.; Tikunov, Y.M.; Bovy, A.G. Screening for pollen tolerance to high temperatures in tomato. Euphytica 2017, 213, 130. [Google Scholar] [CrossRef]
- Rieu, I.; Twell, D.; Firon, N. Pollen development at high temperature: From acclimation to collapse. Plant Physiol. 2017, 173, 1967–1976. [Google Scholar] [PubMed]
- Sato, S.; Peet, M.M.; Thomas, J.F. Physiological factors limit fruit set of tomato (Lycopersicon esculentum Mill.) under chronic, mild heat stress. Plant Cell Environ. 2000, 23, 719–726. [Google Scholar]
- Sato, S.; Kamiyama, M.; Iwata, T.; Makita, N.; Furukawa, H.; Ikeda, H. Moderate increase of mean daily temperature adversely affects fruit set of Lycopersicon esculentum by disrupting specific physiological processes in male reproductive development. Ann. Bot. 2006, 97, 731–738. [Google Scholar]
- Sarma, S.; Pandey, A.K.; Sharma, K.; Ravi, M.; Sreelakshmi, Y.; Sharma, R. MutS-Homolog2 silencing generates tetraploid meiocytes in tomato (Solanum lycopersicum). Plant Direct. 2018, 2, e00017. [Google Scholar] [CrossRef] [PubMed]
- Schindfessel, C.; De Storme, N.; Trinh, H.K.; Geelen, D. Asynapsis and meiotic restitution in tomato male meiosis induced by heat stress. Front. Plant Sci. 2023, 14, 1210092. [Google Scholar] [CrossRef]
- Keller, M.; Simm, S. The coupling of transcriptome and proteome adaptation during development and heat stress response of tomato pollen. BMC Genom. 2018, 19, 447. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Ischebeck, T.; Lichtscheidl, I.; Weckwerth, W. Cell-specific analysis of the tomato pollen proteome from pollen mother cell to mature pollen provides evidence for developmental priming. J. Proteome Res. 2013, 12, 4892–4903. [Google Scholar]
- Li, H.; Qin, Y.; Wu, X.; O’Hair, J.; Potts, J.; Zhou, S.; Yang, Y.; Fish, T.; Thannhauser, T.W. Identification of heat-induced proteomes in meiotic pollen mother cells of tomato ‘Maxifort’ using single-cell-type tandem mass tag (TMT) proteomics. Veg. Res. 2022, 2, 2. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Frank, G.; Pressman, E.; Ophir, R.; Althan, L.; Shaked, R.; Freedman, M.; Shen, S.; Firon, N. Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J. Exp. Bot. 2009, 60, 3891–3908. [Google Scholar] [CrossRef] [PubMed]
- Giorno, F.; Wolters-Arts, M.; Mariani, C.; Rieu, I. Ensuring reproduction at high temperatures: The heat stress response during anther and pollen development. Plants 2013, 2, 489–506. [Google Scholar] [CrossRef]
- Mizoi, J.; Todaka, D.; Imatomi, T.; Kidokoro, S.; Sakurai, T.; Kodaira, K.-S.; Takayama, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. The ability to induce heat shock transcription factor-regulated genes in response to lethal heat stress is associated with thermotolerance in tomato cultivars. Front. Plant Sci. 2023, 14, 1269964. [Google Scholar] [CrossRef]
- Thapa, P.; Shourav, B.R.; Kajol, P.; Jun, G.; Dibya, T.M.; Suping, Z.; Manar, S.; Hui, L.; Sonali, R. Developing new pollen grain count method using machine learning to evaluate heat tolerance of tomatoes. In Proceedings of the 45th Annual University-Wide Research Symposium, Nashville, TN, USA, 28–30 March 2023; Tennessee State University: Nashville, TN, USA, 2023. [Google Scholar]
- Li, H.; Zhu, Y.; Rangu, M.; Wu, X.; Bhatti, S.; Zhou, S.; Yang, Y.; Fish, T.; Thannhauser, T.W. Identification of heat-induced proteomes in tomato microspores using LCM- proteomics analysis. Single Cell Biol. 2018, 7, 173. [Google Scholar] [CrossRef]
- Yang, S.; Li, H.; Bhatti, S.; Zhou, S.; Yang, Y.; Fish, T.; Thannhauser, T.W. The Al-induced proteomes of epidermal and outer cortical cells in root apex of cherry tomato ‘LA 2710’. J. Proteom. 2020, 211, 103560. [Google Scholar] [CrossRef]
- Potts, J.; Li, H.; Qin, Y.; Wu, X.; Hui, D.; Al Nasr, K.; Zhou, S.; Yong, Y.; Fish, T.; Liu, J.; et al. Using single cell type proteomics to identify Al-induced proteomes in outer layer cells and interior tissues in the apical meristem/cell division regions of tomato root-tips. J. Proteom. 2022, 255, 104486. [Google Scholar] [CrossRef]
- Okekeogbu, I.; Ye, Z.; Sangireddy, S.R.; Li, H.; Bhatti, S.; Hui, D.; Zhou, S.; Howe, K.J.; Fish, T.; Yang, Y.; et al. Effect of aluminum treatment on proteomes of radicles of seeds derived from Al-treated tomato plants. Proteomes 2014, 2, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Okekeogbu, I.; Sangireddy, S.; Ye, Z.; Li, H.; Bhatti, S.; Hui, D.; McDonald, D.W.; Yang, Y.; Giri, S.; et al. Proteome modification in tomato plants upon long-term aluminum treatment. J. Proteome Res. 2016, 15, 1670–1684. [Google Scholar]
