Cadmium Highlights Common and Specific Responses of Two Freshwater Sentinel Species, Dreissena polymorpha and Dreissena rostriformis bugensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Organism Collection
2.2. Acclimation
2.3. Exposure Design and Tissue Sampling
2.4. Two-Dimensional Gel Electrophoresis and Mass Spectrometry Identification
2.5. Mass Spectrometry and Protein Identification
2.6. Functional Protein Analysis
3. Results
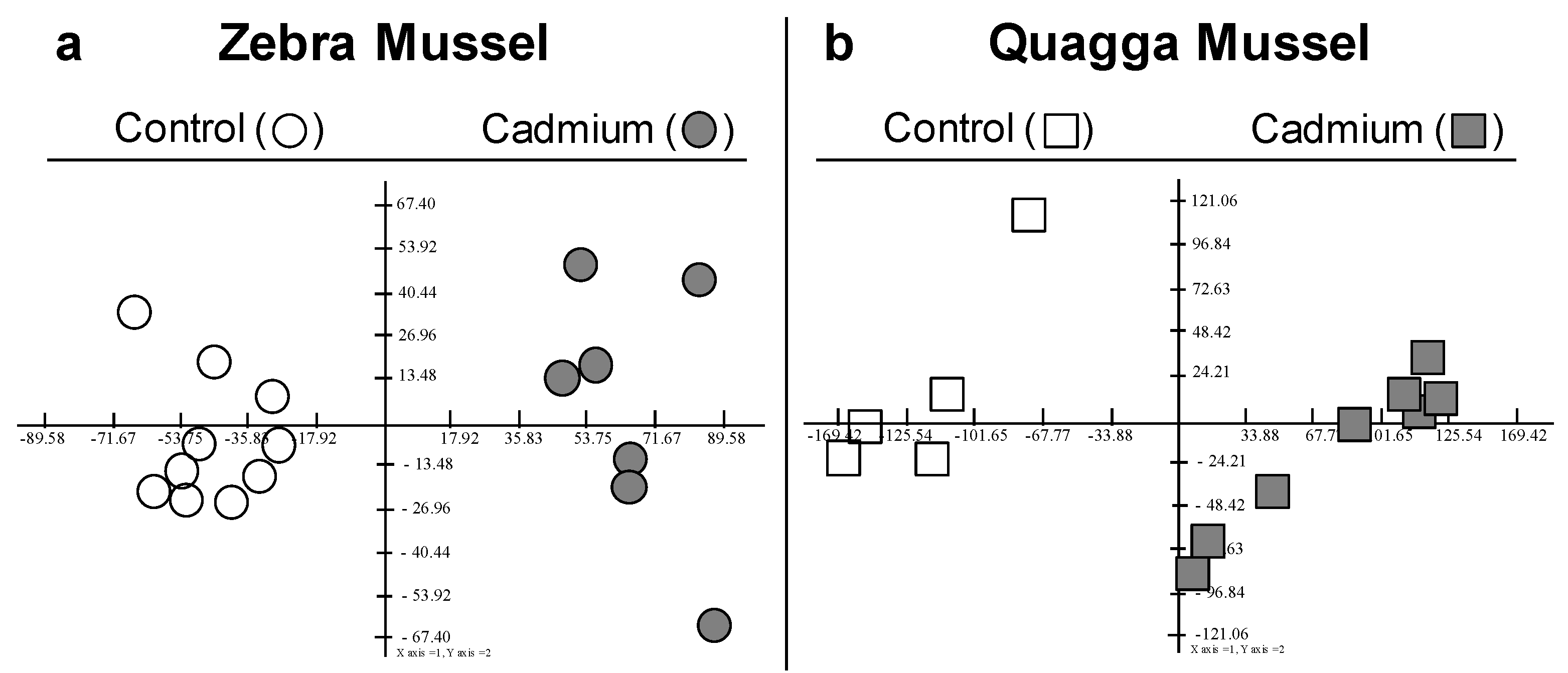
3.1. General Overview
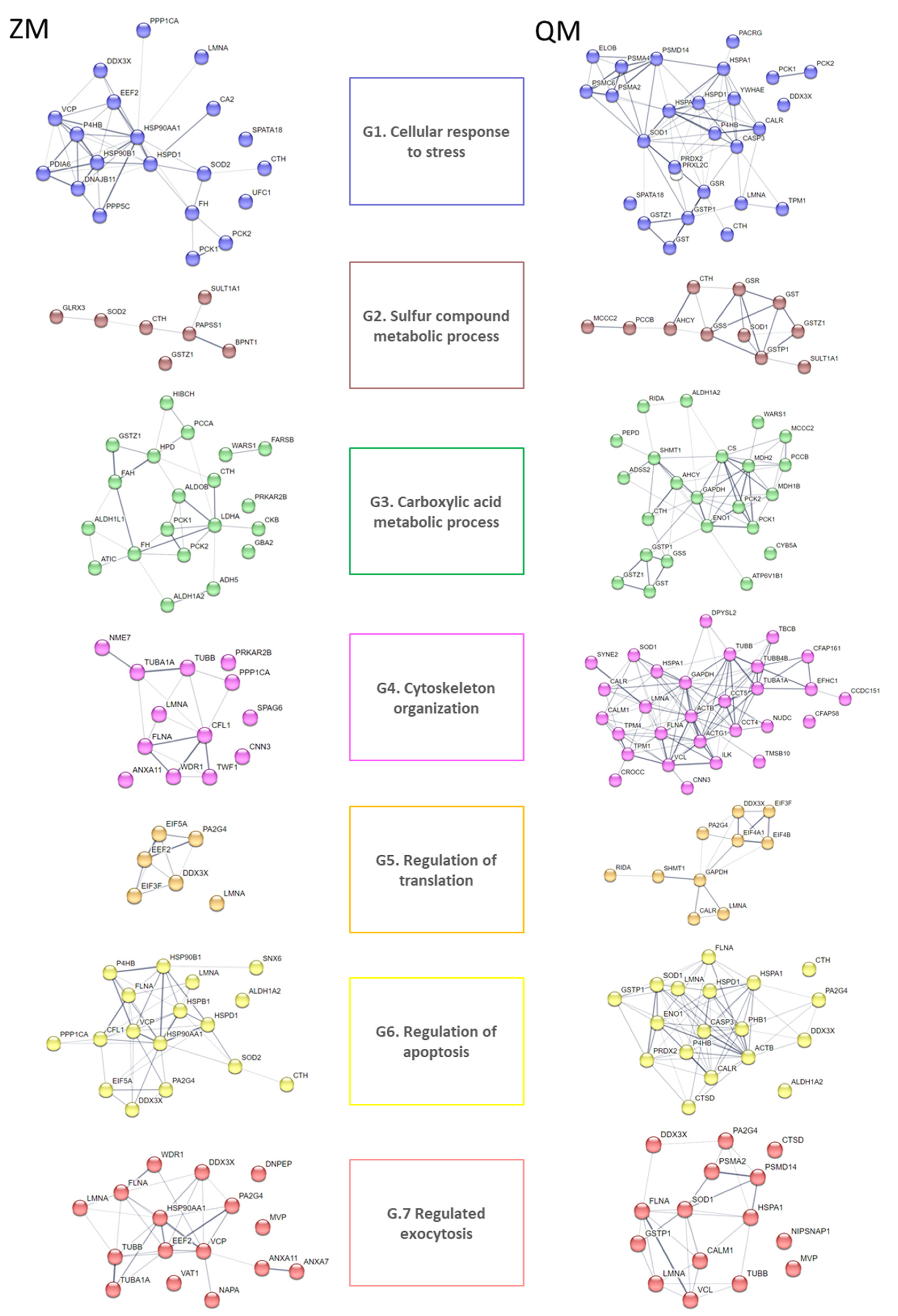
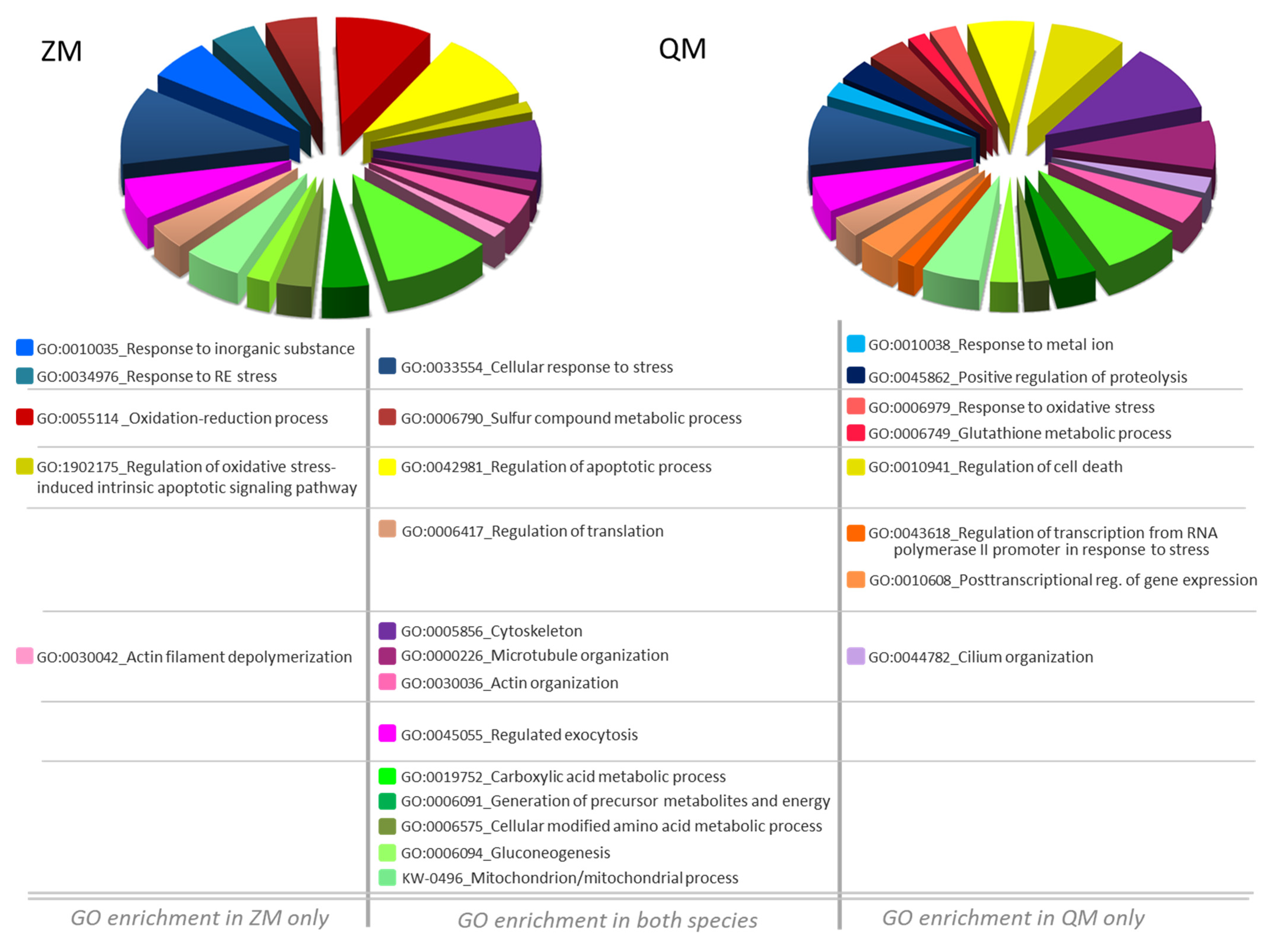
3.2. DAPs Classification and GO Term Enrichment Analysis
3.3. Shared Protein Identity between the Two Species
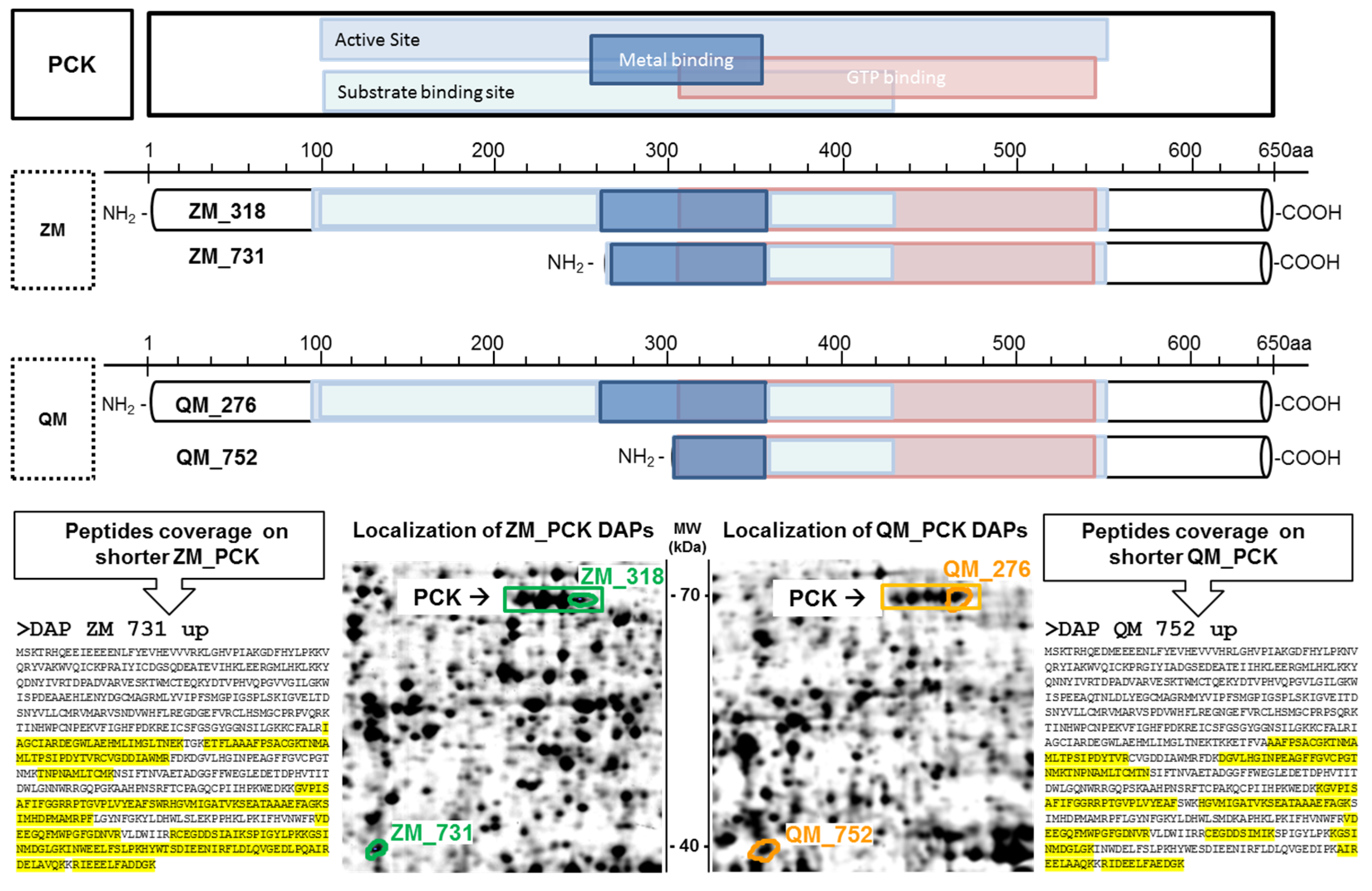
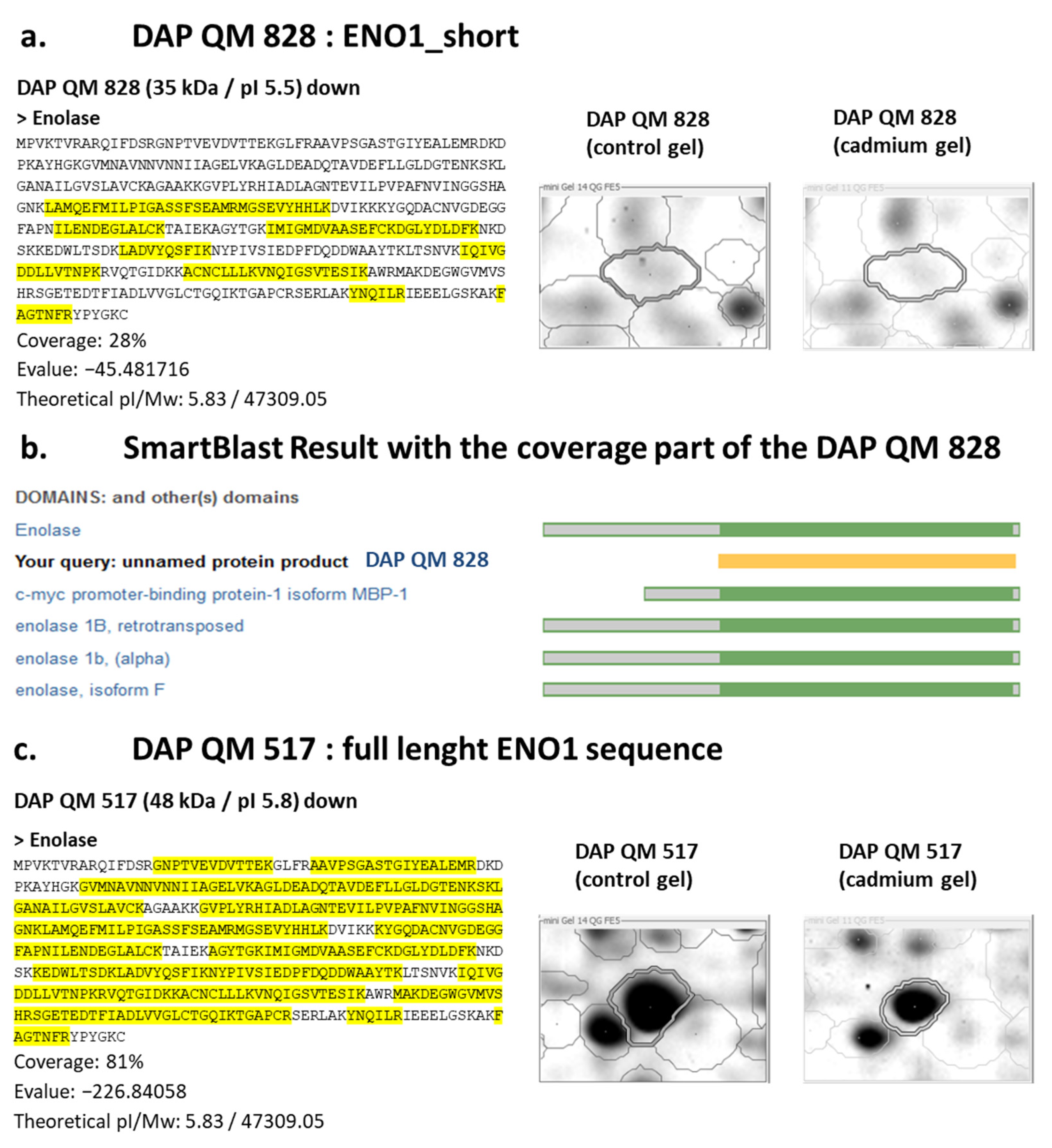
3.4. Evidence of Short Proteoforms
4. Discussion
4.1. DAP Classification and Protein Identification
4.2. Proteome Responses to Cadmium Exposure
4.3. Overlapping Modifications between ZM and QM
4.3.1. Chaperon, Folding, and Degradation
4.3.2. Antioxidant Response and Detoxification
4.3.3. Translation Regulation
4.3.4. Energy Metabolism and Mitochondrial Alterations
4.3.5. Regulation of Apoptotic Process
4.3.6. Cytoskeleton Modifications
4.3.7. Exocytosis
4.4. Focus on Some Species-Specific Modifications
4.4.1. DAPs That Display Common Identity but Opposite Direction of Variation
4.4.2. ZM-Specific Modifications
4.4.3. QM-Specific Modifications
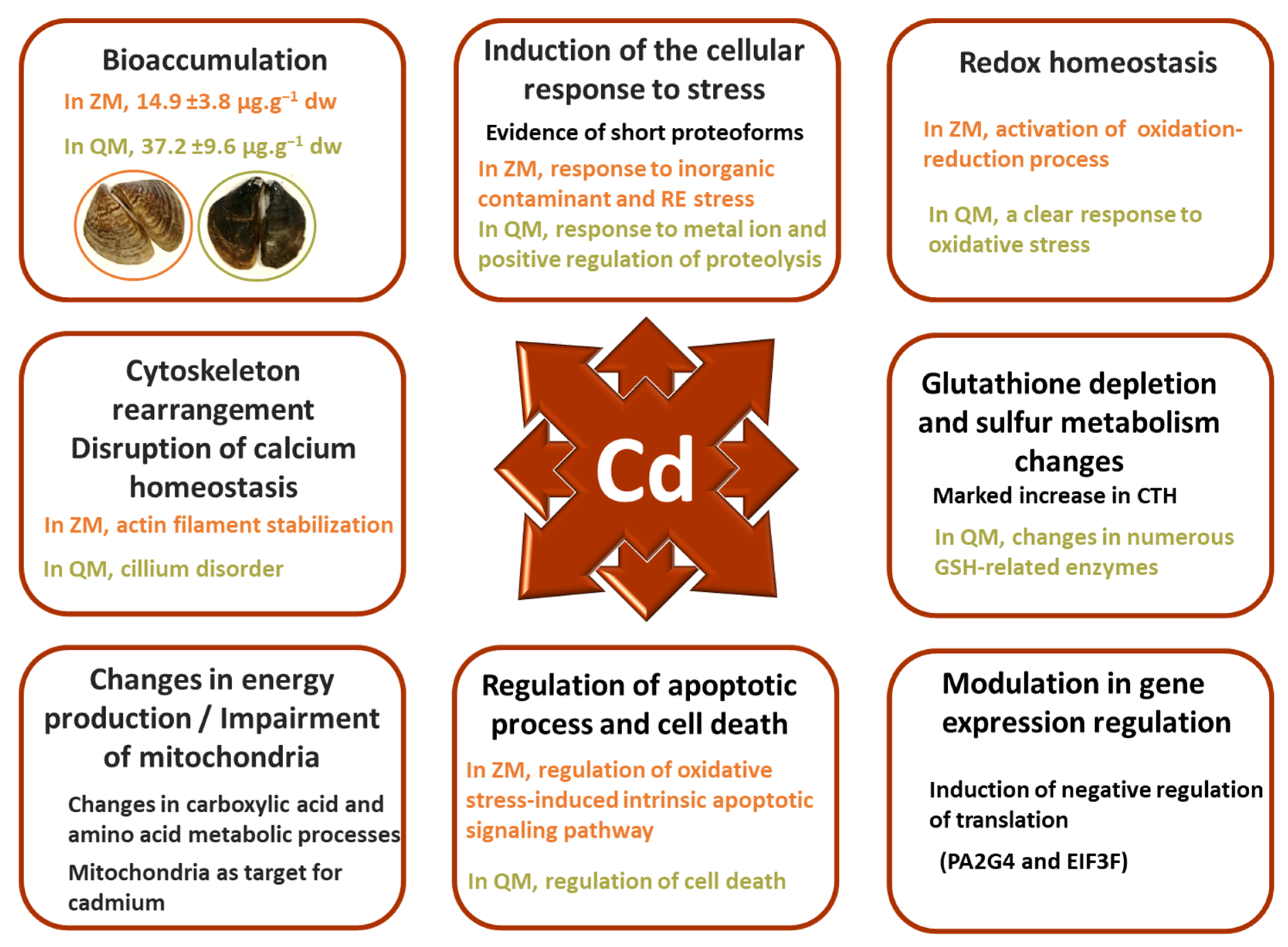
5. Synthesis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Binelli, A.; Della Torre, C.; Parolin, M. Does zebra mussel (Dreissena polymorpha) represent the freshwater counterpart of Mytilus in ecotoxicological studies? A critical review. Environ. Pollut. 2015, 196, 386–403. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.; Rosenberg, G.; Spidle, A.P.; Ludyanskiy, M.; Pligin, Y.; May, B. A review of the biology and ecology of the quagga mussel (Dreissena bugensis), a second species of freshwater Dreissenid introduced to North America. Amer. Zool. 1996, 36, 271–286. [Google Scholar] [CrossRef]
- Bij de Vaate, A.; Beisel, J.-N. Range expansion of the quagga mussel Dreissena rostriformis bugensis (Andrusov, 1897) in Western Europe: First observation from France. Aquat. Invasions 2011, 6, S71–S74. [Google Scholar] [CrossRef]
- Karatayev, A.Y.; Burlakova, L.E. What we know and don’t know about the invasive zebra (Dreissena polymorpha) and quagga (Dreissena rostriformis bugensis) mussels. Hydrobiologia 2022, 13, 1–74. [Google Scholar] [CrossRef]
- Schäfer, S.; Hamer, B.; Treursić, B.; Möhlenkamp, C.; Korlević, M.; Reifferscheid, G.; Claus, E. Comparison of bioaccumulation and biomarker responses in Dreissena polymorpha and D. bugensis after exposure to resuspended sediments. Arch. Environ. Contam. Toxicol. 2012, 62, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Kerambrun, E.; Delahaut, L.; Geffard, A.; David, E. Differentiation of sympatric zebra and quagga mussels in ecotoxicological studies: A comparison of morphometric data, gene expression, and body metal concentrations. Ecotoxicol. Environ. Saf. 2018, 154, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Evariste, L.; David, E.; Cloutier, P.-L.; Brousseau, P.; Auffret, M.; Desrosiers, M.; Groleau, P.E.; Fournier, M.; Betoulle, S. Field biomonitoring using the zebra mussel Dreissena polymorpha and the quagga mussel Dreissena bugensis following immunotoxic reponses. Is there a need to separate the two species? Environ. Pollut. 2018, 238, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Potet, M.; Devin, S.; Pain-Devin, S.; Rousselle, P.; Giambérini, L. Integrated multi-biomarker responses in two dreissenid species following metal and thermal cross-stress. Environ. Pollut. 2016, 218, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Louis, F.; Devin, S.; Giambérini, L.; Potet, M.; David, E.; Pain-Devin, S. Energy allocation in two dreissenid species under metal stress. Environ. Pollut. 2019, 245, 889–897. [Google Scholar] [CrossRef]
- Farkas, A.; Ács, A.; Vehovszky, Á.; Falfusynska, H.; Stoliar, O.; Specziár, A.; Győri, J. Interspecies comparison of selected pollution biomarkers in dreissenid spp. inhabiting pristine and moderately polluted sites. Sci. Total Environ. 2017, 599–600, 760–770. [Google Scholar] [CrossRef]
- Zhang, H.; Reynolds, M. Cadmium exposure in living organisms: A short review. Sci. Total Environ. 2019, 678, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-M.; Yao, L.-A.; Ma, Q.-L.; Zhou, G.-J.; Li Wang, L.; Fang, Q.-L.; Xu, Z.-C. Distribution and ecological risk assessment of cadmium in water and sediment in Longjiang River, China: Implication on water quality management after pollution accident. Chemosphere 2018, 194, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Perera, P.A.C.T.; Sundarabarathy, T.V.; Sivananthawerl, T.; Kodithuwakku, S.P.; Edirisinghe, U. Arsenic and Cadmium Contamination in Water, Sediments and Fish is a Consequence of Paddy Cultivation: Evidence of River Pollution in Sri Lanka. Achiev. Life Sci. 2016, 10, 144–160. [Google Scholar] [CrossRef]
- Amiard, J.-C.; Pineau, A.; Boiteau, H.L.; Métayer, C.; Amiard-Triquet, C. Application of atomique absorption spectrometry using Zeeman effect to the determination of eight trace elements (Ag, Cd, Cr, Cu, Mn, Ni, Pb et Se) in biological materials. Water Res. 1987, 21, 696–697. [Google Scholar] [CrossRef]
- Péden, R.; Rocher, B.; Chan, P.; Vaudry, D.; Poret, A.; Olivier, S.; Le Foll, F.; Bultelle, F. Consequences of acclimation on the resistance to acute thermal stress: Proteomic focus on mussels from pristine site. Mar. Environ. Res. 2016, 121, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Péden, R.; Poupin, P.; Sohm, B.; Flayac, J.; Giambérini, L.; Klopp, C.; Louis, F.; Pain-Devin, S.; Potet, M.; Serre, R.-F.; et al. Environmental Transcriptomes of Invasive Dreissena, a Model Species in Ecotoxicology and Invasion Biology. Sci. Data 2019, 6, 234. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Roseman, E.F.; Mills, E.L.; Rutzke, M.; Gutenmann, W.H.; Lisk, D.J. Absorption of cadmium from water by north american zebra and quagga mussels (bivalvia: Dreissenidae). Chemosphere 1994, 28, 737–743. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, J.; Ding, N.; Zellmer, L.; Zhao, Y.; Liu, S.; Liao, D.J. ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting. Open Life Sci. 2021, 16, 1278–1292. [Google Scholar] [CrossRef]
