Comprehensive Proteome Profiling of a Xanthomonas campestris pv. Campestris B100 Culture Grown in Minimal Medium with a Specific Focus on Nutrient Consumption and Xanthan Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of Xcc
2.2. Xcc Harvesting
2.3. Xanthan Gum, Glucose, and Nitrate Determination
2.4. Whole-Cell Protein Isolation and Digestion
2.5. Sample Preparation for LC-MS/MS Analysis
2.6. LC-MS/MS Analysis
2.7. Data Analysis
3. Results
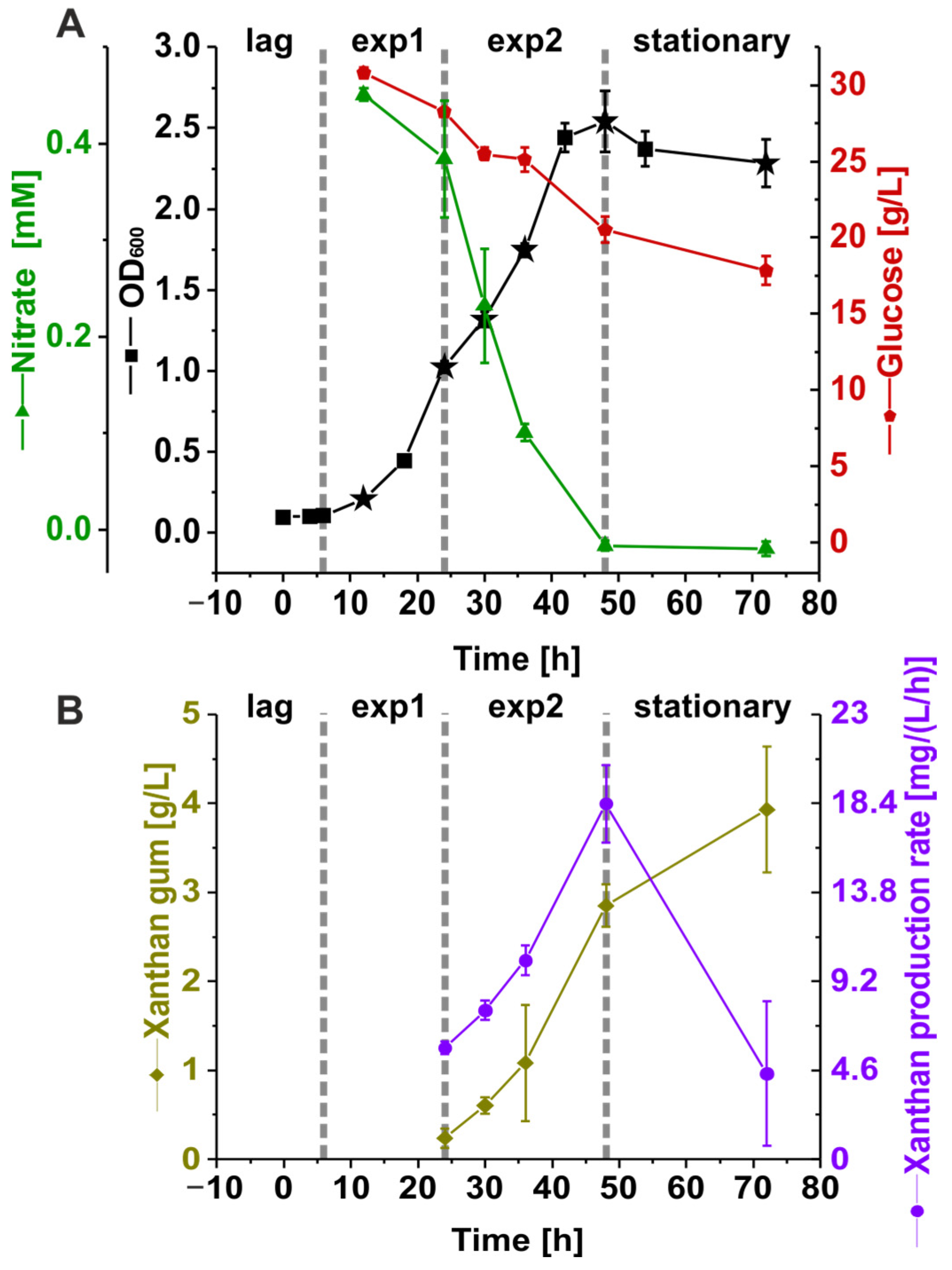