- Fu, Q.; Liu, Z.; Bhawal, R.; Anderson, E.T.; Sherwood, R.W.; Yang, Y.; Thannhauser, T.; Schroyen, M.; Tang, X.; Zhang, H.; et al. Comparison of MS2, synchronous precursor selection MS3, and real-time search MS3 methodologies for lung proteomes of hydrogen sulfide treated swine. Anal. Bioanal Chem. 2021, 413, 419–429. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Miyaji, S.; Ito, T.; Kitaiwa, T.; Nishizono, K.; Agake, S.I.; Harata, H.; Aoyama, H.; Umahashi, M.; Sato, M.; Inaba, J.; et al. N2-Acetylornithine deacetylase functions as a Cys-Gly dipeptidase in the cytosolic glutathione degradation pathway in Arabidopsis thaliana. Plant J. 2024, 118, 1603–1618. [Google Scholar] [CrossRef] [PubMed]
- Jesús-Pires, C.D.; Ferreira-Neto, J.R.C.; Bezerra-Neto, J.P.; Kido, E.A.; de Lima, J.C.; Pandolfi, V.; Benko-Iseppon, A.M. Plant thaumatin-like proteins: Function, evolution, and biotechnological applications. Curr. Protein Pept. Sci. 2020, 21, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Alpdağtaş, S.; Jankowski, N.; Urlacher, V.B.; Koschorreck, K. Identification of redox activators for continuous reactivation of glyoxal oxidase from Trametes versicolor in a two-enzyme reaction cascade. Sci. Rep. 2024, 14, 5932. [Google Scholar] [CrossRef]
- Liu, X.; Keyhani, N.O.; Liu, H.; Zhang, Y.; Xia, Y.; Cao, Y. Glyoxal oxidase-mediated detoxification of reactive carbonyl species contributes to virulence, stress tolerance, and development in a pathogenic fungus. PLoS Pathog. 2024, 20, e1012431. [Google Scholar] [CrossRef]
- Goff, K.E.; Ramonell, K.M. The role and regulation of receptor-like kinases in plant defense. Gene Regul. Syst. Bio. 2007, 1, 167–175. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Phillips, M.J.; Moore, S.; Veit, B. A description of the Mei2-like protein family; structure, phylogenetic distribution and biological context. Dev. Genes Evol. 2004, 214, 149–158. [Google Scholar] [CrossRef]
- Kaur, J.; Sebastian, J.; Siddiqi, I. The Arabidopsis-mei2-like genes play a role in meiosis and vegetative growth in Arabidopsis. Plant Cell 2006, 18, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Brockway, H.; Balukoff, N.; Dean, M.; Alleva, B.; Smolikove, S. The CSN/COP9 signalosome regulates synaptonemal complex assembly during meiotic prophase I of Caenorhabditis elegans. PLoS Genet. 2014, 10, e1004757. [Google Scholar] [CrossRef]
- Horváth, A.; Rona, G.; Pagano, M.; Jordan, P.W. Interaction between NSMCE4A and GPS1 links the SMC5/6 complex to the COP9 signalosome. BMC Mol. Cell Biol. 2020, 21, 36. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Serino, G.; Deng, X.W. Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 2002, 14, 2553–2563. [Google Scholar] [CrossRef]
- Ahmadli, U.; Kalidass, M.; Khaitova, L.C.; Fuchs, J.; Cuacos, M.; Demidov, D.; Zuo, S.; Pecinkova, J.; Mascher, M.; Ingouff, M.; et al. High temperature increases centromere-mediated genome elimination frequency and enhances haploid induction in Arabidopsis. Plant Commun. 2023, 4, 100507. [Google Scholar] [CrossRef] [PubMed]
- Crhak, K.L.; Mikulkova, P.; Pecinkova, J.; Kalidass, M.; Heckmann, S.; Lermontova, I.; Riha, K. Heat stress impairs centromere structure and segregation of meiotic chromosomes in Arabidopsis. eLife 2024, 12, RP90253. [Google Scholar] [CrossRef]
- Lucie, C.K.; Pavlina, M.; Jana, P.; Kalidass, M.; Lermontova, I.; Karel, R. Temperature stress impairs centromere structure and segregation of meiotic chromosomes in Arabidopsis. In Proceedings of the EMBO Workshop: Plant Genome Stability and Change, Olomouc, Czech Republic, 16–19 June 2024; Available online: https://is.muni.cz/publication/2485282/EMBO_Fajk_Fojt_Proch-Sc_Malik-M_Lysak_Xinyi.pdf (accessed on 15 February 2025).
- Ota, S.; Tanaka, Y.; Yasutake, R.; Ikeda, Y.; Yuki, R.; Nakayama, Y.; Saito, Y. Distinct effects of heat shock temperatures on mitotic progression by influencing the spindle assembly checkpoint. Exp. Cell Res. 2023, 429, 113672. [Google Scholar] [CrossRef]
- Quilichini, T.D.; Grienenberger, E.; Douglas, C.J. The biosynthesis, composition, and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 2015, 113, 170–182. [Google Scholar]