- Choong, G.; Liu, Y.; Templeton, D.M. Interplay of calcium and cadmium in mediating cadmium toxicity. Chem. Biol. Interac. 2014, 211, 54–65. [Google Scholar] [CrossRef]
- Péden, R.; Rocher, B.; Chan, P.; Vaudry, D.; Poret, A.; Olivier, S.; Le Foll, F.; Bultelle, F. Highly polluted life history and acute heat stress, a hazardous mix for blue mussels. Mar. Pollut. Bull. 2018, 135, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Leung, P.T.Y.; Wang, Y.; Mak, S.S.T.; Ng, W.C.; Leung, K.M.Y. Differential proteomic responses in hepatopancreas and adductor muscles of the green-lipped mussel Perna viridis to stresses induced by cadmium and hydrogen peroxide. Aquat. Tox. 2011, 105, 49–61. [Google Scholar] [CrossRef]
- Zheng, X.; Gao, Y.; Li, W.; Wang, S. iTRAQ-based quantitative proteomic analysis identified Eno1 as a cadmium stress response gene in Propsilocerus akamusi (Tokunaga) hemolymph. Ecotoxicol. Environ. Saf. 2018, 165, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Dai, Y.; Li, M.; Guo, L.; Cao, C.; Huang, Y.; Ma, R.; Qiu, S.; Su, X.; Zhong, K.; et al. Exposure to cadmium induces neuroinflammation and impairs ciliogenesis in hESC-derived 3D cerebral organoids. Sci. Total Environ. 2021, 797, 149043. [Google Scholar] [CrossRef]
- Zhang, X.; Li, F.; Ji, C.; Wu, H. Toxicological mechanism of cadmium in the clam Ruditapes philippinarum using combined ionomic, metabolomic and transcriptomic analyses. Environ. Pollut. 2023, 323, 121286. [Google Scholar] [CrossRef]
- Zhan, X.; Li, B.; Zhan, X.; Schlüter, H.; Jungblut, P.R.; Coorssen, J.R. Innovating the concept and practice of Two-Dimensional Gel Electrophoresis in the analysis of proteomes at the proteoform level. Proteomes 2019, 7, 36. [Google Scholar] [CrossRef]
- Joseph, P. Mechanisms of cadmium carcinogenesis. Toxicol. Appl. Pharmacol. 2009, 238, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Thévenod, F. Cadmium and cellular signaling cascades: To be or not to be? Toxicol. Appl. Pharmacol. 2009, 238, 221–239. [Google Scholar] [CrossRef]
- Tamás, M.; Fauvet, B.; Christen, P.; Goloubinoff, P. Misfolding and aggregation of nascent proteins: A novel mode of toxic cadmium action in vivo. Curr. Genet. 2018, 64, 177–181. [Google Scholar] [CrossRef]
- Đukic-Cosic, D.; Baralic, K.; Javorac, D.; Djordjevic, A.B.; Bulat, Z. An overview of molecular mechanisms in cadmium toxicity. Curr. Opin. Toxicol. 2020, 19, 56–62. [Google Scholar] [CrossRef]
- Le Saux, A.; David, E.; Betoulle, S.; Bultelle, F.; Rocher, B.; Barjhoux, I.; Cosio, C. New insights into cellular impacts of metals in aquatic animals. Environments 2020, 7, 46. [Google Scholar] [CrossRef]
- Lee, W.-K.; Thévenod, F. Cell organelles as targets of mammalian cadmium toxicity. Arch. Toxicol. 2020, 94, 1017–1049. [Google Scholar] [CrossRef]
- Si, Y.-X.; Lee, J.; Zhao, F.; Yin, S.-J.; Park, Y.-D.; Qian, G.-Y. Effects of cadmium on the cuttlefish Sepia pharaonis’ arginine kinase: Unfolding kinetics integrated with computational simulations. J. Biomol. Struct. Dyn. 2015, 34, 1763–1777. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Yeh, P.-L.; Lee, T.-H. Ionic and amino acid regulation in hard clam (Meretrix lusoria) in response to salinity challenges. Front. Physiol. 2016, 7, 368. [Google Scholar] [CrossRef] [PubMed]
- Harcet, M.; Perina, D.; Pleše, B. Opine Dehydrogenases in Marine Invertebrates. Biochem. Genet. 2013, 51, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Zhang, S.; Dong, Y. Anaerobic metabolism and thermal tolerance: The importance of opine pathways on survival of a gastropod after cardiac dysfunction. Integr. Zool. 2016, 12, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Ivanina, A.V.; Cherkasov, A.S.; Sokolova, I. Effects of cadmium on cellular protein and glutathione synthesis and expression of stress proteins in eastern oysters, Crassostrea virginica Gmelin. J. Exp. Biol. 2008, 211, 577–586. [Google Scholar] [CrossRef]
- Jeppe, K.J.; Carew, M.E.; Long, S.; Lee, S.F.; Pettigrove, V.; Hoffmann, A.A. Genes involved in cysteine metabolism of Chironomus tepperi are regulated differently by copper and by cadmium. Comp. Biochem. Physiol. C 2014, 162, 1–6. [Google Scholar] [CrossRef]
- Kaczor-Kaminska, M.; Sura, P.; Wróbel, M. Multidirectional changes in parameters related to sulfur metabolism in frog tissues exposed to heavy metal-related stress. Biomolecules 2020, 10, 574. [Google Scholar] [CrossRef]
- Xia, L.; Chen, S.; Dahms, H.-U.; Ying, X.; Peng, X. Cadmium induced oxidative damage and apoptosis in the hepatopancreas of Meretrix meretrix. Ecotoxicology 2016, 25, 959–969. [Google Scholar] [CrossRef]
- Huang, Y.; Tang, H.; Jin, J.; Fan, M.; Chang, A.K.; Ying, X. Effects of waterborne cadmium exposure on its internal distribution in Meretrix meretrix and detoxification by metallothionein and antioxidant enzymes. Front. Mar. Sci. 2020, 7, 502. [Google Scholar] [CrossRef]
- Qiu, X.-B.; Shao, Y.-M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Reeg, S.; Jung, T.; Castro, J.P.; Davies, K.J.A.; Henze, A.; Grune, T. The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic Biol Med. 2016, 99, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, F.K.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The contribution of the 20S proteasome to proteostatis. Biomolecules 2020, 9, 190. [Google Scholar] [CrossRef]