3.1. Nutrient Consumption and Xanthan Production of Suspension Cultures of Xcc B100 over a Time Course
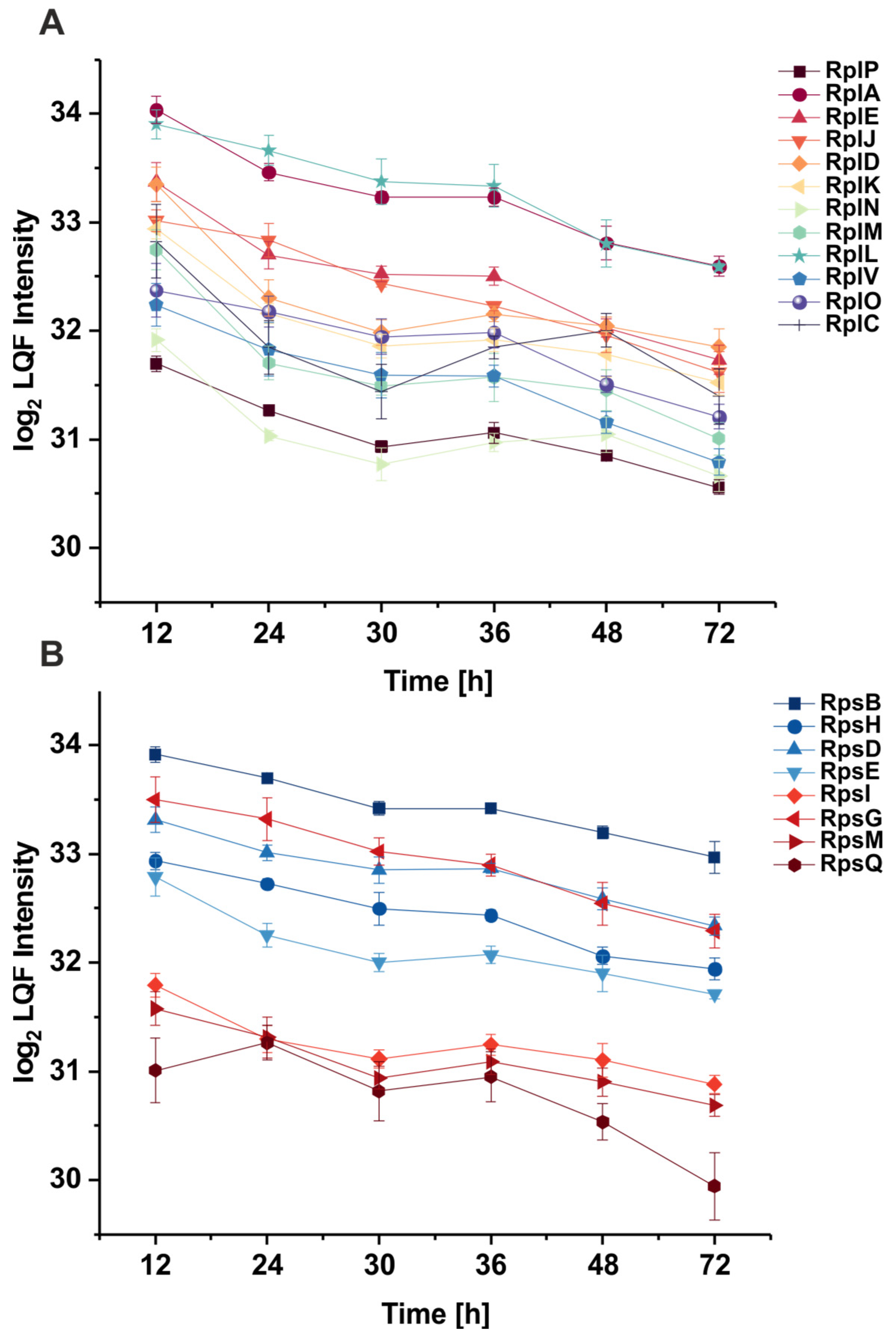
3.2. The Abundance of Ribosomal Proteins Varies Per Growth Phase
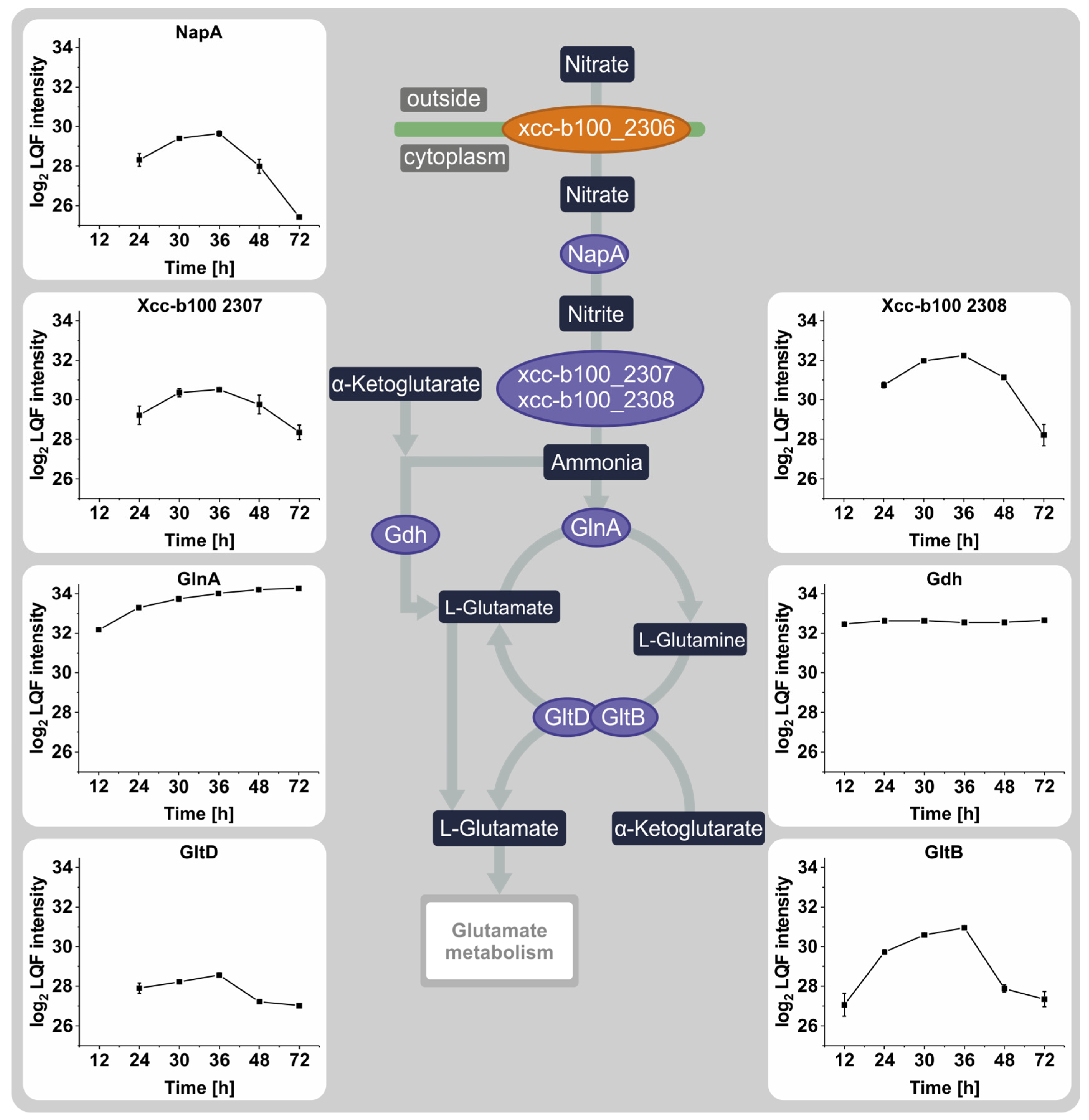
3.3. The Effect of Nitrate Depletion on the Abundance of Proteins Involved in Nitrogen Metabolism
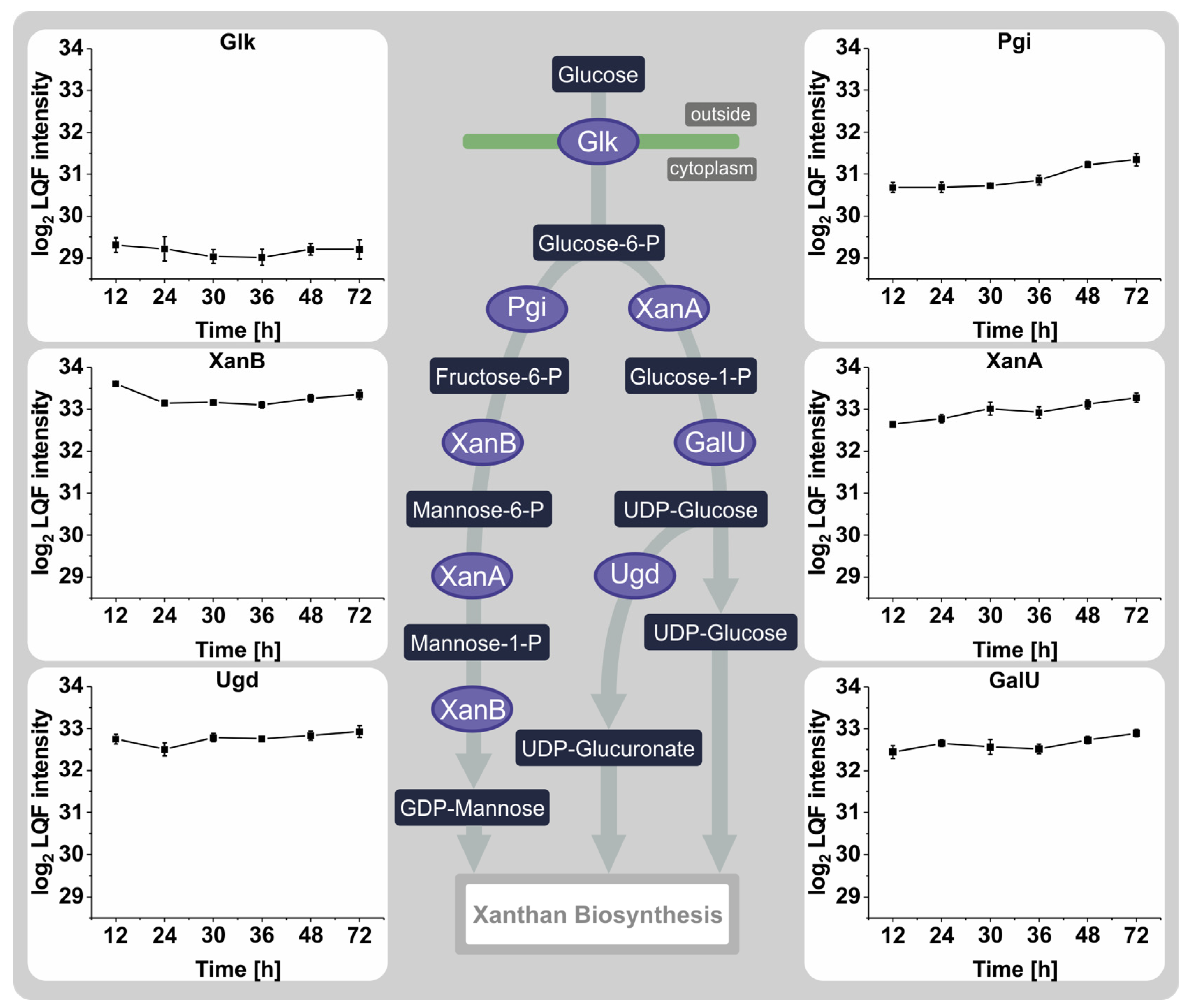
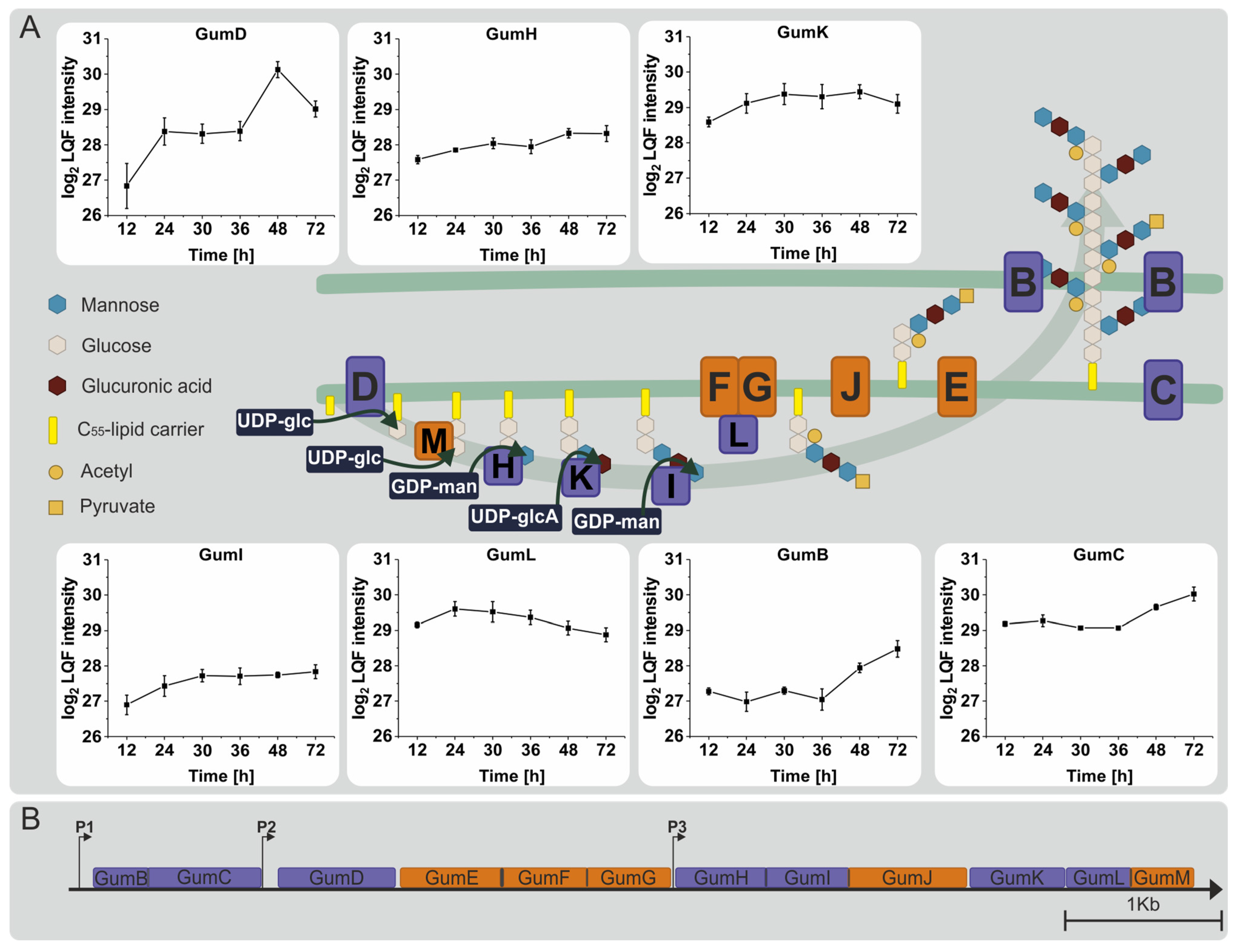
3.4. Proteins Involved in the Synthesis of Sugar Nucleotide Precursors and Xanthan