- Zhang, D.; Wu, S.; Li, C. Lignin biosynthesis and its role in pollen exine formation in Arabidopsis. Plant Cell Rep. 2016, 35, 901–912. [Google Scholar]
- Jaffri, S.R.F.; MacAlister, C.A. Sequential deposition and remodeling of cell wall polymers during tomato pollen development. Front. Plant Sci. 2021, 12, 703713. [Google Scholar] [CrossRef]
- Ma, X.; Wu, Y.; Zhang, G. Formation pattern and regulatory mechanisms of pollen wall in Arabidopsis. J. Plant Physiol. 2021, 260, 153388. [Google Scholar]
- Tian, G.-W.; Chen, M.-H.; Zaltsman, A.; Citovsky, V. Pollen-specific pectin methylesterase involved in pollen tube growth. Dev. Biol. 2006, 294, 83–91. [Google Scholar]
- Advani, V.M.; Ivanov, P. Translation regulation under stress conditions in plants. RNA Biol. 2009, 6, 102–113. [Google Scholar]
- Liu, B.; Qian, S.B. Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA 2014, 5, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Angelos, E.; Ruberti, C.; Kim, S.J.; Brandizzi, F. Maintaining the factory: The roles of the unfolded protein response in cellular homeostasis in plants. Plant J. Cell Mol. Biol. 2017, 90, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Kandel, R.; Jung, J.; Neal, S. Proteotoxic stress and the ubiquitin proteasome system. Semin. Cell Dev. Biol. 2024, 156, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Q.; Bao, Y.; Bassham, D.C. Autophagic degradation of membrane-bound organelles in plants. Biosci. Rep. 2023, 43, BSR20221204. [Google Scholar] [CrossRef]
- Xie, Z.; Zhao, S.; Tu, Y.; Liu, E.; Li, Y.; Wang, X.; Chen, C.; Zhai, S.; Qi, J.; Wu, C.; et al. Proteasome resides in and dismantles plant heat stress granules constitutively. Mol. Cell 2024, 84, 3320–3335.e7. [Google Scholar]
- Beers, E.P.; Woffenden, B.J.; Zhao, C. Plant proteolytic enzymes: Possible roles during programmed cell death. Plant Mol. Biol. 2000, 44, 399–415. [Google Scholar]
- Moloi, S.J.; Ngara, R. The roles of plant proteases and protease inhibitors in drought response: A review. Front. Plant Sci. 2023, 14, 1165845. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.R.; Gao, J.; Lin, H.X.; Lin, Y. The molecular basis of heat stress responses in plants. Mol. Plant 2023, 16, 1612–1634. [Google Scholar]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Front. Plant Sci. 2023, 14, 1110622. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Vu, L.D.; Gevaert, K.; De Smet, I. Protein language: Post-translational modifications talking to each other. Trends Plant Sci. 2018, 23, 1068–1080. [Google Scholar] [PubMed]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Klíma, M.; Renaut, J. Plant proteoforms under environmental stress: Functional proteins arising from a single gene. Front. Plant Sci. 2021, 14, 793113. [Google Scholar]






| Accession | BV 1 (HT/Ctr) 3 | MT 2 (HT/Ctr) | BV/MT (Ctr) | BV/MT (HT) | Protein Name |
|---|---|---|---|---|---|
| Meiosis | |||||
| Solyc06g069480 | −0.8 * | 0.3 | 0.3 | −0.8 * | DExH-box ATP-dependent RNA helicase DExH14 |
| Solyc01g091650 | −0.8 * | −0.1 | 0.0 | −0.7 | COP9 signalosome complex subunit 1 |
| Solyc01g005820 | −0.6 | 0.0 | −0.1 | −0.7 * | Splicing factor arginine/serine-rich 4 |
| Solyc09g072570 | −0.5 * | 0.2 | 0.0 | −0.7 * | Pre-mRNA-splicing factor SLU7-A |
| Solyc05g056230 | 0.6 * | −0.1 | −0.3 | 0.5 * | Calreticulin 2 calcium-binding protein |
| Solyc01g104570 | −0.9 * | 0.5 | 0.4 | −1.0 * | Kinetochore protein ndc80 |
| Solyc06g069480 | −0.8 * | 0.3 | 0.3 | −0.8 * | ATP-dependent RNA helicase |
| Solyc10g080740 | −0.8 | 0.0 | 0.0 | −0.8 * | Smg-7 homolog nonsense-mediated mRNA decay factor |
| Solyc01g091650 | −0.8 * | −0.1 | 0.0 | −0.7 | COP9 signalosome complex subunit 1 |
| Solyc11g070140 | −0.8 * | 0.1 | 0.0 | −0.8 * | Cell division protein kinase 2 |
| Solyc02g084700 | −0.5 * | 0.0 | 0.2 | −0.3 | MORC family CW-type zinc finger 3 |
| Solyc07g064970 | −0.5 * | 0.4 | 0.3 | −0.5 * | Microtubule-associated protein MAP65-1a |
| Solyc11g065180 | −0.5 * | 0.1 | 0.2 | −0.4 | THUMP domain-containing protein |
| Solyc01g079080 | −0.4 | 0.7 * | 0.1 | −1.1 * | Meiosis-specific protein ASY3 |
| Solyc07g018300 | −0.3 | 0.0 | −0.1 | −0.5 * | Single-stranded DNA-binding protein p30 subunit |
| Solyc08g075840 | −0.3 | 0.5 * | 0.4 | −0.3 | Single-stranded DNA-binding replication protein A large subunit |
| Solyc07g045010 | 0.0 | −0.5 * | −0.6 * | −0.2 | Protein JASON |