- Paesano, L.; Perotti, A.; Buschini, A.; Carubbi, C.; Marmiroli, M.; Maestri, E.; Iannotta, S.; Marmiroli, N. Markers for toxicity to HepG2 exposed to cadmium sulphide quantum dots; damage to mitochondria. Toxicology 2016, 374, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.-M.; Rœde, J.R.; Orr, M.; Liang, Y.; Jones, D.P. Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of Cd toxicity. Toxicol. Sci. 2014, 139, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, Q.; Song, R.; Zhang, Y.; Yang, J.; Wang, Y.; Yuan, Y.; Bian, J.; Liu, X.; Gu, J.; et al. Cadmium induced inhibition of autophagy is associated with microtubule disruption and mitochondrial dysfunction in primary rat cerebral cortical neurons. Neurotoxicol. Teratol. 2016, 53, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Deng, W.; Zou, T.; Bai, B.; Chang, A.K.; Ying, X. Cadmium-induced oxidative stress in Meretrix meretrix gills leads to mitochondria-mediated apoptosis. Ecotoxicology 2021, 30, 2011–2023. [Google Scholar] [CrossRef]
- Chen, G.; Wei, T.; Ju, F.; Li, H. Protein quality control and aggregation in the endoplasmic reticulum: From basic to bedside. Front. Cell Dev. Biol. 2023, 11, 1156152. [Google Scholar] [CrossRef]
- Pham, K.; Pal, R.; Qu, Y.; Liu, X.; Yu, H.; Shiao, S.; Wand, X.; O’Brian Smith, E.; Cui, X.; Rodney, G.G.; et al. Nuclear glutaredoxin 3 is critical for protection against oxidative stress-induced cell death. Free Radic. Biol. Med. 2015, 85, 197–206. [Google Scholar] [CrossRef]
- Bao, Y.; Liu, X.; Zhang, W.; Cao, J.; Li, W.; Li, C.; Lin, Z. Identification of a regulation network in response to cadmium toxicity using blood clam Tegillarca granosa as model. Sci. Rep. 2016, 6, 35704. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xu, L.; Ji, C.; Yu, D. Proteomic and metabolomic responses in D-shape larval mussels Mytilus galloprovincialis exposed to cadmium and arsenic. Fish Shellfish Immunol. 2016, 58, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Granger Joly de Boissel, P.; Fournier, M.; Rodriguez-Lecompte, R.C.; McKenna, P.; Kibenge, F.; Siah, A. Functional and molecular responses of the blue mussel Mytilus edulis’hemocytes exposed to cadmium—An in vitro model and transcriptomic approach. Fish Shellfish Immunol. 2017, 67, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wang, W.; Li, L.; Yin, Q.; Zhang, G. Cadmium effects on DNA and protein metabolism in oyster (Crassostrea gigas) revealed by proteomic analyses. Sci. Rep. 2017, 7, 11716. [Google Scholar] [CrossRef] [PubMed]
- Company, R.; Antúnez, O.; Cosson, R.P.; Serafim, A.; Shillito, B.; Cajaraville, M.; Bebianno, M.J.; Torreblanca, A. Protein expression profiles in Bathymodiolus azoricus exposed to cadmium. Ecotoxicol. Environ. Saf. 2019, 171, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.P.; Lima, D.; Paiva, R.; Vilke, J.M.; Mattos, J.J.; Almeida, E.A.; Grott, S.C.; Alves, T.C.; Corrêa, J.N.; Jorge, M.B.; et al. Metal bioaccumulation, oxidative stress and antioxidant responses in oysters Crassostrea gasar transplanted to an estuary in southern Brazil. Sci. Total Environ. 2019, 685, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, X.; Tian, F.; Chen, Q.; Luo, P.; Zhang, F.; Wan, X.; Zhong, Y.; Liu, Q.; Lin, T. Changes in proteome and protein phosphorylation reveal the protective roles of exogenous nitrogen in alleviating cadmium toxicity in poplar plants. Int. J. Mol. Sci. 2020, 21, 278. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, S.; Shan, X.; Ji, C.; Wu, H. Differential biological effects in two pedigrees of clam Ruditapes philippinarum exposed to cadmium using iTRAQ-based proteomics. Environ. Toxicol. Pharmacol. 2019, 65, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Rono, M.K.; Muturi, C.N.; Ochieng, R.; Mwakubabanya, R.; Wachira, F.N.; Mwangangi, J.; Kinyanjui, S.; Njunge, J.; Mireji, P.O. Cadmium tolerance pathway in Anopheles gambiae senso stricto. Acta Tropica 2019, 198, 105033. [Google Scholar] [CrossRef]
- Liu, H.; Tian, X.; Jiang, L.; Han, D.; Hu, S.; Cui, Y.; Jiang, F.; Liu, Y.; Xu, Y.; Li, H. Sources, bioaccumulation, and toxicity mechanisms of cadmium in Chlamys farreri. J. Hazard. Mater. 2023, 453, 131395. [Google Scholar] [CrossRef]
- Cui, Y.; Freedman, J.H. Cadmium induces retinoic acid signaling by regulating retinoic acid metabolic gene expression. J. Biol. Chem. 2009, 284, 24925–24932. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Ke, C.; Guo, X.; Shi, B.; Huang, M. Metal accumulation and differentially expressed proteins in gill of oyster (Crassostrea hongkongensis) exposed to long-term heavy metal-contaminated estuary. Fish Shellfish Immunol. 2014, 38, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ali, A.; Jin, Z.; Pei, Y.; Yang, G. Induction of cystathionine gamma-lyase expression and metallothionein-1 S-sulfhydration alleviate cadmium-induced cell death in myoblast cells. Ecotoxicol. Environ. Saf. 2019, 179, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Dickhout, J.; Carlisle, R.E.; Jerome, D.E.; Mohammed-Ali, Z.; Jiang, H.; Yang, G.; Mani, S.; Garg, S.K.; Banerjee, R.; Kaufman, R.J.; et al. Integrated stress response modulates cellular redox state via induction of cystathionine γ-lyase: Cross-talk between integrated stress response and thiol metabolism. J. Biol. Chem. 2012, 287, 7603–7614. [Google Scholar] [CrossRef] [PubMed]
- Marchione, R.; Leibovitch, S.A.; Lenormand, J.-L. The translational factor eIF3f: The ambivalent eIF3 subunit. Cell. Mol. Life Sci. 2013, 70, 3603–3616. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Lu, Q.; Huang, Q.; Zheng, C.; Chen, B.; Lei, Y. EIF3 regulates migration, invasion and apoptosis in cadmium transformed 16HBE cells and is a novel biomarker of cadmium exposure in a rat model and in workers. Toxicol. Res. 2016, 5, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.; Gorman, M.A.; Koach, J.; Cheung, B.; Marshall, G.M.; Parker, M.W.; Holien, J.K. A structural view of PA2G4 isoforms with opposing functions in cancer. J. Biol. Chem. 2020, 295, 16100–16112. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, I.M.; Sokolov, E.P.; Ponnappa, K.V. Cadmium exposure affects mitochondrial bioenergetics and gene expression of key mitochondrial proteins in the eastern oyster Crassostrea virginica Gmelin (Bivalvia: Ostreidae). Aq. Toxicol. 2005, 73, 242–255. [Google Scholar] [CrossRef]