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ye, F.; Peng, G.; Li, Y. Influences of influent carbon source on extracellular polymeric substances (EPS) and physicochemical properties of activated sludge. Chemosphere 2011, 84, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
- Roberson, E.B.; Firestone, M.K. Relationship between Desiccation and Exopolysaccharide Production in a Soil Pseudomonas sp. Appl. Environ. Microbiol. 1992, 58, 1284–1291. [Google Scholar] [CrossRef]
- Geddie, J.L.; Sutherland, I.W. Uptake of metals by bacterial polysaccharides. J. Appl. Bacteriol. 1993, 74, 467–472. [Google Scholar] [CrossRef]
- Krembs, C.; Eicken, H.; Junge, K.; Deming, J.W. High concentrations of exopolymeric substances in Arctic winter sea ice: Implications for the polar ocean carbon cycle and cryoprotection of diatoms. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2002, 49, 2163–2181. [Google Scholar] [CrossRef]
- Kambourova, M.; Mandeva, R.; Dimova, D.; Poli, A.; Nicolaus, B.; Tommonaro, G. Production and characterization of a microbial glucan, synthesized by Geobacillus tepidamans V264 isolated from Bulgarian hot spring. Carbohydr. Polym. 2009, 77, 338–343. [Google Scholar] [CrossRef]
- Wingender, J.; Neu, T.R.; Flemming, H.-C. (Eds.) Microbial Extracellular Polymeric Substances; Springer: Berlin/Heidelberg, Germany, 1999; ISBN 978-3-642-64277-7. [Google Scholar]
- Future, M.R. Xanthan Gum Market Will Be Worth USD 730.39 Million by 2030. Available online: https://www.marketresearchfuture.com/reports/xanthan-gum-market (accessed on 5 December 2022).