| Solyc04g007120 | 0.5 * | 0.2 | −0.1 | 0.2 | UV excision repair protein RAD23 |
| Heat shock proteins | |||||
| Solyc04g009770 | −0.8 * | −0.1 | 0.1 | −0.5 | DNAJ chaperone |
| Solyc05g055160 | −0.7 * | 0.1 | 0.2 | −0.6 * | DNAJ chaperone |
| Solyc11g006460 | −0.6 * | −0.2 | 0.1 | −0.3 | DNAJ chaperone |
| Solyc03g121640 | 0.2 | 0.1 | 0.4 | 0.5 * | chaperonin |
| Solyc03g116790 | 0.3 | 0.4 | −0.3 | −0.5 * | Chaperone protein dnaJ |
| Solyc10g084300 | 0.3 | 0.6 * | 0.0 | −0.3 | Chaperone protein dnaJ 3 |
| Solyc09g007630 | 0.4 | 0.1 | 0.2 | 0.5 * | Chaperone DnaJ |
| Solyc03g117630 | 1.5 * | 0.8 | −0.4 | 0.3 | Heat shock protein |
| Solyc08g082820 | 0.5 * | 0.1 | −0.2 | 0.2 | Heat shock protein |
| Solyc01g109890 | 0.5 * | 0.4 | 0.2 | 0.3 | Chaperone protein dnaJ 10 |
| Solyc09g005120 | 0.6 * | 0.2 | −0.1 | 0.3 | Chaperone protein DnaJ |
| Solyc11g020040 | 0.6 * | 0.4 | 0.1 | 0.3 | Chaperone DnaK |
| Solyc03g082920 | 0.7 * | 0.2 | −0.2 | 0.3 | Heat shock protein |
| Solyc02g088610 | 0.7 * | 0.4 | −0.2 | 0.1 | ATP-dependent chaperone ClpB |
| Solyc04g081570 | 0.8 * | 0.3 | 0.1 | 0.6 * | Chaperone protein htpG |
| Solyc11g071830 | 1.0 * | 0.0 | −0.1 | 0.8 * | Chaperone protein dnaj |
| Solyc08g062340 | 1.2 * | 0.2 | −0.2 | 0.8 * | Class II small heat shock protein Le-HSP17.6 |
| Solyc06g076520 | 1.2 * | −0.2 | 0.4 | 1.8 * | Class I heat shock protein |
| Solyc03g117630 | 1.5 * | 0.8 * | −0.4 | 0.3 | Heat shock protein |
| Solyc03g123540 | 1.7 * | 0.8 * | 0.9 | 1.8 * | Class II heat shock protein |
| Solyc04g014480 | 2.7 * | 1.1 * | −0.1 | 1.5 * | Class I heat shock protein 3 |
| Accession | BV 1 (HT/Ctr) 3 | MT 2 (HT/Ctr) | BV/MT (Ctr) | BV/MT (HT) | Protein Names |
|---|---|---|---|---|---|
| Phenylpropanoids and pollen wall formation | |||||
| Solyc10g050160 | 2.29 * | 3.02 * | −0.07 | −0.93 | Caffeoyl-CoA 3-O-methyltransferase |
| Solyc07g015960 | 2.46 * | 3.44 * | 0.30 | −0.81 | Hydroxycinnamoyl CoA quinate transferase |
| Solyc12g096770 | −2.06 * | −0.94 | 0.43 | −0.82 | Hydroxycinnamoyl CoA quinate transferase |
| Solyc04g081520 | −1.71 * | −0.87 | 1.31 * | 0.34 | Laccase-16 |
| Solyc12g094460 | 1.30 | 2.43 * | −0.02 | −1.28 | Laccase-2 |
| Solyc05g056170 | 1.45 | 2.13 * | 0.66 | −0.15 | Phenylalanine ammonia-lyase |
| Solyc01g112000 | 2.05 * | 3.03 * | 0.62 | −0.49 | Expansin-like protein |
| Solyc12g015690 | 2.40 * | 1.90 * | 0.20 | 0.57 | Fasciclin-like arabinogalactan protein 10 |
| Solyc11g005490 | −1.83 * | −2.26 * | −0.57 | −0.27 | Fasciclin-like arabinogalactan protein 18 |
| Solyc06g073750 | −2.26 * | −2.84 * | −0.45 | 0.00 | Beta-D-glucosidase |
| Solyc11g072230 | −2.71 * | −1.60 | −0.03 | −1.27 | Glucan endo-1 3-beta-glucosidase 3 |
| Solyc06g076170 | −2.25 * | −3.06 * | −0.99 * | −0.30 | Glucan endo-1 3-beta-glucosidase |
| Solyc11g044910 | 1.68 * | 1.37 | 0.23 | 0.41 | Beta-xylosidase 1 |
| Solyc10g047030 | 1.63 * | 0.89 | 0.90 * | 1.52 * | Beta-xylosidase 4 |
| Solyc01g102330 | −0.26 | −1.92 * | −0.27 | 1.27 | Acetyl xylan esterase A |
| Solyc01g087280 | 1.78 * | 0.92 | −0.88 | −0.15 | Polygalacturonase A |
| Solyc01g097660 | 4.03 * | 4.19 * | −0.02 | −0.31 | Pectinesterase inhibitor |
| Solyc02g093580 | −1.13 | −1.80 * | 0.29 | 0.83 | Pectate lyase |
| Flavonoid/isoflavonoid biosynthesis | |||||
| Solyc02g083860 | 1.98 * | 3.24 * | 1.18 * | −0.21 | Flavanone 3-hydroxylase |
| Solyc01g111070 | 1.20 | 3.25 * | 1.14 * | −1.03 | Chalcone synthase family protein |
| Fatty acid metabolism | |||||
| Solyc09g061840 | 2.17 * | 2.02 * | 0.38 | 0.40 | 3-ketoacyl CoA thiolase 1 |
| Solyc05g056290 | 1.72 * | −0.11 | −1.23 | 0.47 | Acetyl-CoA carboxylase |
| Solyc01g006450 | 1.76 * | 1.34 | 0.01 | 0.31 | Enoyl reductase |
| Solyc12g100120 | 1.47 | 2.00 * | 0.05 | −0.61 | Acetyl-CoA carboxylase-like protein |
| Solyc09g092450 | 1.16 | 2.46 * | 0.51 | −0.91 | Long-chain fatty acid CoA ligase |
| Amino acid metabolism | |||||
| Solyc08g076970 | 1.82 * | 2.34 * | 0.36 | −0.29 | Acetylornithine deacetylase |
| Solyc09g008670 | 1.69 * | 2.91 * | 0.45 | −0.89 | Threonine ammonia-lyase |
| Solyc06g064550 | −1.75 * | −1.37 | −0.55 | −1.06 | Aspartokinase–homoserine dehydrogenase |
| Solyc10g078550 | 2.50 * | 2.52 * | 0.38 | 0.23 | Glutamate dehydrogenase |
| Solyc05g052100 | 0.35 | 1.92 * | 0.33 | −1.37 * | Glutamate dehydrogenase |
| Solyc05g051250 | 2.27 | 1.81 * | −0.21 | 0.12 | Glutamine synthetase |