- Hanana, H.; Kleinert, C.; André, C.; Gagné, F. Influence of cadmium on oxidative stress and NADH oscillations in mussel mitochondria. Comp. Biochem. Physiol. C 2019, 216, 60–66. [Google Scholar] [CrossRef]
- Ji, C.; Lu, Z.; Xu, L.; Li, F.; Cong, M.; Shan, X.; Wu, H. Evaluation of mitochondrial toxicity of cadmium in clam Ruditapes philippinarum using iTRAQ-based proteomics. Environ. Pollut. 2019, 251, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, H.; Yaun, Y.; Wang, Y.; Tian, Z. SPAG6 promotes cell proliferation and inhibits apoptosis through the PTEN/PI3K/AKT pathway in Burkitt lymphoma. Oncol. Rep. 2020, 44, 2021–2030. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Rossi, R.; Milzani, A.; Di Simplicio, P.; Colombo, R. The actin cytoskeleton response to oxidants: From small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic. Biol. Med. 2001, 31, 1624–1632. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, B.; Tyther, R.; Sheehan, D. Carbonylation and glutathionylation of proteins in the blue mussel Mytilus edulis detected by proteomic analysis and Western blotting: Actin as a target for oxidative stress. Aquat. Toxicol. 2005, 73, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Chora, S.; Starita-Geribaldi, M.; Guigonis, J.-M.; Samsom, M.; Roméo, M.; Bebianno, M.J. Effect of cadmium in the clam Ruditapes decussatus assessed by proteomic analysis. Aquat. Toxicol. 2009, 94, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Dailianis, S.; Patetsini, E.; Kaloyanni, M. The role of signalling molecules on actin glutathionylation and protein carbonylation induced by cadmium in haemocytes of mussel Mytilus galloprovincialis (Lmk). J. Exp. Biol. 2009, 212, 3612–3620. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, D.-Z.; Wang, W.-X. Cadmium-induced changes in trace element bioaccumulation and proteomics perspective in four marine bivalves. Environ. Toxicol. Chem. 2012, 31, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.L.; Taylor, D.A.; Nair, S.V.; Birch, G.; Haynes, P.A.; Raftos, D.A. Proteomic discovery of biomarkers of metal contamination in Sydney Rock oysters (Saccostrea glomerata). Aq. Toxicol. 2012, 109, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Melwani, A.R.; Thompson, E.L.; Raftos, D.A. Differential proteomic response of Sydney rock oysters (Saccostrea glomerata) to prolonged environmental stress. Aquat. Toxicol. 2016, 173, 53–62. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, W.; Shinde, M.; Field, J.; Templeton, D.M. Cadmium favors F-actin depolymerization in rat renal mesangial cells by site-specific, disulfide-based dimerization of the CAP1 protein. Arch. Toxicol. 2018, 92, 1049–1064. [Google Scholar] [CrossRef]
- Pizzaia, D.; Nogueira, M.L.; Mondin, M.; Carvalho, M.E.A.; Piotto, F.A.; Rosario, M.F.; Azevedo, R.A. Cadmium toxicity and its relationship with disturbances in the cytoskeleton, cell cycle and chromosome stability. Ecotoxicology 2019, 28, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Perlson, E.; Medzihradszky, K.F.; Darula, Z.; Munno, D.W.; Syed, N.I.; Burlingame, A.L.; Fainzilber, M. Differential proteomics reveals multiple components in retrogradely transported axoplasm after nerve injury. Mol. Cell Proteom. 2004, 3, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Frascotti, G.; Galbiati, E.; Mazzucchelli, M.; Pozzi, M.; Salvioni, L.; Vertemara, J.; Tortora, P. The Vault nanoparticle: A gigantic ribonucleoprotein assembly involved in diverse physiological and pathological phenomena and an Ideal nanovector for drug delivery and therapy. Cancers 2021, 13, 707. [Google Scholar] [CrossRef] [PubMed]
- Grossi, S.; Fenini, G.; Kockmann, T.; Hennig, P.; Di Filippo, M.; Beer, H.D. Inactivation of the cytoprotective major vault protein by caspase-1 and -9 in epithelial cells during apoptosis. J. Investig. Dermatol. 2020, 140, 1335–1345.e10. [Google Scholar] [CrossRef] [PubMed]
- Goswami, P.; Hariharan, G.; Godhantaraman, N.; Munuswamy, N. An integrated use of multiple biomarkers to investigate the individual and combined effect of copper and cadmium on the marine green mussel (Perna viridis). J. Environ. Sci. Health A 2014, 49, 1564–1577. [Google Scholar] [CrossRef] [PubMed]
- Dixit, U.; Liu, Z.; Pandey, A.K.; Kothari, R.; Pandey, V.N. Fuse binding protein antagonizes the transcription activity of tumor suppressor protein p53. BMC Cancer 2014, 14, 925. [Google Scholar] [CrossRef]
- Jang, M.; Park, B.C.; Kang, S.; Chi, S.-W.; Cho, S.; Chung, S.J.; Lee, S.C.; Bae, K.-H.; Park, S.G. Far upstream element-binding protein-1, a novel caspase substrate, acts as a cross-talker between apoptosis and the c-myc oncogene. Oncogene 2009, 12, 1529–1536. [Google Scholar] [CrossRef]
- Zhao, D.; Hu, G.; Chen, R.; Xiao, G.; Teng, S. Molecular cloning, characterization, and tissue distribution of c-Myc from blood clam Tegillarca granosa and its role in cadmium-induced stress response. Gene 2022, 834, 14661. [Google Scholar] [CrossRef]
- Shih, J.-W.; Wang, W.-T.; Tsai, T.-Y.; Kuo, C.-Y.; Li, H.-K.; Wu Lee, Y.-H. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 2012, 441, 119–129. [Google Scholar] [CrossRef]
- Princely Abudu, Y.; Pankiv, S.; Mathai, B.J.; Håkon Lystad, A.; Bindesbøll, C.; Brenne, H.B.; Yoke Wui Ng, M.; Thiede, B.; Yamamoto, A.; Mutugi Nthiga, T.; et al. NIPSNAP1 and NIPSNAP2 Act as “Eat Me” Signals for Mitophagy. Dev. Cell 2019, 49, 509–525.e12. [Google Scholar] [CrossRef]
- Dan, X.; Babbar, M.; Moore, A.; Wechter, N.; Tian, J.; Mohanty, J.G.; Croteau, D.L.; Bohr, V.A. DNA damage invokes mitophagy through a pathway involving Spata18. Nucleic Acids Res. 2020, 48, 6611–6623. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, K.; Yokosawa, T. Chapter Eight-Non-canonical functions of human cytoplasmic tyrosyl-, tryptophanyl- and other aminoacyl-tRNA synthetases. Enzymes 2020, 48, 207–242. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yang, C.; Wang, L.; Wang, X.; Wang, J.; Yue, F.; Liu, R.; Zhang, H.; Song, L. The protein expression profile in hepatopancreas of scallop Chlamys farreri under heat stress and Vibrio anguillarum challenge. Fish Shellfish Immunol. 2014, 36, 252–260. [Google Scholar] [CrossRef]