- Hublik, G. Xanthan. Polymer Science: A Comprehensive Reference; Elsevier: Amsterdam, The Netherlands, 2012; pp. 221–229. ISBN 9780080878621. [Google Scholar]
- Dow, J.M.; Crossman, L.; Findlay, K.; He, Y.-Q.; Feng, J.-X.; Tang, J.-L. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 2003, 100, 10995–11000. [Google Scholar] [CrossRef] [PubMed]
- Crossman, L.; Dow, J.M. Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 2004, 6, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Bhat, I.M.; Wani, S.M.; Mir, S.A.; Masoodi, F.A. Advances in xanthan gum production, modifications and its applications. Biocatal. Agric. Biotechnol. 2022, 42, 102328. [Google Scholar] [CrossRef]
- Jansson, P.-e.; Kenne, L.; Lindberg, B. Structure of the extracellular polysaccharide from xanthomonas campestris. Carbohydr. Res. 1975, 45, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Pieretti, I.; Royer, M.; Barbe, V.; Carrere, S.; Koebnik, R.; Cociancich, S.; Couloux, A.; Darrasse, A.; Gouzy, J.; Jacques, M.-A.; et al. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genom. 2009, 10, 616. [Google Scholar] [CrossRef] [PubMed]
- Vorhölter, F.-J.; Schneiker, S.; Goesmann, A.; Krause, L.; Bekel, T.; Kaiser, O.; Linke, B.; Patschkowski, T.; Rückert, C.; Schmid, J.; et al. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 2008, 134, 33–45. [Google Scholar] [CrossRef]
- Jarman, T.R.; Pace, G.W. Energy requirements for microbial exopolysaccharide synthesis. Arch. Microbiol. 1984, 137, 231–235. [Google Scholar] [CrossRef]
- Shu, C.H.; Yang, S.T. Effects of temperature on cell growth and xanthan production in batch cultures of Xanthomonas campestris. Biotechnol. Bioeng. 1990, 35, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Krishna Leela, J.; Sharma, G. Studies on xanthan production from Xanthomonas campestris. Bioprocess Eng. 2000, 23, 687–689. [Google Scholar] [CrossRef]
- Freitas, F.; Alves, V.D.; Reis, M.A.M. Advances in bacterial exopolysaccharides: From production to biotechnological applications. Trends Biotechnol. 2011, 29, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Roseiro, J.C.; Esgalhado, M.E.; Amaral Collaço, M.T.; Emery, A.N. Medium development for xanthan production. Process Biochem. 1992, 27, 167–175. [Google Scholar] [CrossRef]
- Pielken, P.; Schimz, K.-L.; Eggeling, L.; Sahm, H. Effect of methionine on xanthan formation by Xanthomonas campestris. FEMS Microbiol. Lett. 1987, 44, 27–31. [Google Scholar] [CrossRef]
- Souw, P.; Demain, A.L. Nutritional Studies on Xanthan Production by Xanthomonas campestris NRRL B1459. Appl. Environ. Microbiol. 1979, 37, 1186–1192. [Google Scholar] [CrossRef] [PubMed]
- Letisse, F.; Chevallereau, P.; Simon, J.L.; Lindley, N.D. Kinetic analysis of growth and xanthan gum production with Xanthomonas campestris on sucrose, using sequentially consumed nitrogen sources. Appl. Microbiol. Biotechnol. 2001, 55, 417–422. [Google Scholar] [CrossRef]
- Seviour, R.J.; McNeil, B.; Fazenda, M.L.; Harvey, L.M. Operating bioreactors for microbial exopolysaccharide production. Crit. Rev. Biotechnol. 2011, 31, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Freitas, F.; Torres, C.A.V.; Reis, M.A.M. Engineering aspects of microbial exopolysaccharide production. Bioresour. Technol. 2017, 245, 1674–1683. [Google Scholar] [CrossRef] [PubMed]
- Rosalam, S.; England, R. Review of xanthan gum production from unmodified starches by Xanthomonas comprestris sp. Enzym. Microb. Technol. 2006, 39, 197–207. [Google Scholar] [CrossRef]
- Alkhateeb, R.S.; Rückert, C.; Rupp, O.; Pucker, B.; Hublik, G.; Wibberg, D.; Niehaus, K.; Pühler, A.; Vorhölter, F.-J. Refined annotation of the complete genome of the phytopathogenic and xanthan producing Xanthomonas campestris pv. campestris strain B100 based on RNA sequence data. J. Biotechnol. 2017, 253, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Alkhateeb, R.S.; Vorhölter, F.-J.; Steffens, T.; Rückert, C.; Ortseifen, V.; Hublik, G.; Niehaus, K.; Pühler, A. Comparative transcription profiling of two fermentation cultures of Xanthomonas campestris pv. campestris B100 sampled in the growth and in the stationary phase. Appl. Microbiol. Biotechnol. 2018, 102, 6613–6625. [Google Scholar] [CrossRef]