| Solyc01g080280 | 1.23 | 2.25 * | 0.44 | −0.71 | Glutamine synthetase |
| Solyc04g014510 | 1.52 | 2.63 * | 0.67 | −0.56 | Glutamine synthetase |
| Protease activity | |||||
| Solyc00g187050 | 1.75 * | 1.99 * | 0.43 | 0.06 | Leucyl aminopeptidase |
| Solyc12g010020 | 2.11 * | 2.18 * | 0.32 | 0.13 | Leucyl aminopeptidase |
| Solyc02g080880 | 1.76 * | 2.13 * | −0.12 | −0.62 | Aspartic proteinase |
| Solyc06g069220 | 4.60 * | 5.98 * | 1.61 | 0.10 | Aspartic proteinase nepenthesin I |
| Solyc01g107760 | 2.98 * | 2.73 * | −0.18 | −0.06 | Cathepsin B-like cysteine proteinase |
| Solyc03g111730 | 2.41 * | 4.10 * | 0.22 | −1.59 | Cathepsin B-like cysteine proteinase |
| Solyc07g053460 | 3.73 * | 4.38 * | 0.67 | −0.12 | Cathepsin B-like cysteine proteinase |
| Solyc07g041920 | 3.68 * | 3.16 * | 0.09 | 0.48 | Cathepsin L-like cysteine proteinase |
| Solyc01g110110 | 3.16 * | 5.28 * | 1.38 * | −0.86 | Cysteine proteinase cathepsin F |
| Solyc11g008240 | 3.27 * | 2.74 * | −0.70 | −0.29 | SKP1-like protein |
| Solyc09g084490 | 3.38 * | 3.32 * | 1.80 | 1.73 * | Proteinase inhibitor I |
| Solyc03g098680 | 3.30 * | 4.51 * | 0.71 | −0.64 | Kunitz trypsin inhibitor |
| Solyc04g081810 | 3.40 * | 1.92 * | 0.99 | 2.35 * | Subtilisin-like protease |
| Solyc02g092670 | 2.81 * | 2.91 * | 0.54 | 0.31 | Subtilisin-like protease |
| Solyc07g041900 | 1.85 * | 1.19 | −0.40 | 0.13 | Cathepsin L-like cysteine proteinase |
| Solyc03g098780 | 1.89 * | 0.06 | −0.06 | 1.64 * | Kunitz-type protease inhibitor |
| Solyc03g098790 | 2.26 * | −0.04 | 0.16 | 2.34 * | Kunitz-type protease inhibitor |
| Solyc03g098760 | 2.83 * | 0.79 | 0.91 | 2.81 * | Kunitz-type protease inhibitor-like protein |
| Solyc03g098710 | 2.36 * | 1.25 | 1.24 * | 2.22 * | Kunitz-type proteinase inhibitor A4 |
| Solyc02g091410 | 1.63 * | 0.72 | −0.68 | 0.09 | Pro-apoptotic serine protease nma111-like protein |
| Solyc09g084480 | 2.26 * | 1.59 | 1.49 * | 2.02 * | Proteinase inhibitor I |
| Solyc03g020030 | 2.34 * | 1.08 | 0.54 | 1.68 * | Proteinase inhibitor II |
| Solyc04g079440 | 1.62 * | 0.73 | 0.16 | 0.93 | Serpin |
| Solyc00g071180 | 3.89 * | 1.14 | 2.16 | 4.78 * | Cysteine proteinase inhibitor |
| Solyc06g072570 | −2.16 * | −0.54 | −0.44 | −2.18 * | Ubiquitin-conjugating enzyme E2 1 |
| Solyc01g096450 | −1.03 | −1.87 * | −0.42 | 0.29 | Aspartic proteinase nepenthesin-1 |
| Solyc11g020960 | −1.02 | −2.45 * | −0.14 | 1.16 | Proteinase inhibitor II |
| Solyc01g111400 | −1.43 | −1.96 * | −0.44 | −0.04 | Subtilisin-like protease |
| Solyc11g065210 | −0.91 | −2.43 * | 0.17 | 1.56 | Ubiquitin carboxyl-terminal hydrolase |
| Solyc06g074850 | 0.02 | 2.11 * | 0.53 | −1.69 | Serine carboxypeptidase |
| Solyc08g079910 | 1.27 | 2.89 * | 0.27 | −1.47 | Subtilisin-like protease |
| Transcription factors | |||||
| Solyc04g071620 | 3.47 * | 0.65 | −1.47 | 1.22 | ASR4 |
| Solyc02g062210 | 1.98 * | 1.46 | 1.57 * | 1.96 * | NAC transcription factor |
| Solyc01g100750 | 2.05 * | 1.75 | 0.21 | 0.38 | Susceptibility homeodomain transcription factor |
| Mitosis | |||||
| Solyc02g062410 | 3.04 * | 3.94 * | 0.72 | −0.31 | Nuclear movement protein nudc |
| Solyc06g084440 | 1.88 * | 1.66 | 0.08 | 0.17 | Nuclear protein localization 4 |
| Solyc04g007330 | −1.84 * | −2.35 * | 0.56 | 0.94 | HORMA domain-containing protein expressed |
| Solyc06g083870 | −1.78 * | −1.49 | 0.29 | −0.13 | Structural maintenance of the chromosome 1 protein |
| Solyc08g078320 | −1.01 | −1.82 * | −0.01 | 0.67 | E3 ubiquitin–protein ligase HOS1 |
| Solyc02g078290 | −1.98 * | −2.67 * | 0.26 | 0.82 | Myosin-like protein |
| Responses to stress | |||||
| Solyc06g076520 | −1.47 | −2.35 * | 0.64 | 1.39 | Class I heat shock protein |
| Solyc06g076570 | −1.18 | −1.82 * | 0.53 | 1.05 | Class I heat shock protein |
| Solyc01g099660 | 2.08 * | 0.84 | 0.01 | 1.11 | Heat shock protein |
| Solyc03g111860 | 1.02 | 2.25 * | 0.50 | −0.86 | Glyoxal oxidase |
| Solyc11g069040 | 1.61 * | 1.26 | −0.22 | 0.00 | Lactoylglutathione lyase |
| Solyc04g074640 | 1.24 | 2.16 * | 0.80 | −0.25 | L-ascorbate peroxidase |
| Solyc11g066390 | 1.63 * | 0.58 | 0.10 | 1.02 | Superoxide dismutase |
| Solyc03g062890 | 1.84 * | 1.25 | 0.38 | 0.83 | Superoxide dismutase |
| Solyc04g079890 | 3.69 * | 4.23 * | 0.99 | 0.33 | Thaumatin-like protein |
| Solyc07g007730 | 2.65 * | 3.00 * | 0.86 | 0.38 | Defensin protein |
| Solyc07g053360 | 2.69 * | 2.39 * | 0.96 | 1.13 | Late embryogenesis-abundant protein |