- Zhang, Z.; Miao, L.; Xin, X.; Zhang, J.; Yang, S.; Miao, M.; Kong, X.; Jia, B. Underexpressed CNDP2 participates in gastric cancer growth inhibition through activating the MAPK signaling pathway. Mol. Med. 2014, 20, 17–28. [Google Scholar] [CrossRef]
- van Vliet, A.R.; Giordano, F.; Gerlo, S.; Segura, I.; Van Eygen, S.; Molenberghs, G.; Rocha, S.; Houcine, A.; Derua, R.; Verfaillie, T.; et al. The ER Stress Sensor PERK Coordinates ER-Plasma Membrane Contact Site Formation through Interaction with Filamin-A and F-Actin Remodeling. Mol. Cell 2017, 65, 885–899.e6. [Google Scholar] [CrossRef]
- Lewinska, A.; Klukowska-Rötzlerb, J.; Deregowskad, A.; Adamczyk-Grochalaa, J.; Wnuk, M. c-Myc activation promotes cofilin-mediated remodeling and telomere homeostasis as a response to oxidant-based DNA damage in medulloblastoma cells. Redox Biol. 2019, 24, 101163. [Google Scholar] [CrossRef]
- Devin, S.; Potet, M.; Louis, F.; Pauly, D.; Rocher, B.; Wagner, P.; Giambérini, L.; Pain-Devin, S. Spatial and seasonal use of biomarkers in dreissenids: Implications for biomonitoring. Environ. Sci. Pollut. Res. 2023. [Google Scholar] [CrossRef]
- Potet, M.; Giambérini, L.; Pain-Devin, S.; Louis, F.; Bertrand, C.; Devin, S. Differential tolerance to nickel between Dreissena polymorpha and Dreissena rostriformis bugensis populations. Sci. Rep. 2018, 8, 700. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhu, J.; Zhang, K.; Jiang, C.; Wang, Y.; Yuan, Y.; Bian, J.; Liu, X.; Gu, J.; Liu, Z. Induction of cytoprotective autophagy in PC-12 cells by cadmium. Biochem. Biophys. Res. Commun. 2013, 438, 186–192. [Google Scholar] [CrossRef]
- Quinta, H.R.; Galigniana, N.M.; Erlejman, A.G.; Lagadari, M.; Piwien-Pilipuk, G.; Galigniana, M.D. Management of cytoskeleton architecture by molecular chaperones and immunophilins. Cell. Signal. 2011, 23, 1907–1920. [Google Scholar] [CrossRef]
- Kung, C.-P.; Wu, Y.-R.; Chuang, H.-W. Expression of a dye-decolorizing peroxidase results in hypersensitive response to cadmium stress through reducing the ROS signal in Arabidopsis. Environ. Exp. Bot. 2014, 101, 47–55. [Google Scholar] [CrossRef]
- Antonioli, M.; Albiero, F.; Piacentini, M.; Fimia, G.M. Temporal regulation of autophagy response by the CULLIN 4-AMBRA1-CULLIN 5 axis. Mol. Cell Oncol. 2015, 3, e10083042016. [Google Scholar] [CrossRef]
- Díaz-Ramos, À.; Roig-Borrellas, A.; García-Melero, A.; López-Alemany, R. α-Enolase, a Multifunctional Protein: Its Role on Pathophysiological Situations. J. Biomed. Biotechnol. 2012, 2012, 1567952012. [Google Scholar] [CrossRef]
- Zandberg, L.; van Dyk, H.C.; van der Westhuizen, F.H.; van Dijk, A.A. A 3-methylcrotonyl-CoA carboxylase deficient human skin fibroblast transcriptome reveals underlying mitochondrial dysfunction and oxidative stress. Int. J. Biochem. Cell Biol. 2016, 78, 116–129. [Google Scholar] [CrossRef]
- Zhang, H.-L.; Liu, Z.-H.; Luo, Q.; Wang, Y.; Zhao, Z.-H. Abnormal expression of NSF, α-SNAP and SNAP23 in pulmonary arterial hypertension in rats treated with monocrotaline. Int. J. Clin. Exp. Med. 2015, 8, 1834–1843. [Google Scholar]
- Muralidharan, S.; Thompson, E.; Raftos, D.; Birch, G.; Haynes, P.A. Quantitative proteomics of heavy metal stress responses in Sydney rock oysters. Proteomics 2012, 12, 906–921. [Google Scholar] [CrossRef]
- Latorre-Muro, P.; Baeza, J.; Armstrong, E.; Hurtado-Guerrero, R.; Corzana, F.; Wu, L.; Sinclair, D.A.; Lopez-Buesa, P.; Carrodeguas, J.A.; Denu, J.M. Dynamic acetylation of phosphoenolpyruvate carboxykinase toggles enzyme activity between gluconeogenic and anaplerotic reactions. Mol. Cell 2018, 71, 718–732. [Google Scholar] [CrossRef] [PubMed]
- Mori, C.; Lee, J.-Y.; Tokumoto, M.; Satoh, M. Cadmium toxicity is Regulated by peroxisome proliferator-activated receptor δ in human proximal tubular cells. Int. J. Mol. Sci. 2022, 23, 8652. [Google Scholar] [CrossRef] [PubMed]
- Muthukumar, K.; Nachiappan, V. Phosphatidylethanolamine from phosphatidylserine decarboxylase2 is essential for autophagy under cadmium stress in Saccharomyces cerevisiae. Cell Biochem. Biophys. 2013, 67, 1353–1363. [Google Scholar] [CrossRef]
- Adle, D.J.; Wei, W.; Smith, N.; Bies, J.J.; Lee, J. Cadmium-mediated rescue from ER-associated degradation induces expression of its exporter. Proc. Natl. Acad. Sci. USA 2008, 106, 10189–10194. [Google Scholar] [CrossRef]
- Song, C.; Xiao, Z.; Nagashima, K.; Li, C.-C.H.; Lockett, S.J.; Dai, R.-M.; Cho, E.H.; Conrads, T.P.; Veenstra, T.D.; Colburn, N.H.; et al. The heavy metal cadmium induces valosin-containing protein (VCP)-mediated aggresome formation. Toxicol. Appl. Pharmacol. 2008, 228, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Pennington, K.L.; Chan, T.Y.; Torres, M.P.; Andersen, J.L. The dynamic and stress-adaptive signaling hub of 14-3-3: Emerging mechanisms of regulation and context-dependent protein–protein interactions. Oncogene 2018, 37, 5587–5604. [Google Scholar] [CrossRef] [PubMed]


| Part A. Chaperone/processing/folding/degradation (GO:0033554_Cellular response to stress in bold) | ||||||||
| Sp. | label | abbrev. | Mr | pI | Var. | Identification | G. | Ca |
| ZM | 44,479 | COL12A1 | 83,379 | 5.6 | 2.07 | collagen alpha-1(XII) chain | ||
| ZM | 346 | HSPA12A | 69,540 | 6.6 | 1.83 | heat shock 70 kDa protein 12A | ||
| ZM | 645 | PDIA6 | 44,461 | 5.6 | 1.68 | protein disulfide-isomerase A6 | 1 | |
| ZM | 8514 | NAPA | 24,787 | 5.0 | 1.64 | alpha-soluble NSF attachment protein | 7 | |
| ZM | 901 | TGFBI | 34,177 | 7.4 | 1.52 | transforming growth factor-beta-induced protein ig-h3 | ||
| ZM | 434 | PPP5C | 56,199 | 6.0 | 1.49 | serine/threonine-protein phosphatase 5 | 1 | |
| ZM | 2519 | HSPD1 | 54,076 | 5.4 | 1.46 | heat shock protein 60, mitochondrial | 1; 6 | |
| ZM | 102,964 | UFC1 | 23,929 | 6.8 | 1.39 | ubiquitin-fold modifier-conjugating enzyme 1 | 1 | |
| ZM | 163 | RRBP1 | 89,856 | 5.6 | 1.35 | ribosome-binding protein 1 | ||
| ZM | 984 | CA2 | 29,793 | 6.1 | 1.34 | carbonic anhydrase II | 1 | |