- Chung, W.-J.; Shu, H.-Y.; Lu, C.-Y.; Wu, C.-Y.; Tseng, Y.-H.; Tsai, S.-F.; Lin, C.-H. Qualitative and comparative proteomic analysis of Xanthomonas campestris pv. campestris 17. Proteomics 2007, 7, 2047–2058. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, V.K.; Vorhölter, F.-J.; Niehaus, K.; Watt, S.A. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol. 2008, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Tortosa, M.; Velasco, P.; Rodríguez, V.M.; Cartea, M.E. Changes in Brassica oleracea Leaves Infected with Xanthomonas campestris pv. campestris by Proteomics Analysis. Front. Plant Sci. 2021, 12, 781984. [Google Scholar] [CrossRef] [PubMed]
- Villeth, G.R.; Reis, F.B.; Tonietto, A.; Huergo, L.; de Souza, E.M.; Pedrosa, F.O.; Franco, O.L.; Mehta, A. Comparative proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the susceptible and the resistant cultivars of Brassica oleracea. FEMS Microbiol. Lett. 2009, 298, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.E.; Silva, L.P.; Pereira, J.L.; Noronha, E.F.; Reis, F.B.; Bloch, C.; dos Santos, M.F.; Domont, G.B.; Franco, O.L.; Mehta, A. In vivo proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the host plant Brassica oleracea. FEMS Microbiol. Lett. 2008, 281, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.; Koch, D.; Niehaus, K.; Ortseifen, V. Analysis of Gum proteins involved in xanthan biosynthesis throughout multiple cell fractions in a “single-tube”. J. Proteom. 2022, 257, 104513. [Google Scholar] [CrossRef] [PubMed]
- Schatschneider, S.; Persicke, M.; Watt, S.A.; Hublik, G.; Pühler, A.; Niehaus, K.; Vorhölter, F.-J. Establishment, in silico analysis, and experimental verification of a large-scale metabolic network of the xanthan producing Xanthomonas campestris pv. campestris strain B100. J. Biotechnol. 2013, 167, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qian, W.-J.; Mottaz, H.M.; Clauss, T.R.W.; Anderson, D.J.; Moore, R.J.; Camp, D.G.; Khan, A.H.; Sforza, D.M.; Pallavicini, M.; et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J. Proteome Res. 2005, 4, 2397–2403. [Google Scholar] [CrossRef] [PubMed]
- Dilillo, M.; de Graaf, E.L.; Yadav, A.; Belov, M.E.; McDonnell, L.A. Ultraviolet Photodissociation of ESI- and MALDI-Generated Protein Ions on a Q-Exactive Mass Spectrometer. J. Proteome Res. 2019, 18, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Brink, B.G.; Seidel, A.; Kleinbölting, N.; Nattkemper, T.W.; Albaum, S.P. Omics Fusion—A Platform for Integrative Analysis of Omics Data. J. Integr. Bioinform. 2016, 13, 296. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Scott, M.; Klumpp, S.; Mateescu, E.M.; Hwa, T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 2014, 10, 747. [Google Scholar] [CrossRef]
- Scott, M.; Gunderson, C.W.; Mateescu, E.M.; Zhang, Z.; Hwa, T. Interdependence of cell growth and gene expression: Origins and consequences. Science 2010, 330, 1099–1102. [Google Scholar] [CrossRef] [PubMed]
- Spahn, C.M.; Beckmann, R.; Eswar, N.; Penczek, P.A.; Sali, A.; Blobel, G.; Frank, J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 2001, 107, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, S.; Ben-Shem, A.; Garreau de Loubresse, N.; Jenner, L.; Yusupova, G.; Yusupov, M. One core, two shells: Bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012, 19, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. Is proteomics the new genomics? Cell 2007, 130, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Claassen, M.; Aebersold, R. Comprehensive proteomics. Curr. Opin. Biotechnol. 2011, 22, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jang, W.E.; Park, J.; Kim, M.-S.; Kim, J.-G.; Kang, L.-W. Combined Analysis of the Time-Resolved Transcriptome and Proteome of Plant Pathogen Xanthomonas oryzae pv. oryzae. Front. Microbiol. 2021, 12, 664857. [Google Scholar] [CrossRef] [PubMed]
- Soares, M.R.; Facincani, A.P.; Ferreira, R.M.; Moreira, L.M.; de Oliveira, J.C.; Ferro, J.A.; Ferro, M.I.; Meneghini, R.; Gozzo, F.C. Proteome of the phytopathogen Xanthomonas citri subsp. citri: A global expression profile. Proteome Sci. 2010, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Malmström, J.; Beck, M.; Schmidt, A.; Lange, V.; Deutsch, E.W.; Aebersold, R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 2009, 460, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Malmström, J.A.; Lange, V.; Schmidt, A.; Deutsch, E.W.; Aebersold, R. Visual proteomics of the human pathogen Leptospira interrogans. Nat. Methods 2009, 6, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Bosdriesz, E.; Molenaar, D.; Teusink, B.; Bruggeman, F.J. How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J. 2015, 282, 2029–2044. [Google Scholar] [CrossRef]