| Accession | BV 1 Microspore | BV PMC 3 | MT 2 Microspore | MT PMC | Description |
|---|---|---|---|---|---|
| Variety specific | |||||
| Solyc04g080880.2.1 | 1.37 * | 0.82 * | 0.60 | −0.19 | Cathepsin B-like cysteine proteinase |
| Solyc06g074850.2.1 | 0.02 | 0.41 | 2.11 * | 1.03 * | Serine carboxypeptidase |
| Solyc06g068140.2.1 | 1.18 * | 0.97 * | 0.51 | −0.17 | Methyl-binding domain protein |
| Solyc05g013720.2.1 | 1.38 | 0.26 | 2.26 * | 1.24 | Alpha-galactosidase |
| Solyc01g090600.2.1 | 1.01 | 0.43 | 1.55 * | 1.41 * | Chalcone synthase 3 protein |
| Solyc01g111070.2.1 | 1.20 | −0.15 | 3.25 * | 0.77 * | Chalcone synthase |
| Solyc01g010900.2.1 | 0.86 | 0.60 | 1.19 * | 1.10 * | Cytochrome P450 |
| Solyc01g107910.2.1 | 0.24 | 0.23 | 1.64 * | 1.23 * | Caffeoyl CoA 3-O-methyltransferase |
| Solyc01g099580.1.1 | 0.96 | 0.10 | 4.08 * | 2.41 * | Desiccation-related protein PCC13-62 |
| Solyc07g006660.1.1 | 1.19 | 0.50 | 0.96 * | 1.21 * | Glyoxal oxidase |
| Solyc05g050700.1.1 | 0.55 | 0.40 | 1.09 * | 0.73 * | LRR receptor-like serine/threonine–protein kinase, RLP |
| Solyc03g044150.2.1 | −0.66 | 0.37 | −1.17 * | −0.72 * | Subtilisin-like protease |
| Pollen developmental stage-specific | |||||
| Solyc04g014480.2.1 | 0.16 | 2.68 * | −0.27 | 1.06 * | Class I heat shock protein 3 |
| Solyc03g123540.2.1 | −0.22 | 1.69 * | 0.00 | 0.77 * | Class II heat shock protein |
| Solyc03g111730.2.1 | 2.41 * | 0.21 | 4.10 * | 0.50 | Cathepsin B-like cysteine proteinase |
| Solyc04g081810.2.1 | 3.40 * | 0.44 | 1.92 * | 0.40 | Subtilisin-like protease |
| Solyc02g082300.2.1 | 1.38 * | 0.42 | 1.97 * | 0.55 | AT-hook motif nuclear-localized protein 2 |
| Solyc02g062210.1.1 | 1.98 * | −0.04 | 1.46 * | −0.01 | NAC transcription factor |
| Solyc06g084440.2.1 | 1.88 * | −0.05 | 1.66 * | 0.20 | Nuclear protein localization 4 |
| Solyc01g100750.2.1 | 2.05 * | −0.08 | 1.75 * | −0.30 | Susceptibility homeodomain transcription factor |
| Solyc09g061230.2.1 | 2.07 * | 0.14 | 1.61 * | 0.06 | Cytochrome b5 |
| Solyc02g083860.2.1 | 1.98 * | −0.16 | 3.24 * | 0.61 | Flavanone 3-hydroxylase |
| Solyc01g058390.2.1 | 1.33 * | −0.06 | 2.37 * | 0.10 | Galactokinase |
| Solyc05g012540.2.1 | 1.54 * | 0.23 | 2.22 * | 0.02 | Heparanase |
| Solyc11g066290.1.1 | 2.24 * | 0.40 | 2.18 * | 0.35 | Icc family phosphohydrolase |
| Solyc05g051850.2.1 | 2.31 * | 0.07 | 1.18 * | −0.03 | Inositol-3-phosphate synthase |
| Solyc01g110060.2.1 | 2.34 * | 0.01 | 2.52 * | −0.37 | Purple acid phosphatase |
| Solyc00g058900.1.1 | 1.68 * | 0.64 | 3.56 * | 0.35 | GDSL esterase/lipase |
| Solyc11g071800.1.1 | 0.84 * | 0.55 | 0.97 * | 0.57 | Strictosidine synthase family protein |
| Solyc07g042220.1.1 | 2.62 * | −0.29 | 3.51 * | 0.18 | Beta-galactosidase |
| Solyc05g054590.2.1 | 1.73 * | 0.29 | 1.64 * | −0.10 | Bifunctional polymyxin resistance protein ArnA |
| Solyc05g051250.2.1 | 2.27 * | −0.05 | 1.81 * | −0.36 | Glutamine synthetase |
| Solyc11g069040.2.1 | 1.61 * | 0.35 | 1.26 * | 0.26 | Lactoylglutathione lyase |
| Solyc04g074640.2.1 | 1.24 * | 0.52 | 2.16 * | 0.21 | L-ascorbate peroxidase |
| Solyc11g073210.1.1 | 1.39 * | −0.47 | 3.37 * | 0.09 | Legumin 11S-globulin |
| Solyc07g008710.2.1 | 2.79 * | −0.40 | 2.10 * | 0.23 | Major latex-like protein |
| Solyc01g068040.1.1 | −2.36 * | 0.14 | −1.70 * | −0.35 | 3-5 exonuclease/nucleic acid binding protein |
| Solyc03g115360.2.1 | −1.80 * | −0.44 | −1.55 * | −0.30 | 40S ribosomal protein S19-like |
| Solyc02g021400.1.1 | −1.25 * | −0.58 | −1.92 * | −0.45 | 40S ribosomal protein S28 |
| Solyc12g096150.1.1 | −1.66 * | −0.23 | −0.96 * | −0.28 | 60S ribosomal protein L13 |
| Solyc09g066430.2.1 | −1.77 * | −0.42 | −0.91 * | −0.27 | 60S ribosomal protein L14 |
| Solyc12g008720.1.1 | −1.69 * | 0.00 | −1.08 * | −0.46 | 60S ribosomal protein L31 |
| Solyc04g007330.1.1 | −1.84 * | 0.17 | −2.35 * | 0.01 | DNA-binding HORMA |
| Solyc09g008620.1.1 | −1.57 * | −0.34 | −1.91 * | 0.10 | Polyadenylate-binding protein |
| Solyc08g061320.2.1 | −1.90 * | 0.36 | −2.26 * | −0.41 | Smr domain-containing protein |
| Solyc06g083870.2.1 | −1.78 * | −0.09 | −1.49 * | 0.07 | Structural maintenance of the chromosome 1 protein |
| Solyc01g103420.2.1 | −1.08 * | −0.23 | −1.96 * | 0.18 | Upf3 regulator of nonsense transcripts-like protein B |
| Solyc06g064480.2.1 | −2.06 * | 0.01 | −2.04 * | 0.40 | Protein TAP1 |