| ZM | 817 | HSPB1 | 38,223 | 6.5 | 1.28 | small heat shock protein p36 | 6 | |
| ZM | 705 | DNAJB11 | 42,109 | 5.7 | 1.26 | dnaJ homolog subfamily B member 11 | 1 | |
| ZM | 211 | HSP90AA1 | 85,659 | 5.1 | 0.78 | heat shock protein 90 | 1; 6; 7 | |
| ZM | 170 | MVP | 89,818 | 5.7 | 0.74 | major vault protein | 7 | |
| ZM | 207 | VCP | 85,723 | 5.0 | 0.65 | transitional endoplasmic reticulum ATPase | 1; 6; 7 | |
| ZM | 159 | HSP90B1 | 89,954 | 5.1 | 0.61 | endoplasmin | 1; 6 | |
| ZM | 655 | P4HB | 44,315 | 4.4 | 0.56 | protein disulfide-isomerase | 1; 6 | |
| ZM | 334 | STIP1 | 73,026 | 5.8 | 0.55 | stress-induced-phosphoprotein 1 | ||
| QM | 583 | CCT5 | 45,634 | 6.1 | 3.34 | T-complex protein 1 subunit epsilon | 4 | |
| QM | 382 | HSPA1 | 58,954 | 5.4 | 1.81 | heat shock protein 70, partial | 1; 4; 6; 7 | |
| QM | 851 | PSMD14 | 34,207 | 6.1 | 1.81 | 26S proteasome non-ATPase regulatory subunit 14 | 1; 7 | |
| QM | 673 | PSMC6 | 41,340 | 6.2 | 1.71 | 26S proteasome regulatory subunit 10B | 1 | |
| QM | 341 | ATP6V1A | 63,911 | 5.4 | 1.69 | V-type proton ATPase catalytic subunit A | ||
| QM | 546 | HSPD1 | 46,798 | 5.5 | 1.61 | heat shock protein 60, mitochondrial | 1; 6 | |
| QM | 1058 | PSMA4 | 28,393 | 5.5 | 1.56 | proteasome subunit alpha type-4 | 1 | |
| QM | 625 | CASP3 | 43,567 | 4.8 | 1.55 | caspase 3 like | 1; 6 | |
| QM | 1066 | YWHAB | 28,086 | 4.9 | 1.47 | 14-3-3 protein 1 | ||
| QM | 839 | PEPD | 34,135 | 5.7 | 0.74 | proline iminopeptidase | 3 | |
| QM | 424 | CCT4 | 57,096 | 6.2 | 0.70 | T-complex protein 1 subunit delta | 4 | |
| QM | 1026 | CTSL | 28,816 | 4.6 | 0.69 | cathepsin L | ||
| QM | 291 | HSPA1 | 68,940 | 5.4 | 0.67 | heat shock protein 70 | 1; 4; 6; 7 | |
| QM | 1137 | PSMA2 | 27,167 | 5.5 | 0.66 | proteasome subunit alpha type-2 | 1; 7 | |
| QM | 437 | ATP6V1B1 | 55,137 | 5.7 | 0.65 | V-type proton ATPase subunit B | 3 | |
| QM | 299 | HSPA5 | 66,677 | 5.2 | 0.62 | Endoplasmic reticulum chaperone BiP GRP78 | 1 | |
| QM | 620 | CTSD | 43,053 | 4.6 | 0.61 | cathepsin D | 6; 7 | |
| QM | 4174 | PACRG | 27,552 | 7.0 | 0.59 | parkin coregulated gene protein homolog | 1 | |
| QM | 748 | P4HB | 38,485 | 4.7 | 0.58 | protein disulfide-isomerase | 1; 6 | |
| QM | 100 | MVP | 94,623 | 5.7 | 0.56 | major vault protein | 7 | |
| QM | 985 | YWHAE | 29,752 | 4.7 | 0.53 | 14-3-3 protein epsilon | 1 | |
| QM | 151 | MVP | 88,509 | 5.7 | 0.52 | major vault protein, partial | 7 | |
| Part B. Calcium and cytoskeleton alterations (GO:0005856 Cytoskeleton in bold) | ||||||||
| Sp. | label | abbrev. | Mr | pI | Var. | Identification | G. | Ca |
| ZM | 191 | FLNA | 85,827 | 6.0 | 1.63 | filamin-A, partial | 4; 6; 7 | |
| ZM | 834 | PPP1CA | 38,250 | 5.8 | 1.63 | serine/threonine-protein phosphatase alpha-2 isoform | 1; 4; 6 | |
| ZM | 244 | ANXA7 | 78,074 | 7.6 | 1.62 | annexin A7 | 7 | Ca |
| ZM | 597 | PRKAR2B | 45,791 | 4.7 | 1.51 | cAMP-dependent protein kinase type II regulatory subunit | 3; 4 | |
| ZM | 722 | NME7 | 41,220 | 6.3 | 1.50 | nucleoside diphosphate kinase 7 | 4 | |
| ZM | 83,183 | IFI44 | 47,224 | 6.6 | 1.45 | microtubule-associated protein 44 | ||
| ZM | 932 | ANXA11 | 33,143 | 5.8 | 1.43 | annexin A11 | 4; 7 | Ca |
| ZM | 82,002 | WDR1 | 64,919 | 6.6 | 1.43 | WD repeat-containing protein 1 | 4; 7 | Ca |
| ZM | 723 | TWF1 | 41,814 | 5.8 | 1.40 | twinfilin-1 | 4 | |
| ZM | 766 | SPAG6 | 40,347 | 5.9 | 1.39 | sperm-associated antigen 6 | 4 | |
| ZM | 679 | DNPEP | 42,701 | 6.3 | 1.29 | aspartyl aminopeptidase | 7 | |
| ZM | 1334 | CFL1 | 13,142 | 4.6 | 1.25 | cofilin | 4; 6 | |
| ZM | 550 | SNX6 | 47,482 | 5.7 | 1.22 | sorting nexin-6 | 6 | |
| ZM | 1039 | TUBB | 28,478 | 5.3 | 0.80 | tubulin beta, partial | 4; 7 | |
| ZM | 610 | VAT1 | 45,137 | 5.7 | 0.73 | synaptic vesicle membrane protein VAT-1 homolog | 7 | Ca |
| ZM | 20,235 | TUBA1A | 46,039 | 5.8 | 0.68 | tubulin alpha-1A chain | 4; 7 | |
| ZM | 1173 | CAPS | 25,562 | 5.4 | 0.68 | calcyphosin protein | Ca | |
| ZM | 110 | FLNA | 94,619 | 5.7 | 0.65 | filamin A_B_C partial | 4; 6; 7 | |
| ZM | 9159 | CNN3 | 22,873 | 5.1 | 0.56 | calponin homolog, protein unc-87 | 4 | Ca |
| ZM | 319 | LMNA | 75,067 | 5.2 | 0.55 | 60 kDa neurofilament protein-like | 1; 4; 5; 6; 7 | Ca |
| QM | 182 | CFAP58 | 83,075 | 6.6 | 3.75 | cilia- and flagella-associated protein 58 | ||
| QM | 177 | PEFLIN | 83,987 | 6.9 | 2.82 | peflin | Ca | |
| QM | 1116 | EFHD2 | 27,319 | 5.5 | 1.89 | EF-hand domain-containing protein D2 | Ca | |
| QM | 632 | CROCC | 43,108 | 6.1 | 1.87 | rootletin | 4 | |
| QM | 992 | CNN3 | 29,595 | 5.3 | 1.70 | calponin homolog, protein unc-87 | 4 | Ca |
| QM | 188 | FLNA | 83,094 | 5.9 | 1.65 | filamin-A, partial | 4; 6; 7 | |
| QM | 198 | VCL | 80,926 | 5.6 | 1.64 | vinculin | 4; 7 | |
| QM | 1119 | CALM3 | 27,412 | 5.2 | 1.62 | calmodulin protein 3 | Ca | |
| QM | 718 | CALR | 39,142 | 4.8 | 1.60 | calreticulin | 1; 4; 5; 6 | Ca |
| QM | 508 | TUBB4B | 48,029 | 5.4 | 1.59 | tubulin beta-4B chain | 4 | |
| QM | 495 | SYNE2 | 49,936 | 5.6 | 1.53 | nesprin-2 | 4 | |
| QM | 1090 | EFHC1 | 27,602 | 5.2 | 1.52 | EF-hand calcium-binding domain-containing protein 1 | 4 | Ca |
| QM | 926 | TBCB | 31,668 | 4.8 | 1.42 | tubulin-folding cofactor B | 4 | |
| QM | 1219 | TMSB10 | 27,111 | 5.3 | 1.39 | thymosin beta | 4 | |
| QM | 26,826 | TUBB | 27,900 | 4.7 | 1.37 | tubulin beta, partial | 4; 7 | |
| QM | 1360 | ACTB | 14,615 | 5.0 | 1.21 | actin, cytoplasmic, partial | 4; 6 | |
| QM | 821 | TPM1 | 34,308 | 4.6 | 1.18 | tropomyosin-1 | 1; 4 | Ca |
| QM | 869 | CFAP161 | 33,433 | 6.0 | 0.73 | cilia- and flagella-associated protein 161 | 4 | |
| QM | 1349 | CALM1 | 17,827 | 4.1 | 0.73 | calmodulin | 4; 7 | Ca |
| QM | 946 | TPM4 | 30,326 | 4.7 | 0.70 | tropomyosin-4 | 4 | Ca |
| QM | 435 | LMNA | 55,491 | 5.3 | 0.61 | 60 kDa neurofilament protein-like | 1; 4; 5; 6; 7 | Ca |
| QM | 14,602 | NUDC | 44,488 | 5.1 | 0.61 | nuclear migration protein nudC | 4 | |
| QM | 638 | ACTG1 | 41,797 | 5.2 | 0.60 | actin beta/gamma 1 | 4 | |
| QM | 1331 | TUBB | 19,149 | 4.7 | 0.58 | tubulin beta, first part | 4; 7 | |