- Belliveau, N.M.; Chure, G.; Hueschen, C.L.; Garcia, H.G.; Kondev, J.; Fisher, D.S.; Theriot, J.A.; Phillips, R. Fundamental limits on the rate of bacterial growth and their influence on proteomic composition. Cell Syst. 2021, 12, 924–944.e2. [Google Scholar] [CrossRef] [PubMed]
- Sujithra, B.; Deepika, S.; Akshaya, K.; Ponnusami, V. Production and optimization of xanthan gum from three-step sequential enzyme treated cassava bagasse hydrolysate. Biocatal. Agric. Biotechnol. 2019, 21, 101294. [Google Scholar] [CrossRef]
- Yan, D. Protection of the glutamate pool concentration in enteric bacteria. Proc. Natl. Acad. Sci. USA 2007, 104, 9475–9480. [Google Scholar] [CrossRef] [PubMed]
- Merrick, M.J.; Edwards, R.A. Nitrogen control in bacteria. Microbiol. Rev. 1995, 59, 604–622. [Google Scholar] [CrossRef]
- Pietack, N.; Becher, D.; Schmidl, S.R.; Saier, M.H.; Hecker, M.; Commichau, F.M.; Stülke, J. In vitro phosphorylation of key metabolic enzymes from Bacillus subtilis: PrkC phosphorylates enzymes from different branches of basic metabolism. J. Mol. Microbiol. Biotechnol. 2010, 18, 129–140. [Google Scholar] [CrossRef]
- Foor, F.; Janssen, K.A.; Magasanik, B. Regulation of synthesis of glutamine synthetase by adenylylated glutamine synthetase. Proc. Natl. Acad. Sci. USA 1975, 72, 4844–4848. [Google Scholar] [CrossRef] [PubMed]
- Musa, Y.R.; Bäsell, K.; Schatschneider, S.; Vorhölter, F.-J.; Becher, D.; Niehaus, K. Dynamic protein phosphorylation during the growth of Xanthomonas campestris pv. campestris B100 revealed by a gel-based proteomics approach. J. Biotechnol. 2013, 167, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Chou, F.L.; Chou, H.C.; Lin, Y.S.; Yang, B.Y.; Lin, N.T.; Weng, S.F.; Tseng, Y.H. The Xanthomonas campestris gumD gene required for synthesis of xanthan gum is involved in normal pigmentation and virulence in causing black rot. Biochem. Biophys. Res. Commun. 1997, 233, 265–269. [Google Scholar] [CrossRef]
- Galván, E.M.; Ielmini, M.V.; Patel, Y.N.; Bianco, M.I.; Franceschini, E.A.; Schneider, J.C.; Ielpi, L. Xanthan chain length is modulated by increasing the availability of the polysaccharide copolymerase protein GumC and the outer membrane polysaccharide export protein GumB. Glycobiology 2013, 23, 259–272. [Google Scholar] [CrossRef]
- Alkhateeb, R.S.; Vorhölter, F.-J.; Rückert, C.; Mentz, A.; Wibberg, D.; Hublik, G.; Niehaus, K.; Pühler, A. Genome wide transcription start sites analysis of Xanthomonas campestris pv. campestris B100 with insights into the gum gene cluster directing the biosynthesis of the exopolysaccharide xanthan. J. Biotechnol. 2016, 225, 18–28. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Struck, B.; Wiersma, S.J.; Ortseifen, V.; Pühler, A.; Niehaus, K. Comprehensive Proteome Profiling of a Xanthomonas campestris pv. Campestris B100 Culture Grown in Minimal Medium with a Specific Focus on Nutrient Consumption and Xanthan Biosynthesis. Proteomes 2024, 12, 12. https://doi.org/10.3390/proteomes12020012
Struck B, Wiersma SJ, Ortseifen V, Pühler A, Niehaus K. Comprehensive Proteome Profiling of a Xanthomonas campestris pv. Campestris B100 Culture Grown in Minimal Medium with a Specific Focus on Nutrient Consumption and Xanthan Biosynthesis. Proteomes. 2024; 12(2):12. https://doi.org/10.3390/proteomes12020012
Chicago/Turabian StyleStruck, Ben, Sanne Jitske Wiersma, Vera Ortseifen, Alfred Pühler, and Karsten Niehaus. 2024. "Comprehensive Proteome Profiling of a Xanthomonas campestris pv. Campestris B100 Culture Grown in Minimal Medium with a Specific Focus on Nutrient Consumption and Xanthan Biosynthesis" Proteomes 12, no. 2: 12. https://doi.org/10.3390/proteomes12020012
APA StyleStruck, B., Wiersma, S. J., Ortseifen, V., Pühler, A., & Niehaus, K. (2024). Comprehensive Proteome Profiling of a Xanthomonas campestris pv. Campestris B100 Culture Grown in Minimal Medium with a Specific Focus on Nutrient Consumption and Xanthan Biosynthesis. Proteomes, 12(2), 12. https://doi.org/10.3390/proteomes12020012