| Solyc06g051010.1.1 | −1.51 * | 0.35 | −2.08 * | −0.21 | Asparagine amidase A |
| Solyc01g111400.2.1 | −1.43 * | 0.08 | −1.96 * | −0.17 | Subtilisin-like protease |
| Solyc01g096450.2.1 | −1.03 * | 0.59 | −1.87 * | −0.58 | Aspartic proteinase nepenthesin-1 |
| Solyc08g078320.2.1 | −1.01 * | −0.66 | −1.82 * | 0.16 | E3 ubiquitin–protein ligase HOS1 |
| Solyc07g045010.2.1 | −1.93 * | 0.02 | −1.98 * | −0.46 | Dentin sialophosphoprotein-like |
| Solyc11g066130.1.1 | −1.82 * | 0.60 | −2.19 * | −0.54 | Thaumatin-like protein |
| Solyc01g111280.2.1 | −1.17 * | −0.14 | −2.17 * | −0.37 | Cold shock protein-1 |
| Solyc11g072230.1.1 | −2.71 * | 0.46 | −1.60 * | −0.11 | Glucan endo-1 3-beta-glucosidase 3 |
| Solyc06g076170.2.1 | −2.25 * | −0.39 | −3.06 * | −0.20 | Glucan endo-1 3-beta-glucosidase |
| Solyc12g096770.1.1 | −2.06 * | 0.09 | −0.94 * | 0.19 | Hydroxycinnamoyl CoA quinate transferase |
| Solyc11g005490.1.1 | −1.83 * | 0.05 | −2.26 * | −0.50 | Fasciclin-like arabinogalactan protein |
| Solyc06g064550.2.1 | −1.75 * | 0.04 | −1.37 * | 0.25 | Aspartokinase |
| Solyc07g040720.2.1 | −1.55 * | 0.01 | −2.08 * | −0.12 | Serine/threonine kinase |
| Solyc10g051090.1.1 | −2.37 * | 0.39 | −2.82 * | −0.04 | Serine/threonine–protein phosphatase |
| Variety- and cell-type-specific | |||||
| Solyc06g074720.2.1 | 2.15 * | −0.44 | 0.90 | −0.24 | FHA domain-interacting nucleolar phosphoprotein |
| Solyc09g005910.2.1 | 1.87 * | −0.12 | 0.66 | 0.47 | Mg2+-dependent phosphatidate |
| Solyc10g083650.1.1 | 0.89 * | 0.13 | 0.05 | −0.79 * | Peroxiredoxin ahpC/TSA family |
| Solyc04g082010.1.1 | 2.11 * | 0.19 | 0.62 | −0.23 | Plastocyanin |
| Solyc11g066390.1.1 | 1.63 * | 0.22 | 0.58 | −0.03 | Superoxide dismutase |
| Solyc08g005060.2.1 | −0.12 | 0.89 * | −0.32 | −0.57 | SnRK1-interacting protein 1 |
| Solyc06g034120.2.1 | −0.96 | 0.92 * | −0.01 | −0.36 | Alcohol dehydrogenase |
| Solyc11g069180.1.1 | 0.55 | 0.86 * | 0.80 | −0.11 | Isovaleryl-CoA dehydrogenase |
| Solyc01g111660.2.1 | −0.30 | 0.94 * | 0.02 | −0.38 | Aquaporin-like protein |
| Solyc08g075870.2.1 | −0.04 | 0.84 * | −0.03 | 0.67 | Dehydration-responsive family protein-like |
| Solyc01g081270.2.1 | 0.62 | 0.87 * | −0.17 | 0.35 | Glutathione S-transferase |
| Solyc00g009040.2.1 | 1.50 | −0.33 | 2.40 * | 0.41 | Polyadenylate-binding protein 2 |
| Solyc01g110210.1.1 | −0.11 | −0.22 | 1.11 * | 0.98 | Terminal ear1-like 2 protein |
| Solyc06g073190.2.1 | 0.91 | −0.10 | 2.82 * | 0.12 | Fructokinase-like |
| Solyc11g073200.1.1 | 1.59 | 0.11 | 3.55 * | 0.02 | Legumin 11S-globulin |
| Solyc10g085650.1.1 | 0.77 | 0.25 | 1.89 * | 0.16 | Beta-fructofuranosidase insoluble isoenzyme 2 |
| Solyc02g062460.2.1 | 0.58 | 0.06 | 1.98 * | 0.12 | 2-oxoglutarate-dependent dioxygenase |
| Solyc09g065840.2.1 | −0.53 | −0.24 | −0.11 | 0.73 * | Single-stranded DNA-binding protein |
| Solyc10g009390.2.1 | 0.77 | 0.14 | 0.13 | 0.89 * | Cytochrome P450 |
| Solyc01g088430.2.1 | 0.26 | 0.17 | 0.11 | 0.71 * | Fatty acid hydroxylase |
| Solyc04g005030.2.1 | −0.68 | 0.72 | −0.03 | 0.79 * | Phosphoglucomutase |
| Solyc10g078490.1.1 | 0.20 | 0.40 | −0.15 | 0.86 * | Aquaporin |
| Solyc09g075120.2.1 | 0.85 | −0.13 | 0.47 | 0.70 * | Bifunctional polymyxin resistance protein ArnA |
| Solyc05g015390.2.1 | −0.26 | 0.58 | −0.65 | 0.81 * | REF-like stress-related protein 1/Rubber elongation factor |
| Solyc12g096540.1.1 | −1.66 * | −0.52 | 0.08 | −0.58 | 40S ribosomal protein S10-like |
| Solyc07g055230.2.1 | −2.01 * | −0.15 | −0.46 | −0.31 | 50S ribosomal protein L5 |
| Solyc05g053670.2.1 | −1.95 * | −0.08 | −0.47 | −0.43 | 60S ribosomal protein L13a-like protein |
| Solyc12g098330.1.1 | −2.83 * | −0.08 | −0.24 | −0.25 | 60S ribosomal protein L13a-like protein |
| Solyc06g073300.1.1 | −1.70 * | −0.57 | −0.62 | 0.23 | 60S ribosomal protein L27 |
| Solyc03g112360.1.1 | −1.75 * | −0.56 | −0.64 | −0.66 | 60S ribosomal protein L27A |
| Solyc07g008370.2.1 | −1.68 * | 0.11 | −0.62 | 0.01 | 60S ribosomal protein L7 |
| Solyc06g072570.2.1 | −2.16 * | 0.01 | −0.54 | −0.14 | Ubiquitin-conjugating enzyme E2 1 |
| Solyc12g062250.1.1 | −1.61 * | −0.16 | −0.34 | 0.20 | 5′-AMP-activated protein kinase |
| Solyc04g008810.2.1 | −0.49 | −0.91 * | 0.00 | −0.20 | 40S ribosomal protein S26 |
| Solyc04g009770.2.1 | 0.43 | −0.81 * | 0.67 | −0.14 | DNAJ chaperone |
| Solyc09g098280.2.1 | −0.57 | −1.31 * | −0.08 | 0.03 | Eukaryotic translation initiation factor 3 subunit 3 |