| QM | 624 | ACTG1 | 42,449 | 4.6 | 0.56 | actin beta/gamma 1 | 4 | |
| QM | 431 | LMNA | 55,663 | 5.3 | 0.52 | 60 kDa neurofilament protein-like | 1; 4; 5; 6; 7 | Ca |
| QM | 820 | ANXA13 | 34,910 | 4.7 | 0.51 | annexin A13 | Ca | |
| QM | 570 | ILK | 46,517 | 6.4 | 0.51 | integrin-linked protein kinase | 4 | |
| QM | 815 | TUBB | 34,832 | 5.1 | 0.51 | tubulin beta, partial | 4; 7 | |
| QM | 2254 | CCDC151 | 56,609 | 5.7 | 0.49 | coiled-coil domain-containing protein 151 | 4 | |
| QM | 663 | TUBA1A | 41,535 | 5.7 | 0.48 | tubulin alpha-1A chain | 4 | |
| QM | 1256 | DPYSL2 | 27,335 | 6.3 | 0.44 | dihydropyrimidinase-like, partial | 4 | |
| QM | 1011 | ACTB | 29,008 | 5.4 | 0.42 | actin, cytoplasmic, partial | 4; 6 | |
| QM | 133 | CLCA4 | 93,882 | 5.8 | 0.36 | calcium-activated chloride channel regulator 4A | Ca | |
| Part C. Redox and detoxification (GO:0006790_ Sulphur compound metabolic process in bold) | ||||||||
| Sp. | label | abbrev. | Mr | pI | Var. | Identification | G. | Ca |
| ZM | 4237 | TALDO1 | 38,290 | 4.8 | 2.61 | transaldolase | ||
| ZM | 4204 | GLRX3 | 39,207 | 4.8 | 2.47 | glutaredoxin-3 | 2 | |
| ZM | 686 | CTH | 42,496 | 6.0 | 1.54 | cystathionine gamma-lyase | 1; 2; 3; 6 | Ca |
| ZM | 81,959 | PAPSS1 | 67,505 | 6.6 | 1.50 | bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase | 2 | |
| ZM | 644 | ALDH1A2 | 44,166 | 5.7 | 1.47 | retinal dehydrogenase 2 | 3; 6 | |
| ZM | 830 | SULT1A1 | 37,942 | 5.5 | 1.32 | sulfotransferase family 1A member 1 | 2 | |
| ZM | 81,979 | PAPSS1 | 65,696 | 6.7 | 1.28 | bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase | 2 | |
| ZM | 650 | ADH5 | 44,380 | 4.67 | 1.25 | NADP-dependent alcohol dehydrogenase C | 3 | |
| ZM | 639 | BPNT1 | 44,175 | 5.8 | 1.23 | 3′(2′),5′-bisphosphate nucleotidase 1 | 2 | |
| ZM | 1176 | SOD2 | 24,887 | 8.5 | 0.86 | superoxide dismutase [Mn], mitochondrial | 1; 2; 6 | |
| ZM | 1105 | GSTZ1 | 26,883 | 5.7 | 0.75 | maleylacetoacetate isomerase | 2; 3 | |
| ZM | 517 | CNDP2 | 48,800 | 5.5 | 0.68 | cytosolic non-specific dipeptidase | ||
| QM | 1127 | CNDP2 | 27,369 | 4.7 | 1.66 | cytosolic non-specific dipeptidase | ||
| QM | 518 | ALDH1A2 | 48,664 | 6.1 | 1.59 | retinal dehydrogenase 2 | 3; 6 | |
| QM | 2530 | SULT1A1 | 33,864 | 5.9 | 1.48 | sulfotransferase family 1A member 1 | 2 | |
| QM | 1099 | GST | 27,684 | 5.9 | 1.38 | glutathione S-transferase-like | 1; 2; 3 | |
| QM | 669 | CTH | 41,119 | 6.1 | 1.37 | cystathionine gamma-lyase | 1; 2; 3; 6 | Ca |
| QM | 694 | CTH | 40,865 | 6.2 | 1.35 | cystathionine gamma-lyase | 1; 2; 3; 6 | Ca |
| QM | 1061 | GSTZ1 | 27,873 | 5.4 | 1.23 | maleylacetoacetate isomerase | 1; 2; 3 | |
| QM | 1303 | SOD1 | 21,470 | 5.9 | 0.87 | superoxide dismutase [Cu/Zn] | 1; 2; 4; 6; 7 | |
| QM | 558 | GSS | 45,618 | 5.8 | 0.77 | glutathione synthetase | 2; 3 | |
| QM | 1353 | CYB5A | 18,002 | 4.7 | 0.73 | cytochrome b5 | 3 | |
| QM | 1351 | NENF | 17,075 | 4.7 | 0.71 | neudesin | ||
| QM | 1028 | PRDX2 | 28,889 | 6.3 | 0.70 | peroxiredoxin-like | 1; 6 | |
| QM | 840 | GSR | 34,032 | 5.9 | 0.65 | glutathionyl-hydroquinone reductase YqjG | 1; 2 | |
| QM | 1321 | AHCY | 20,643 | 6.3 | 0.64 | adenosylhomocysteinase B | 2; 3 | |
| QM | 1184 | PRXL2C | 27,023 | 5.7 | 0.63 | dye-decolourizing peroxidase YfeX | 1 | |
| QM | 736 | GSTP1 | 38,905 | 5.8 | 0.48 | glutathione S-transferase P 1 | 1; 2; 3; 6; 7 | |
| QM | 578 | ADSS2 | 46,200 | 6.3 | 0.47 | adenylosuccinate synthetase | 3 | |
| Part D. Energy and metabolism (GO:0019752_ Carboxylic acid metabolic process in bold) | ||||||||
| Sp. | label | abbrev. | Mr | pI | Var. | Identification | G. | Ca |
| ZM | 12,452 | NTPCR | 26,212 | 5.7 | 2.46 | cancer-related nucleoside-triphosphatase | ||
| ZM | 1159 | GPD2 | 26,110 | 6.6 | 2.01 | glycerol-3-phosphate dehydrogenase, mitochondrial | Ca | |
| ZM | 71,084 | ATIC | 66,279 | 5.9 | 1.65 | bifunctional purine biosynthesis protein PURH | 3 | |
| ZM | 242 | PCCA | 76,341 | 6.7 | 1.49 | propionyl-CoA carboxylase alpha chain, mitochondrial | 3 | |
| ZM | 20,205 | FAH | 44,598 | 5.6 | 1.46 | fumarylacetoacetase | 3 | |
| ZM | 731 | PCK1_2 | 41,321 | 5.9 | 1.46 | phosphoenolpyruvate carboxykinase [GTP], partial | 1; 3 | |
| ZM | 607 | FH | 45,269 | 6.0 | 1.45 | fumarate hydratase, mitochondrial | 1; 3 | |
| ZM | 14,678 | ALDOB | 28,111 | 5.7 | 1.42 | fructose-1, 6-bisphosphate aldolase | 3 | |
| ZM | 114 | ALDH1L1 | 92,374 | 6.0 | 1.41 | cytosolic 10-formyltetrahydrofolate dehydrogenase | 3 | |
| ZM | 1670 | SPATA18 | 70,773 | 5.5 | 1.41 | mitochondria-eating protein | 1 | |
| ZM | 1339 | ATP5F1D | 12,048 | 4.4 | 1.23 | ATP synthase delta chain, mitochondrial | ||
| ZM | 729 | HPD | 41,675 | 5.7 | 1.23 | 4-hydroxyphenylpyruvate dioxygenase | 3 | |
| ZM | 720 | MYG1 | 41,996 | 5.5 | 1.21 | UPF0160 protein MYG1, mitochondrial | ||
| ZM | 812 | HIBCH | 38,744 | 6.2 | 0.78 | 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial | 3 | |
| ZM | 641 | CKB | 43,718 | 7.0 | 0.71 | arginine kinase | 3 | |
| ZM | 532 | PCYT2 | 47,622 | 6.3 | 0.70 | ethanolamine-phosphate cytidylyltransferase | ||
| ZM | 318 | PCK1 | 71,984 | 6.7 | 0.66 | phosphoenolpyruvate carboxykinase, cytosolic [GTP] | 1; 3 | |
| ZM | 20,062 | LDHB | 42,852 | 5.6 | 0.64 | opine/octopine dehydrogenase, tauropine dehydrogenase | 3 | |
| ZM | 272 | GBA2 | 79,092 | 4.6 | 0.62 | glucosidase 2 subunit beta | 3 | |
| QM | 609 | TKTL2_short | 44,391 | 6.1 | 2.67 | transketolase protein 2, partial | ||
| QM | 2367 | CS | 40,656 | 6.4 | 2.47 | citrate synthase, mitochondrial | 3 | |
| QM | 276 | PCK1 | 69,690 | 6.4 | 2.25 | phosphoenolpyruvate carboxykinase [GTP] | 1; 3 | |
| QM | 611 | AK5 | 43,717 | 5.8 | 2.24 | adenylate kinase isoenzyme 5 | ||
| QM | 811 | MDH1B | 35,684 | 6.3 | 2.06 | malate dehydrogenase, cytosolic | 3 | |
| QM | 1217 | MDH1B | 27,069 | 4.7 | 1.96 | malate dehydrogenase, cytosolic | 3 | |
| QM | 792 | GAPDH | 35,711 | 6.2 | 1.84 | glyceraldehyde-3-phosphate dehydrogenase | 3; 4; 5 | |
| QM | 420 | MCCC2 | 56,870 | 6.7 | 1.76 | 3-methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | 2; 3 | |