| Solyc02g064670.1.1 | −0.37 | −1.08 * | −0.03 | 0.11 | Ribosomal protein L26-like 1 |
| Solyc05g053780.2.1 | −0.18 | −1.03 * | −0.56 | −0.43 | RNA-binding protein-like protein |
| Solyc06g008130.2.1 | 0.35 | −0.99 * | 0.07 | −0.28 | SET domain protein SUVR2 |
| Solyc10g080180.1.1 | 0.08 | −0.83 * | 0.37 | 0.13 | U3 small nucleolar RNA-associated protein 18 |
| Solyc04g010030.1.1 | 0.23 | −0.83 * | 0.71 | 0.08 | Thioredoxin-like protein 1 |
| Solyc12g056220.1.1 | 0.87 | 0.93 * | −0.59 | −0.38 | Aquaporin |
| Solyc09g083380.2.1 | −0.90 | −1.04 | −0.93 * | −0.50 | Histone H1 |
| Solyc11g007930.1.1 | −0.68 | −0.76 | −1.80 * | −0.13 | Histone H2B |
| Solyc10g081030.1.1 | −0.38 | −0.46 | −0.86 * | −0.68 | Nascent polypeptide-associated complex alpha subunit |
| Solyc08g016420.2.1 | −0.37 | −0.70 | −2.14 * | −0.46 | Prefoldin subunit 6 |
| Solyc03g121310.2.1 | 0.07 | −0.81 | −1.03 * | 0.01 | RWD domain-containing protein |
| Solyc03g119380.1.1 | −0.08 | −0.28 | −0.06 | −0.79 * | 50S ribosomal protein L14 |
| Solyc09g091740.2.1 | −0.01 | 0.10 | 0.29 | −0.97 * | 60S ribosomal protein L13a-like protein |
| Solyc01g098000.2.1 | −0.25 | 0.06 | −0.65 | −0.72 * | Elongation factor-like protein |
| Solyc01g109660.2.1 | 0.05 | −0.63 | −0.56 | −0.68 * | Glycine-rich RNA-binding protein |
| Solyc05g051290.2.1 | 0.23 | −0.23 | −0.41 | −0.74 * | High-mobility group family |
| Solyc02g083470.2.1 | −0.02 | −1.08 | −0.11 | −1.08 * | Pre-rRNA-processing protein ESF1 |
| Solyc11g005170.1.1 | −0.25 | −0.49 | 0.25 | −1.00 * | RNA-splicing factor |
| Solyc01g095050.2.1 | −0.39 | −0.63 | −2.00 * | −0.44 | Negatively light-regulated protein |
| Solyc12g014420.1.1 | 0.11 | 0.20 | 0.76 | 0.77 * | Glucan endo-1 3-beta-glucosidase A6 |
| Proteins with consistent HS responses | |||||
| Solyc06g069220.1.1 | 4.60 * | 0.82 * | 5.98 * | 1.10 * | Aspartic proteinase nepenthesin I |
| Solyc03g098710.1.1 | 2.36 * | 1.10 * | 1.25 * | 1.01 * | Kunitz-type proteinase inhibitor A4 |
| Solyc00g187050.2.1 | 1.75 * | 1.15 * | 1.99 * | 1.07 * | Leucyl aminopeptidase |
| Solyc12g010020.1.1 | 2.11 * | 1.30 * | 2.18 * | 1.09 * | Leucyl aminopeptidase |
| Solyc08g079910.1.1 | 1.27 * | 0.71 * | 2.89 * | 1.07 * | Subtilisin-like protease |
| Solyc07g064600.2.1 | 2.95 * | 0.81 * | 2.23 * | 1.10 * | Endoribonuclease L-PSP family protein |
| Solyc08g076970.2.1 | 1.82 * | 0.91 * | 2.34 * | 1.52 * | Acetylornithine deacetylase |
| Solyc01g066460.2.1 | 0.90 * | 3.68 * | 1.77 * | 1.39 * | Epoxide hydrolase |
| Solyc09g008670.2.1 | 1.69 * | 0.90 * | 2.91 * | 0.73 * | Threonine ammonia-lyase biosynthetic |
| Solyc01g010390.2.1 | 3.03 * | 0.82 * | 3.56 * | 1.31 * | Beta-glucosidase |
| Solyc12g094460.1.1 | 1.30 * | 0.73 * | 2.43 * | 1.40 * | Laccase-2 |
| Solyc04g079890.2.1 | 3.69 * | 1.17 * | 4.23 * | 1.41 * | Thaumatin-like protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thapa, P.; Guo, J.; Pradhan, K.; Thapa, D.; Madhavarapu, S.; Zou, J.; Potts, J.; Li, H.; O’Hair, J.; Wang, C.; et al. Cell-Type-Specific Heat-Induced Changes in the Proteomes of Pollen Mother Cells and Microspores Provide New Insights into Tomato Pollen Production Under Elevated Temperature. Proteomes 2025, 13, 13. https://doi.org/10.3390/proteomes13020013
Thapa P, Guo J, Pradhan K, Thapa D, Madhavarapu S, Zou J, Potts J, Li H, O’Hair J, Wang C, et al. Cell-Type-Specific Heat-Induced Changes in the Proteomes of Pollen Mother Cells and Microspores Provide New Insights into Tomato Pollen Production Under Elevated Temperature. Proteomes. 2025; 13(2):13. https://doi.org/10.3390/proteomes13020013
Chicago/Turabian StyleThapa, Priya, Jun Guo, Kajol Pradhan, Dibya Thapa, Sudhakar Madhavarapu, Jing Zou, Jesse Potts, Hui Li, Joshua O’Hair, Chen Wang, and et al. 2025. "Cell-Type-Specific Heat-Induced Changes in the Proteomes of Pollen Mother Cells and Microspores Provide New Insights into Tomato Pollen Production Under Elevated Temperature" Proteomes 13, no. 2: 13. https://doi.org/10.3390/proteomes13020013
APA StyleThapa, P., Guo, J., Pradhan, K., Thapa, D., Madhavarapu, S., Zou, J., Potts, J., Li, H., O’Hair, J., Wang, C., Zhou, S., Yang, Y., Fish, T., & Thannhauser, T. W. (2025). Cell-Type-Specific Heat-Induced Changes in the Proteomes of Pollen Mother Cells and Microspores Provide New Insights into Tomato Pollen Production Under Elevated Temperature. Proteomes, 13(2), 13. https://doi.org/10.3390/proteomes13020013