| QM | 752 | PCK1_2 | 37,948 | 5.8 | 1.69 | phosphoenolpyruvate carboxykinase [GTP] | 1; 3 | |
| QM | 1385 | NIPSNAP1 | 14,360 | 6.5 | 1.49 | protein NipSnap | 7 | Ca |
| QM | 513 | SHMT1 | 49,003 | 6.5 | 1.47 | serine hydroxymethyltransferase, cytosolic | 3; 5 | |
| QM | 855 | IMPA1 | 33,752 | 5.4 | 1.40 | inositol monophosphatase | ||
| QM | 1358 | COX6B1 | 16,778 | 6.0 | 0.78 | cytochrome c oxidase subunit 6B1 | ||
| QM | 932 | PHB1 | 31,386 | 5.4 | 0.76 | prohibitin | 6 | |
| QM | 517 | ENO1 | 48,104 | 5.8 | 0.76 | enolase | 3; 6 | |
| QM | 616 | SPATA18 | 43,782 | 5.3 | 0.69 | mitochondria-eating protein | 1 | |
| QM | 1408 | ATP5F1D | 12,863 | 4.6 | 0.65 | ATP synthase delta chain, mitochondrial | ||
| QM | 828 | ENO1_short | 34,988 | 5.5 | 0.64 | enolase-short form alias c-myc promoter-binding protein-1 | 3; 6 | |
| QM | 1281 | MDH2 | 24,327 | 6.4 | 0.64 | malate dehydrogenase, mitochondrial | 3 | |
| QM | 501 | ATP5F1B | 50,871 | 5.1 | 0.61 | ATP synthase subunit beta, mitochondrial | ||
| QM | 7190 | ENO1 | 39,080 | 5.5 | 0.58 | enolase | 3; 6 | |
| QM | 314 | TKTL2 | 65,910 | 6.0 | 0.56 | transketolase protein 2 | ||
| QM | 655 | PCCB | 41,805 | 5.3 | 0.45 | propionyl-CoA carboxylase beta chain, mitochondrial | 2; 3 | |
| Part E. Transcription/translation (GO:0006417_ Regulation of translation in bold) | ||||||||
| Sp. | label | abbrev. | Mr | pI | Var. | Identification | G. | Ca |
| ZM | 995 | EIF3F | 29,818 | 5.4 | 1.71 | eukaryotic translation initiation factor 3 subunit F | 5 | |
| ZM | 352 | DDX3X | 66,641 | 6.1 | 1.69 | ATP-dependent RNA helicase DDX3X | 1; 5; 6; 7 | |
| ZM | 819 | EEF2_short | 38,350 | 5.9 | 1.58 | elongation factor 2, partial | 1; 5; 7 | |
| ZM | 19,977 | FAM172A | 44,808 | 5.8 | 1.54 | cotranscriptional regulator FAM172A | ||
| ZM | 474 | U2AF2 | 53,138 | 5.6 | 1.48 | splicing factor U2AF 50 kDa subunit | ||
| ZM | 356 | FARSB | 69,076 | 5.4 | 1.46 | phenylalanine-tRNA ligase beta subunit | 3 | |
| ZM | 629 | PA2G4 | 44,025 | 6.3 | 1.41 | proliferation-associated protein 2G4 | 5; 6; 7 | |
| ZM | 260 | FUBP1 | 76,901 | 6.0 | 1.27 | far upstream element-binding protein 1 | ||
| ZM | 1259 | EIF5A | 20,524 | 5.0 | 1.18 | eukaryotic translation initiation factor 5A | 5; 6 | |
| ZM | 649 | WARS1 | 43,779 | 5.96 | 0.70 | tryptophanyl-tRNA synthetase, cytoplasmic | 3 | |
| ZM | 431 | NAP1L1 | 60,086 | 4.3 | 0.68 | nucleosome assembly protein 1 1 | ||
| ZM | 81 | EEF2 | 93,962 | 7.5 | 0.65 | elongation factor 2 | 1; 5 | |
| QM | 644 | WARS1 | 42,768 | 5.8 | 2.44 | tryptophanyl-tRNA synthetase, cytoplasmic | 3 | |
| QM | 710 | EIF4A1 | 39,889 | 5.8 | 1.60 | eukaryotic initiation factor 4A-I | 5 | |
| QM | 991 | EIF4B | 29,725 | 5.6 | 1.44 | eukaryotic translation initiation factor 4B | 5 | |
| QM | 614 | PA2G4 | 43,956 | 6.3 | 1.39 | proliferation-associated protein 2G4 | 3; 5; 7 | |
| QM | 952 | EIF3F | 30,865 | 5.5 | 1.28 | eukaryotic translation initiation factor 3 subunit F | 5 | |
| QM | 261 | FUBP1 | 70,697 | 5.7 | 0.68 | far upstream element-binding protein 1 | ||
| QM | 780 | DDX3X | 37,130 | 6.3 | 0.66 | ATP-dependent RNA helicase DDX3X | 1; 5; 6; 7 | |
| QM | 1559 | ELOB | 21,107 | 4.6 | 0.65 | elongin B | 1 | |
| QM | 1422 | RIDA | 11,327 | 6.0 | 0.40 | 2-iminobutanoate/2-iminopropanoate deaminase | 3; 5 | |
| Variations | Abbreviation | Protein | |
|---|---|---|---|
| Abundances change in the same direction | Up | ALDH1A2 | retinal dehydrogenase 2 |
| CTH | cystathionine gamma-lyase | ||
| EIF3F | eukaryotic translation initiation factor 3 subunit F | ||
| FLNA | filamin-A, partial | ||
| HSPD1 | heat shock protein 60, mitochondrial | ||
| PA2G4 | proliferation-associated protein 2G4 | ||
| PCK1 | phosphoenolpyruvate carboxykinase [GTP], partial | ||
| SULT1A1 | sulfotransferase family 1A member 1 | ||
| Down | LMNA | 60 kDa neurofilament protein-like | |
| MVP | major vault protein | ||
| P4HB | protein disulfide-isomerase | ||
| TUBA | tubulin alpha subunit | ||
| Abundances change in opposite directions | ZM Up QM Down | ATP5F1D | ATP synthase delta chain, mitochondrial |
| DDX3X | ATP-dependent RNA helicase DDX3X | ||
| FUBP1 | far upstream element-binding protein 1 | ||
| SPATA18 | mitochondria-eating protein | ||
| ZM Down QM Up | CNDP2 | cytosolic non-specific dipeptidase | |
| CNN3 | calponin homolog, protein unc-87 | ||
| GSTZ1 | maleylacetoacetate isomerase alias GST Z | ||
| WARS1 | tryptophanyl-tRNA synthetase, cytoplasmic | ||
| PCK1 | phosphoenolpyruvate carboxykinase, cytosolic [GTP] | ||
| TUBB | tubulin beta, partial | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bultelle, F.; Le Saux, A.; David, E.; Tanguy, A.; Devin, S.; Olivier, S.; Poret, A.; Chan, P.; Louis, F.; Delahaut, L.; et al. Cadmium Highlights Common and Specific Responses of Two Freshwater Sentinel Species, Dreissena polymorpha and Dreissena rostriformis bugensis. Proteomes 2024, 12, 10. https://doi.org/10.3390/proteomes12020010
Bultelle F, Le Saux A, David E, Tanguy A, Devin S, Olivier S, Poret A, Chan P, Louis F, Delahaut L, et al. Cadmium Highlights Common and Specific Responses of Two Freshwater Sentinel Species, Dreissena polymorpha and Dreissena rostriformis bugensis. Proteomes. 2024; 12(2):10. https://doi.org/10.3390/proteomes12020010
Chicago/Turabian StyleBultelle, Florence, Aimie Le Saux, Elise David, Arnaud Tanguy, Simon Devin, Stéphanie Olivier, Agnès Poret, Philippe Chan, Fanny Louis, Laurence Delahaut, and et al. 2024. "Cadmium Highlights Common and Specific Responses of Two Freshwater Sentinel Species, Dreissena polymorpha and Dreissena rostriformis bugensis" Proteomes 12, no. 2: 10. https://doi.org/10.3390/proteomes12020010
APA StyleBultelle, F., Le Saux, A., David, E., Tanguy, A., Devin, S., Olivier, S., Poret, A., Chan, P., Louis, F., Delahaut, L., Pain-Devin, S., Péden, R., Vaudry, D., Le Foll, F., & Rocher, B. (2024). Cadmium Highlights Common and Specific Responses of Two Freshwater Sentinel Species, Dreissena polymorpha and Dreissena rostriformis bugensis. Proteomes, 12(2), 10. https://doi.org/10.3390/proteomes12020010









